MOB: Pivotal Conserved Proteins in Cytokinesis, Cell Architecture and Tissue Homeostasis
Abstract
Simple Summary
Abstract
1. Introduction
2. MOB Proteins in Multicellular Eukaryotes
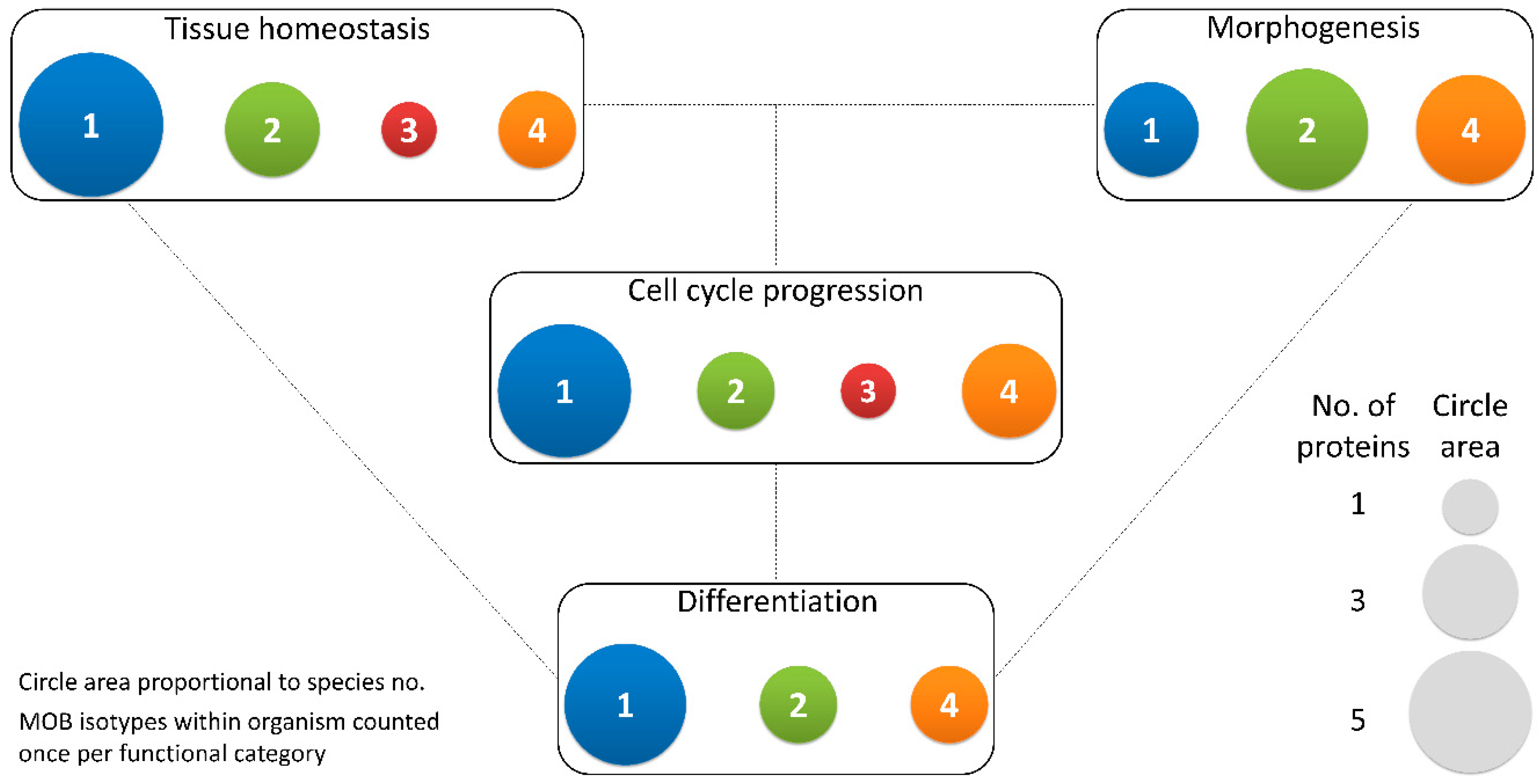
2.1. Functions of MOB Proteins
2.1.1. Tissue Homeostasis: MOB as Regulators of Cell Proliferation and Apoptosis
2.1.2. Morphogenesis: A MOB Function in Various Species and Cell Types
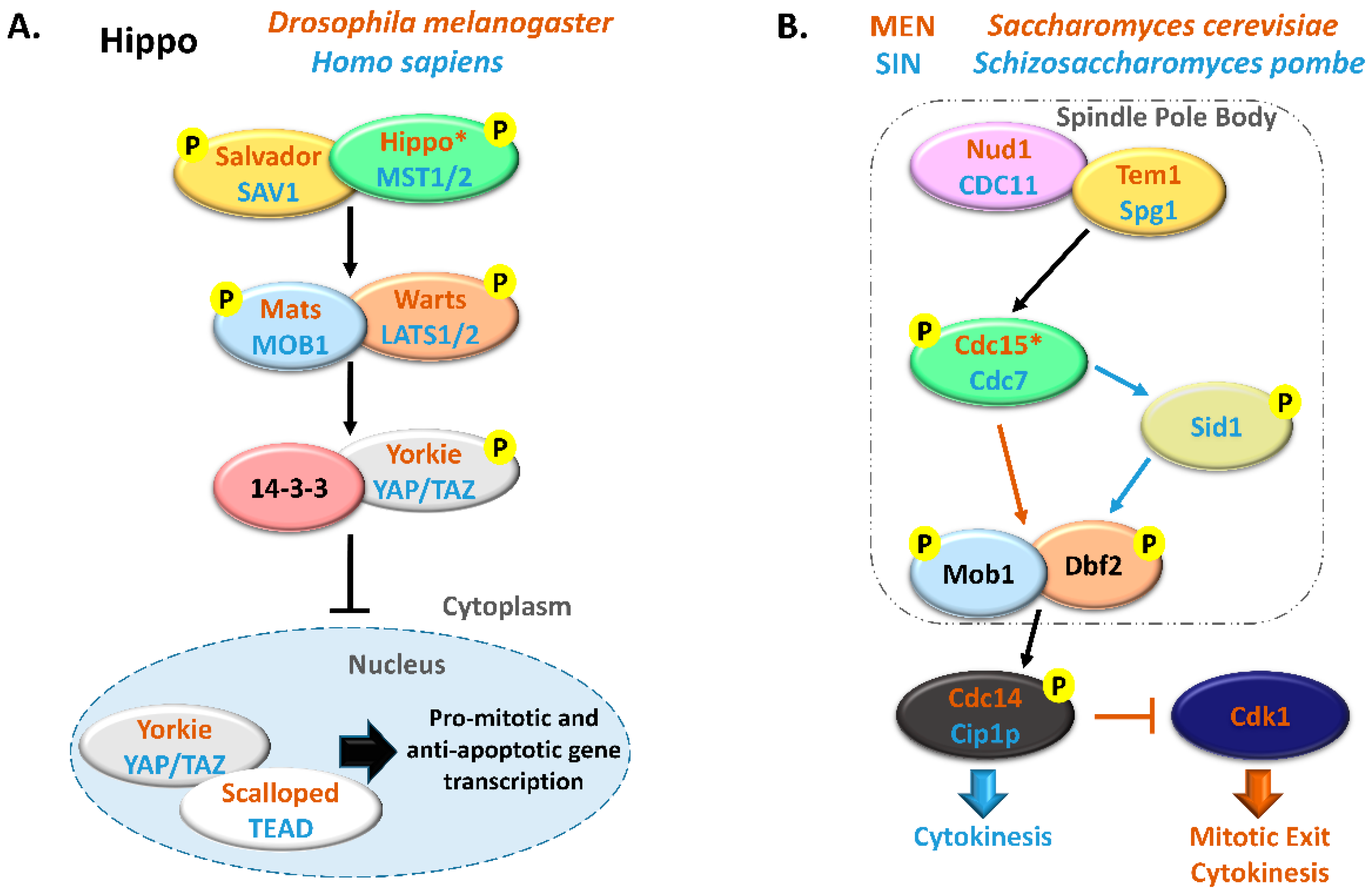
2.1.3. Cell Cycle Progression: MOBs as Regulators of Mitosis, Cytokinesis, and Centrosome Biology
2.1.4. Differentiation and Stem Cell Maintenance: MOB in a Crossroad between Morphogenesis, Tissue Homeostasis, and Cell Cycle Regulation
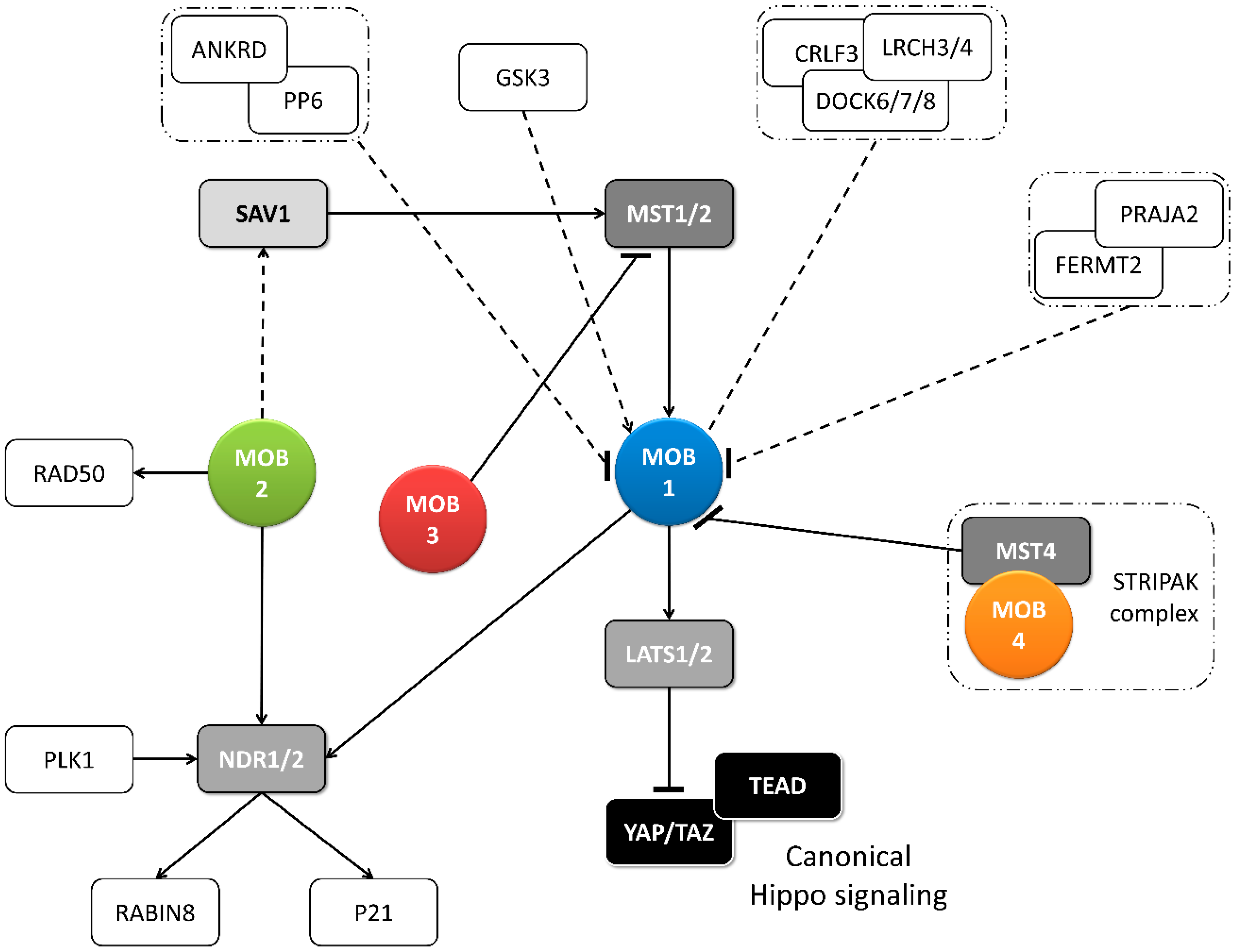
2.2. Regulation of MOB Proteins
2.2.1. Post-Translation Modifications of MOBs
2.2.2. Transcriptional and Post-Transcriptional Regulation of MOBs
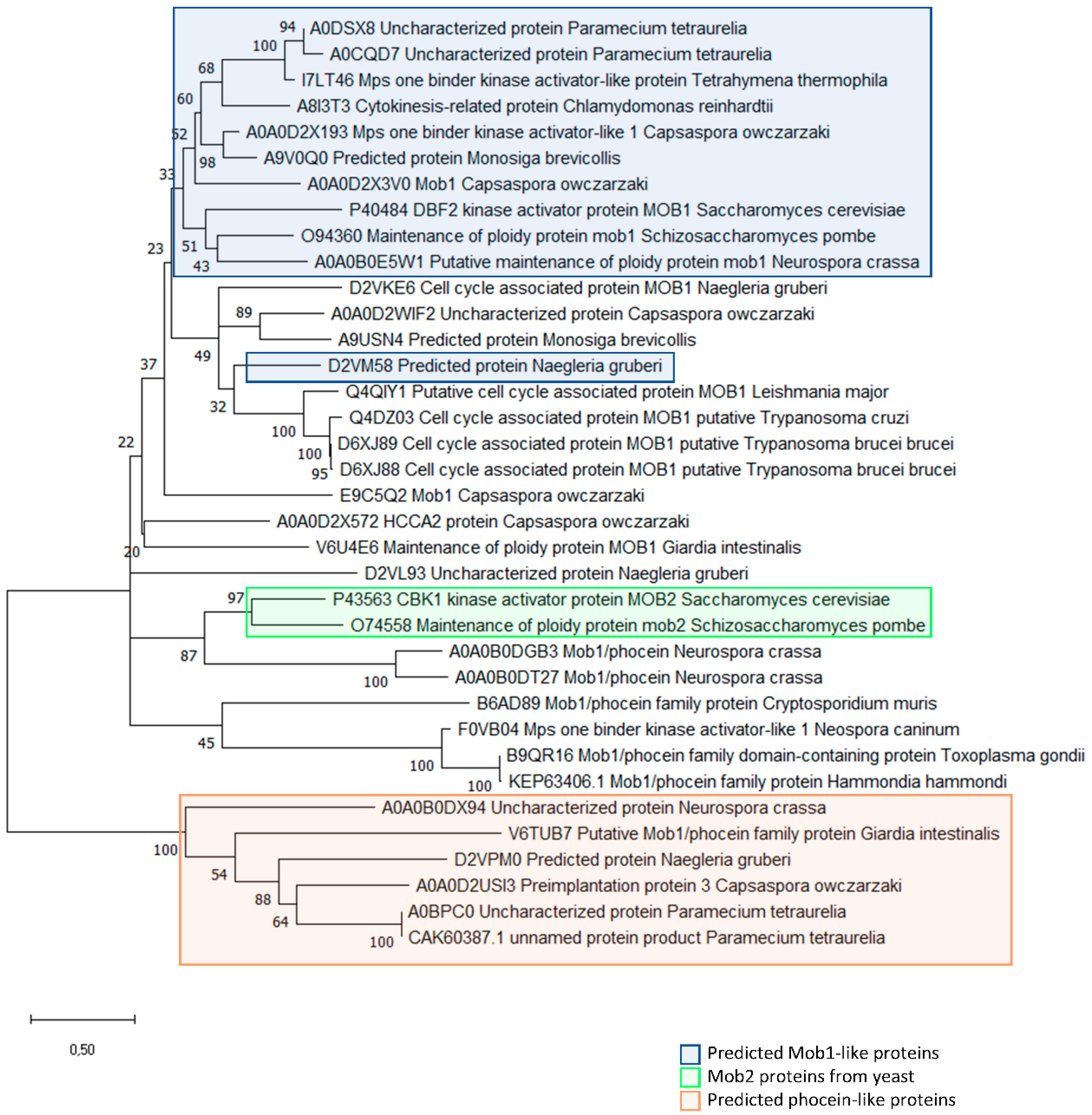
3. MOB Proteins in Unicellular Organisms: The Roots of Multicellularity?
3.1. MOB Proteins in Yeast: From Cell Division to Signaling Pathways
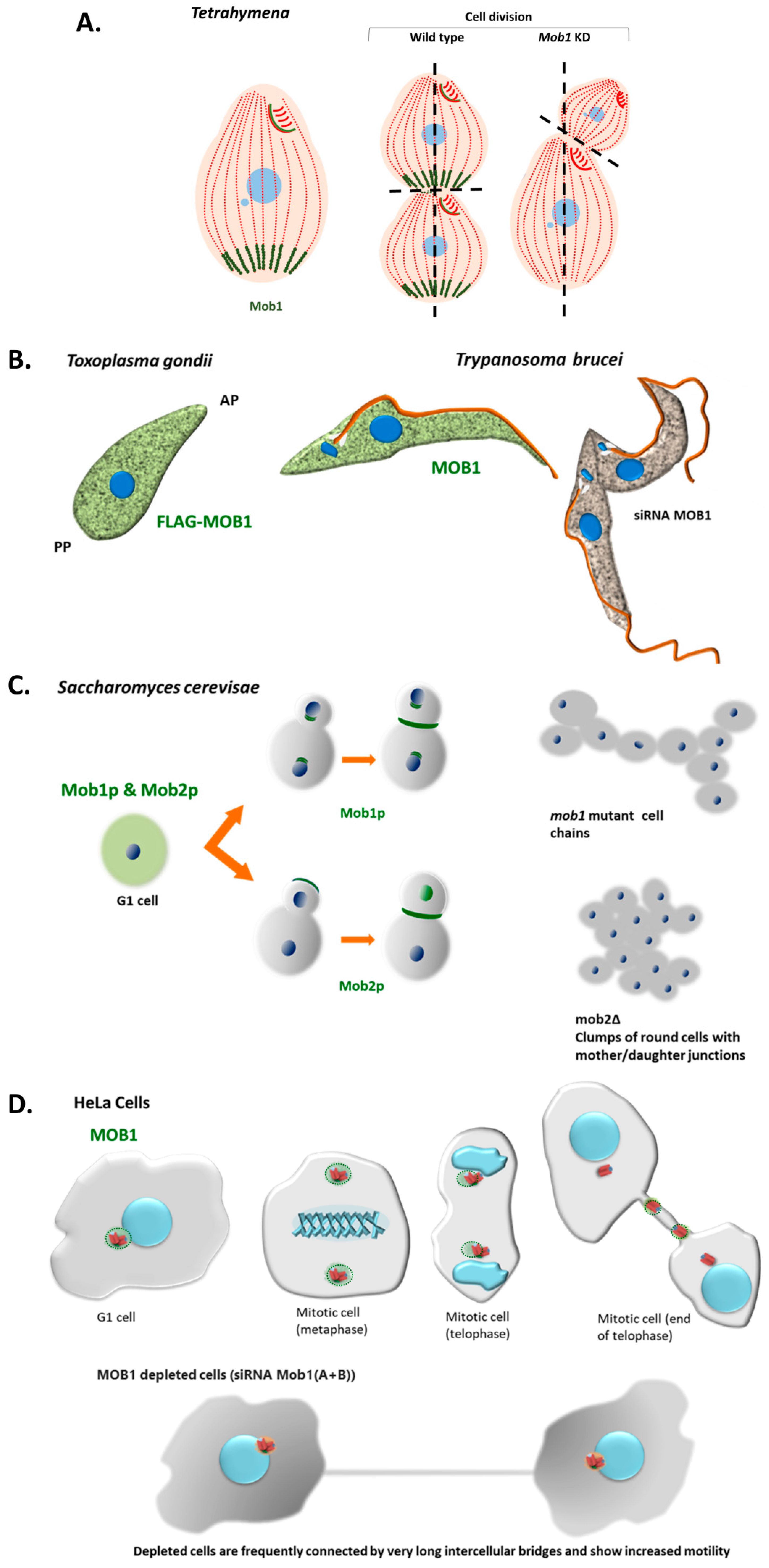
3.2. MOB Proteins in the Alveolate: From Cytokinesis to Morphology
3.2.1. MOB in Ciliates
3.2.2. MOB in Apicomplexa
3.3. Non-Alveolate Protozoa
3.4. MOBs: From Unicellular to Multicellular Organisms
4. Concluding Remarks
4.1. The Ancient MOB: In the Crossroad of Accurate Cell Division, Cytokinesis, and Morphogenesis
4.2. MOB: New Roles, the Same Function?
Author Contributions
Funding
Conflicts of Interest
References
- Luca, F.C.; Winey, M. MOB1, an essential yeast gene required for completion of mitosis and maintenance of ploidy. Mol. Biol. Cell 1998, 9, 29–46. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lai, Z.-C.; Wei, X.; Shimizu, T.; Ramos, E.; Rohrbaugh, M.; Nikolaidis, N.; Ho, L.-L.; Li, Y. Control of Cell Proliferation and Apoptosis by Mob as Tumor Suppressor, Mats. Cell 2005, 120, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Maerz, S.; Dettmann, A.; Ziv, C.; Liu, Y.; Valerius, O.; Yarden, O.; Seiler, S. Two NDR kinase-MOB complexes function as distinct modules during septum formation and tip extension in Neurospora crassa. Mol. Microbiol. 2009, 74, 707–723. [Google Scholar] [CrossRef] [PubMed]
- Schmidpeter, J.; Dahl, M.; Hofmann, J.; Koch, C. ChMob2 binds to ChCbk1 and promotes virulence and conidiation of the fungal pathogen Colletotrichum higginsianum. BMC Microbiol. 2017, 17, 1–15. [Google Scholar] [CrossRef]
- Duhart, J.C.; Raftery, L.A. Mob Family Proteins: Regulatory Partners in Hippo and Hippo-Like Intracellular Signaling Pathways. Front. Cell Dev. Biol. 2020, 8, 1–22. [Google Scholar] [CrossRef]
- Chalker, D.L.; Frankel, J. Morphogenesis: A mob rules from the rear. Curr. Biol. 2014, 24, R700–R702. [Google Scholar] [CrossRef][Green Version]
- Tavares, A.; Gonçalves, J.; Florindo, C.; Tavares, A.A.; Soares, H. Mob1: Defining cell polarity for proper cell division. J. Cell Sci. 2012, 125, 516–527. [Google Scholar] [CrossRef]
- Shimobayashi, M.; Hall, M.N. Making new contacts: The mTOR network in metabolism and signalling crosstalk. Nat. Rev. Mol. Cell Biol. 2014, 15, 155–162. [Google Scholar] [CrossRef]
- Hwang, J.; Pallas, D.C. STRIPAK complexes: Structure, biological function, and involvement in human diseases. Int. J. Biochem. Cell Biol. 2014, 47, 118–148. [Google Scholar] [CrossRef]
- Bae, J.S.; Jeon, Y.; Kim, S.M.; Jang, J.Y.; Park, M.K.; Kim, I.H.; Hwang, D.S.; Lim, D.S.; Lee, H. Depletion of MOB1A/B causes intestinal epithelial degeneration by suppressing Wnt activity and activating BMP/TGF-β signaling. Cell Death Dis. 2018, 9. [Google Scholar] [CrossRef]
- Dong, J.; Feldmann, G.; Huang, J.; Wu, S.; Zhang, N.; Comerford, S.A.; Gayyed, M.F.F.; Anders, R.A.; Maitra, A.; Pan, D. Elucidation of a Universal Size-Control Mechanism in Drosophila and Mammals. Cell 2007, 130, 1120–1133. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Smolen, G.A.; Haber, D.A. Negative regulation of YAP by LATS1 underscores evolutionary conservation of the Drosophila Hippo pathway. Cancer Res. 2008, 68, 2789–2794. [Google Scholar] [CrossRef] [PubMed]
- Bothos, J.; Tuttle, R.L.; Ottey, M.; Luca, F.C.; Halazonetis, T.D. Human LATS1 is a mitotic exit network kinase. Cancer Res. 2005, 65, 6568–6575. [Google Scholar] [CrossRef] [PubMed]
- Hergovich, A.; Kohler, R.S.; Schmitz, D.; Vichalkovski, A.; Cornils, H.; Hemmings, B.A. The MST1 and hMOB1 tumor suppressors control human centrosome duplication by regulating NDR kinase phosphorylation. Curr. Biol. 2009, 19, 1692–1702. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Ho, L.-L.; Lai, Z.-C. The mob as tumor suppressor gene is essential for early development and regulates tissue growth in Drosophila. Genetics 2008, 178, 957–965. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hergovich, A.; Hemmings, B.A. Seminars in Cell & Developmental Biology Hippo signalling in the G2 / M cell cycle phase: Lessons learned from the yeast MEN and SIN pathways. Semin. Cell Dev. Biol. 2012, 23, 794–802. [Google Scholar] [CrossRef] [PubMed]
- Hergovich, A. Hippo Signaling in Mitosis: An Updated View in Light of the MEN Pathway. Methods Mol. Biol. 2017, 1505, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Gundogdu, R.; Hergovich, A. MOB (Mps one Binder) Proteins in the Hippo Pathway and Cancer. Cells 2019, 8, 569. [Google Scholar] [CrossRef]
- Ma, S.; Meng, Z.; Chen, R.; Guan, K.-L. The Hippo Pathway: Biology and Pathophysiology. Annu. Rev Biochem. 2019, 88, 577–604. [Google Scholar] [CrossRef]
- Bardin, A.J.; Amon, A. Men and sin: What’s the difference? Nat. Rev. Mol. Cell Biol. 2001, 2, 815–826. [Google Scholar] [CrossRef]
- Hotz, M.; Barral, Y. The Mitotic Exit Network: New turns on old pathways. Trends Cell Biol. 2014, 24, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Elramli, N.; Karahoda, B.; Sarikaya-Bayram, O.; Frawley, D.; Ulas, M.; Oakley, C.E.; Oakley, B.R.; Seiler, S.; Bayram, O. Assembly of a heptameric STRIPAK complex is required for coordination of light-dependent multicellular fungal development with secondary metabolism in aspergillus nidulans. PLoS Genet. 2019, 15, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Bernhards, Y.; Pöggeler, S. The phocein homologue SmMOB3 is essential for vegetative cell fusion and sexual development in the filamentous ascomycete Sordaria macrospora. Curr. Genet. 2011, 57, 133–149. [Google Scholar] [CrossRef] [PubMed]
- Dettmann, A.; Heilig, Y.; Ludwig, S.; Schmitt, K.; Illgen, J.; Fleißner, A.; Valerius, O.; Seiler, S. HAM-2 and HAM-3 are central for the assembly of the NeurosporaSTRIPAK complex at the nuclear envelope and regulate nuclear accumulation of the MAP kinase MAK-1 in a MAK-2-dependent manner. Mol. Microbiol. 2013, 90, 796–812. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Musante, V.; Zhou, W.; Picciotto, M.R.; Nairn, A.C. Striatin-1 is a B subunit of protein phosphatase PP2A that regulates dendritic arborization and spine development in striatal neurons. J. Biol. Chem. 2018, 293, 11179–11194. [Google Scholar] [CrossRef]
- Vrabioiu, A.M.; Struhl, G. Fat/Dachsous Signaling Promotes Drosophila Wing Growth by Regulating the Conformational State of the NDR Kinase Warts. Dev. Cell. 2015, 35, 737–749. [Google Scholar] [CrossRef]
- Praskova, M.; Xia, F.; Avruch, J. MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr. Biol. 2008, 18, 311–321. [Google Scholar] [CrossRef]
- Kulaberoglu, Y.; Lin, K.; Holder, M.; Gai, Z.; Gomez, M.; Assefa Shifa, B.; Mavis, M.; Hoa, L.; Sharif, A.A.D.; Lujan, C.; et al. Stable MOB1 interaction with Hippo/MST is not essential for development and tissue growth control. Nat. Commun. 2017, 8, 695. [Google Scholar] [CrossRef]
- Vichalkovski, A.; Gresko, E.; Cornils, H.; Hergovich, A.; Schmitz, D.; Hemmings, B.A. NDR Kinase Is Activated by RASSF1A/MST1 in Response to Fas Receptor Stimulation and Promotes Apoptosis. Curr. Biol. 2008, 18, 1889–1895. [Google Scholar] [CrossRef]
- Zhou, D.; Conrad, C.; Xia, F.; Park, J.-S.; Payer, B.; Yin, Y.; Lauwers, G.Y.; Thasler, W.; Lee, J.T.; Avruch, J.; et al. Mst1 and Mst2 Maintain Hepatocyte Quiescence and Suppress Hepatocellular Carcinoma Development through Inactivation of the Yap1 Oncogene. Cancer Cell 2009, 16, 425–438. [Google Scholar] [CrossRef]
- Schumacher, F.R.; Al Olama, A.A.; Berndt, S.I.; Benlloch, S.; Ahmed, M.; Saunders, E.J.; Dadaev, T.; Leongamornlert, D.; Anokian, E.; Cieza-Borrella, C.; et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat. Genet. 2018, 50, 928–936. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.-Y.; Maier, W.; Baumeister, R.; Joachimiak, E.; Ruan, Z.; Kannan, N.; Clarke, D.; Louka, P.; Guha, M.; Frankel, J.; et al. Two Antagonistic Hippo Signaling Circuits Set the Division Plane at the Medial Position in the Ciliate Tetrahymena. Genetics 2019, 211, 651–663. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Shen, J.; Gu, F.; Zhang, Y.; Wu, W.; Weng, J.; Liao, Y.; Deng, Z.; Yuan, Q.; Zheng, L.; et al. Monopolar spindle-one-binder protein 2 regulates the activity of large tumor suppressor/yes-associated protein to inhibit the motility of SMMC-7721 hepatocellular carcinoma cells. Oncol. Lett. 2018, 15, 5375–5383. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhang, H.; Shi, Z.; Li, Y.; Zhang, X.; Gao, Z.; Zhou, L.; Ma, J.; Xu, Q.; Guan, J.; et al. The MST4-MOB4 complex disrupts the MST1-MOB1 complex in the Hippo-YAP pathway and plays a pro-oncogenic role in pancreatic cancer. J. Biol. Chem. 2018, 293, 14455–14469. [Google Scholar] [CrossRef]
- Gordon, J.; Hwang, J.; Carrier, K.J.; Jones, C.A.; Kern, Q.L.; Moreno, C.S.; Karas, R.H.; Pallas, D.C. Protein phosphatase 2a (PP2A) binds within the oligomerization domain of striatin and regulates the phosphorylation and activation of the mammalian Ste20-Like kinase Mst3. BMC Biochem. 2011, 12, 54. [Google Scholar] [CrossRef]
- Tang, F.; Zhang, L.; Xue, G.; Hynx, D.; Wang, Y.; Cron, P.D.; Hundsrucker, C.; Hergovich, A.; Frank, S.; Hemmings, B.A.; et al. hMOB3 modulates MST1 apoptotic signaling and supports tumor growth in glioblastoma multiforme. Cancer Res. 2014, 74, 3779–3789. [Google Scholar] [CrossRef]
- Nishio, M.; Hamada, K.; Kawahara, K.; Sasaki, M.; Noguchi, F.; Chiba, S.; Mizuno, K.; Suzuki, S.O.; Dong, Y.; Tokuda, M.; et al. Cancer susceptibility and embryonic lethality in Mob1a/1b double-mutant mice. J. Clin. Investig. 2012, 122, 4505–4518. [Google Scholar] [CrossRef]
- Nishio, M.; Sugimachi, K.; Goto, H.; Wang, J.; Morikawa, T.; Miyachi, Y.; Takano, Y.; Hikasa, H.; Itoh, T.; Suzuki, S.O.; et al. Dysregulated YAP1/TAZ and TGF-β signaling mediate hepatocarcinogenesis in Mob1a/1b-deficient mice. Proc. Natl. Acad. Sci. USA 2016, 113, E71–80. [Google Scholar] [CrossRef]
- Kato, W.; Nishio, M.; To, Y.; Togashi, H.; Mak, T.W.; Takada, H.; Ohga, S.; Maehama, T.; Suzuki, A. MOB1 regulates thymocyte egress and T-cell survival in mice in a YAP1-independent manner. Genes Cells 2019, 24, 485–495. [Google Scholar] [CrossRef]
- Song, J.; Wang, T.; Chi, X.; Wei, X.; Xu, S.; Yu, M.; He, H.; Ma, J.; Li, X.; Du, J.; et al. Kindlin-2 Inhibits the Hippo Signaling Pathway by Promoting Degradation of MOB1. Cell Rep. 2019, 29, 3664–3677. [Google Scholar] [CrossRef]
- Gardiner, K.L.; Downs, L.; Berta-Antalics, A.I.; Santana, E.; Aguirre, G.D.; Genini, S. Photoreceptor proliferation and dysregulation of cell cycle genes in early onset inherited retinal degenerations. BMC Genom. 2016, 17, 221. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Z.; Qin, N.; Tyasi, T.L.; Zhu, H.; Liu, D.; Yuan, S.; Xu, R. The Hippo/MST pathway member SAV1 plays a suppressive role in development of the prehierarchical follicles in hen ovary. PLoS ONE 2016, 11, e0160896. [Google Scholar] [CrossRef] [PubMed]
- Galla, G.; Zenoni, S.; Marconi, G.; Marino, G.; Botton, A.; Pinosa, F.; Citterio, S.; Ruperti, B.; Palme, K.; Albertini, E.; et al. Sporophytic and gametophytic functions of the cell cycle-associated Mob1 gene in Arabidopsis thaliana L. Gene 2011, 484, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Guo, Z.; Song, L.; Wang, Y.; Cheng, Y. NCP1/AtMOB1A Plays Key Roles in Auxin-Mediated Arabidopsis Development. PLoS Genet. 2016, 12, e1005923. [Google Scholar] [CrossRef]
- Xiong, J.; Cui, X.; Yuan, X.; Yu, X.; Sun, J.; Gong, Q. The Hippo/STE20 homolog SIK1 interacts with MOB1 to regulate cell proliferation and cell expansion in Arabidopsis. J. Exp. Bot. 2016, 67, 1461–1475. [Google Scholar] [CrossRef]
- Guo, Z.; Yue, X.; Cui, X.; Song, L.; Cheng, Y. AtMOB1 Genes Regulate Jasmonate Accumulation and Plant Development. Plant Physiol. 2020, 182, 1481–1493. [Google Scholar] [CrossRef]
- Citterio, S.; Albertini, E.; Varotto, S.; Feltrin, E.; Soattin, M.; Marconi, G.; Sgorbati, S.; Lucchin, M.; Barcaccia, G. Alfalfa Mob 1-like genes are expressed in reproductive organs during meiosis and gametogenesis. Plant Mol. Biol. 2005, 58, 789–807. [Google Scholar] [CrossRef]
- Yan, M.; Chu, L.; Qin, B.; Wang, Z.; Liu, X.; Jin, C.; Zhang, G.; Gomez, M.; Hergovich, A.; Chen, Z.; et al. Regulation of NDR1 activity by PLK1 ensures proper spindle orientation in mitosis. Sci. Rep. 2015, 5, 10449. [Google Scholar] [CrossRef]
- Cornils, H.; Kohler, R.S.; Hergovich, A.; Hemmings, B.A. Human NDR kinases control G(1)/S cell cycle transition by directly regulating p21 stability. Mol. Cell. Biol. 2011, 31, 1382–1395. [Google Scholar] [CrossRef]
- Chiba, S.; Amagai, Y.; Homma, Y.; Fukuda, M.; Mizuno, K. NDR2-mediated Rabin8 phosphorylation is crucial for ciliogenesis by switching binding specificity from phosphatidylserine to Sec15. EMBO J. 2013, 32, 874–885. [Google Scholar] [CrossRef]
- Couzens, A.L.; Knight, J.D.R.; Kean, M.J.; Teo, G.; Weiss, A.; Dunham, W.H.; Lin, Z.-Y.; Bagshaw, R.D.; Sicheri, F.; Pawson, T.; et al. Protein interaction network of the mammalian Hippo pathway reveals mechanisms of kinase-phosphatase interactions. Sci. Signal. 2013, 6, rs15. [Google Scholar] [CrossRef] [PubMed]
- Xiong, S.; Couzens, A.L.; Kean, M.J.; Mao, D.Y.; Guettler, S.; Kurinov, I.; Gingras, A.-C.; Sicheri, F. Regulation of Protein Interactions by Mps One Binder (MOB1) Phosphorylation. Mol. Cell. Proteom. 2017, 16, 1111–1125. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Han, X.; Zou, H.; Zhang, B.; Ding, Y.; Xu, X.; Zeng, J.; Liu, J.; Gong, A. PTEN-GSK3β-MOB1 axis controls neurite outgrowth in vitro and in vivo. Cell. Mol. Life Sci. 2018, 75, 4445–4464. [Google Scholar] [CrossRef] [PubMed]
- Gomez, V.; Gundogdu, R.; Gomez, M.; Hoa, L.; Panchal, N.; O’Driscoll, M.; Hergovich, A. Regulation of DNA damage responses and cell cycle progression by hMOB2. Cell. Signal. 2015, 27, 326–339. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.S.; Kim, S.M.; Jeon, Y.; Sim, J.; Jang, J.Y.; Son, J.; Hong, W.; Park, M.K.; Lee, H. Loss of Mob1a/b impairs the differentiation of mouse embryonic stem cells into the three germ layer lineages. Exp. Mol. Med. 2019, 51. [Google Scholar] [CrossRef] [PubMed]
- Otsubo, K.; Goto, H.; Nishio, M.; Kawamura, K.; Yanagi, S.; Nishie, W.; Sasaki, T.; Maehama, T.; Nishina, H.; Mimori, K.; et al. MOB1-YAP1/TAZ-NKX2.1 axis controls bronchioalveolar cell differentiation, adhesion and tumour formation. Oncogene 2017, 36, 4201–4211. [Google Scholar] [CrossRef]
- He, Y.; Emoto, K.; Fang, X.; Ren, N.; Tian, X.; Jan, Y.-N.; Adler, P.N. Drosophila Mob family proteins interact with the related tricornered (Trc) and warts (Wts) kinases. Mol. Biol. Cell 2005, 16, 4139–4152. [Google Scholar] [CrossRef]
- Liu, L.Y.; Lin, C.H.; Fan, S.S. Function of Drosophila mob2 in photoreceptor morphogenesis. Cell Tissue Res. 2009, 338, 377–389. [Google Scholar] [CrossRef]
- Campbell, M.; Ganetzky, B. Identification of Mob2, a novel regulator of larval neuromuscular junction morphology, in natural populations of Drosophila melanogaster. Genetics 2013, 195, 915–926. [Google Scholar] [CrossRef]
- Schulte, J.; Sepp, K.J.; Jorquera, R.A.; Wu, C.; Song, Y.; Hong, P.; Troy Littleton, J. DMob4/Phocein regulates synapse formation, axonal transport, and microtubule organization. J. Neurosci. 2010, 30, 5189–5203. [Google Scholar] [CrossRef]
- Trammell, M.A.; Mahoney, N.M.; Agard, D.A.; Vale, R.D. Mob4 plays a role in spindle focusing in Drosophila S2 cells. J. Cell Sci. 2008, 121, 1284–1292. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Neisch, A.L.; Neufeld, T.P.; Hays, T.S. A STR IPAK complex mediates axonal transport of autophagosomes and dense core vesicles through PP2A regulation. J. Cell Biol. 2017, 216, 441–461. [Google Scholar] [CrossRef] [PubMed]
- Florindo, C.; Perdigão, J.; Fesquet, D.; Schiebel, E.; Pines, J.; Tavares, A.A. Human Mob1 proteins are required for cytokinesis by controlling microtubule stability. J. Cell Sci. 2012, 125, 3085–3090. [Google Scholar] [CrossRef] [PubMed]
- Wilmeth, L.J.; Shrestha, S.; Montan, G.; Rashe, J.; Shuster, C.B. Mutual Dependence of Mob1 and the Chromosomal Passenger Complex for Localization during Mitosis. Mol. Biol. Cell 2010, 21, 380–392. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mardin, B.R.; Lange, C.; Baxter, J.E.; Hardy, T.; Scholz, S.R.; Fry, A.M.; Schiebel, E. Components of the Hippo pathway cooperate with Nek2 kinase to regulate centrosome disjunction. Nat. Cell Biol. 2010, 12, 1166–1176. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Yao, G.; Hu, L.; Yan, Y.; Liu, J.; Shi, J.; Chang, Y.; Zhang, Y.; Liang, D.; Shen, D.; et al. MOB2 suppresses GBM cell migration and invasion via regulation of FAK/Akt and cAMP/PKA signaling. Cell Death Dis. 2020, 11. [Google Scholar] [CrossRef]
- O’Neill, A.C.; Kyrousi, C.; Einsiedler, M.; Burtscher, I.; Drukker, M.; Markie, D.M.; Kirk, E.P.; Götz, M.; Robertson, S.P.; Cappello, S. Mob2 insufficiency disrupts neuronal migration in the developing cortex. Front. Cell. Neurosci. 2018, 12, 1–13. [Google Scholar] [CrossRef]
- Kohler, R.S.; Schmitz, D.; Cornils, H.; Hemmings, B.A.; Hergovich, A. Differential NDR/LATS Interactions with the Human MOB Family Reveal a Negative Role for Human MOB2 in the Regulation of Human NDR Kinases. Mol. Cell. Biol. 2010, 30, 4507–4520. [Google Scholar] [CrossRef]
- Hergovich, A.; Bichsel, S.J.; Hemmings, B.A. Human NDR Kinases Are Rapidly Activated by MOB Proteins through Recruitment to the Plasma Membrane and Phosphorylation. Mol. Cell. Biol. 2005, 25, 8259–8272. [Google Scholar] [CrossRef]
- Mou, F.; Praskova, M.; Xia, F.; Van Buren, D.; Hock, H.; Avruch, J.; Zhou, D. The Mst1 and Mst2 kinases control activation of rho family GTPases and thymic egress of mature thymocytes. J. Exp. Med. 2012, 209, 741–759. [Google Scholar] [CrossRef]
- Lin, C.H.; Hsieh, M.; Fan, S.S. The promotion of neurite formation in Neuro2A cells by mouse Mob2 protein. FEBS Lett. 2011, 585, 523–530. [Google Scholar] [CrossRef][Green Version]
- Citterio, S.; Piatti, S.; Albertini, E.; Aina, R.; Varotto, S.; Barcaccia, G.; Scienza, P. Alfalfa Mob1-like proteins are involved in cell proliferation and are localized in the cell division plane during cytokinesis. Exp. Cell Res. 2005, 312, 1050–1064. [Google Scholar] [CrossRef] [PubMed]
- Cappello, S.; Gray, M.J.; Badouel, C.; Lange, S.; Einsiedler, M.; Srour, M.; Chitayat, D.; Hamdan, F.F.; Jenkins, Z.A.; Morgan, T.; et al. Mutations in genes encoding the cadherin receptor-ligand pair DCHS1 and FAT4 disrupt cerebral cortical development. Nat. Genet. 2013, 45, 1300–1310. [Google Scholar] [CrossRef] [PubMed]
- Nagini, S.; Sophia, J.; Mishra, R. Glycogen synthase kinases: Moonlighting proteins with theranostic potential in cancer. Semin. Cancer Biol. 2019, 56, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015, 43, D512–D520. [Google Scholar] [CrossRef]
- Feng, X.; Arang, N.; Rigiracciolo, D.C.; Lee, J.S.; Yeerna, H.; Wang, Z.; Lubrano, S.; Kishore, A.; Pachter, J.A.; König, G.M.; et al. A Platform of Synthetic Lethal Gene Interaction Networks Reveals that the GNAQ Uveal Melanoma Oncogene Controls the Hippo Pathway through FAK. Cancer Cell 2019, 35, 457–472. [Google Scholar] [CrossRef]
- Wong, Y.-H.; Lee, T.-Y.; Liang, H.-K.; Huang, C.-M.; Wang, T.-Y.; Yang, Y.-H.; Chu, C.-H.; Huang, H.-D.; Ko, M.-T.; Hwang, J.-K. KinasePhos 2.0: A web server for identifying protein kinase-specific phosphorylation sites based on sequences and coupling patterns. Nucleic Acids Res. 2007, 35, W588–W594. [Google Scholar] [CrossRef]
- O’Donnell, L.; Panier, S.; Wildenhain, J.; Tkach, J.M.; Al-Hakim, A.; Landry, M.C.; Escribano-Diaz, C.; Szilard, R.K.; Young, J.T.F.; Munro, M.; et al. The MMS22L-TONSL complex mediates recovery from replication stress and homologous recombination. Mol. Cell 2010, 40, 619–631. [Google Scholar] [CrossRef]
- Paulsen, R.D.; Soni, D.V.; Wollman, R.; Hahn, A.T.; Yee, M.; Hesley, J.A.; Miller, S.C.; Cromwell, E.F.; Solow-cordero, D.E.; Meyer, T.; et al. Processes and Pathways that Mediate Genome Stability. Mol. Cell. 2009, 35, 228–239. [Google Scholar] [CrossRef]
- Adamson, B.; Smogorzewska, A.; Sigoillot, F.D.; King, R.W.; Elledge, S.J. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat. Cell Biol. 2012, 14, 318–328. [Google Scholar] [CrossRef]
- Wagner, S.A.; Beli, P.; Weinert, B.T.; Schölz, C.; Kelstrup, C.D.; Young, C.; Nielsen, M.L.; Olsen, J.V.; Brakebusch, C.; Choudhary, C. Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues. Mol. Cell. Proteom. 2012, 11, 1578–1585. [Google Scholar] [CrossRef] [PubMed]
- Akimov, V.; Barrio-Hernandez, I.; Hansen, S.V.F.; Hallenborg, P.; Pedersen, A.-K.; Bekker-Jensen, D.B.; Puglia, M.; Christensen, S.D.K.; Vanselow, J.T.; Nielsen, M.M.; et al. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat. Struct. Mol. Biol. 2018, 25, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Deol, K.K.; Lorenz, S.; Strieter, E.R. Enzymatic Logic of Ubiquitin Chain Assembly. Front. Physiol. 2019, 10, 835. [Google Scholar] [CrossRef] [PubMed]
- Quan, M.; Chen, Z.; Jiao, F.; Xiao, X.; Xia, Q.; Chen, J.; Chao, Q.; Li, Y.; Gao, Y.; Yang, H.; et al. Lysine demethylase 2 (KDM2B) regulates hippo pathway via MOB1 to promote pancreatic ductal adenocarcinoma (PDAC) progression. J. Exp. Clin. Cancer Res. 2020, 39, 13. [Google Scholar] [CrossRef] [PubMed]
- Medina-Aguilar, R.; Pérez-Plasencia, C.; Marchat, L.A.; Gariglio, P.; García Mena, J.; Rodríguez Cuevas, S.; Ruíz-García, E.; Astudillo-de la Vega, H.; Hernández Juárez, J.; Flores-Pérez, A.; et al. Methylation Landscape of Human Breast Cancer Cells in Response to Dietary Compound Resveratrol. PLoS ONE 2016, 11, e0157866. [Google Scholar] [CrossRef] [PubMed]
- Haldrup, C.; Mundbjerg, K.; Vestergaard, E.M.; Lamy, P.; Wild, P.; Schulz, W.A.; Arsov, C.; Visakorpi, T.; Borre, M.; Høyer, S.; et al. DNA methylation signatures for prediction of biochemical recurrence after radical prostatectomy of clinically localized prostate cancer. J. Clin. Oncol. 2013, 31, 3250–3258. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wu, Y.; Ma, J. Interaction of non-coding RNAs and Hippo signaling: Implications for tumorigenesis. Cancer Lett. 2020, 493, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Astamal, R.V.; Maghoul, A.; Taefehshokr, S.; Bagheri, T.; Mikaeili, E.; Derakhshani, A.; Delashoub, M.; Taefehshokr, N.; Isazadeh, A.; Hajazimian, S.; et al. Regulatory role of microRNAs in cancer through Hippo signaling pathway. Pathol.-Res. Pract. 2020, 216, 153241. [Google Scholar] [CrossRef]
- Wang, L.; Li, B.; Zhang, L.; Li, Q.; He, Z.; Zhang, X.; Huang, X.; Xu, Z.; Xia, Y.; Zhang, Q.; et al. miR-664a-3p functions as an oncogene by targeting Hippo pathway in the development of gastric cancer. Cell Prolif. 2019, 52, e12567. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-W.; Chang, Y.-L.; Chang, Y.-C.; Lin, J.-C.; Chen, C.-C.; Pan, S.-H.; Wu, C.-T.; Chen, H.-Y.; Yang, S.-C.; Hong, T.-M.; et al. MicroRNA-135b promotes lung cancer metastasis by regulating multiple targets in the Hippo pathway and LZTS1. Nat. Commun. 2013, 4, 1877. [Google Scholar] [CrossRef]
- Chen, M.; Wang, M.; Xu, S.; Guo, X.; Jiang, J. Upregulation of miR-181c contributes to chemoresistance in pancreatic cancer by inactivating the Hippo signaling pathway. Oncotarget 2015, 6, 44466–44479. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.-C.; Kuang, M.-J.; Kang, J.-Y.; Zhao, J.; Ma, J.-X.; Ma, X.-L. Human umbilical cord mesenchymal stem cell-derived exosomes act via the miR-1263/Mob1/Hippo signaling pathway to prevent apoptosis in disuse osteoporosis. Biochem. Biophys. Res. Commun. 2020, 524, 883–889. [Google Scholar] [CrossRef] [PubMed]
- Hilman, D.; Gat, U. The evolutionary history of YAP and the hippo/YAP pathway. Mol. Biol. Evol. 2011, 28, 2403–2417. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Zhou, Z.; Wang, D.; Liu, W.; Zhu, H. Hippo pathway genes developed varied exon numbers and coevolved functional domains in metazoans for species specific growth control. BMC Evol. Biol. 2013, 13, 76. [Google Scholar] [CrossRef] [PubMed]
- Sebé-Pedrós, A.; Zheng, Y.; Ruiz-Trillo, I.; Pan, D. Premetazoan origin of the hippo signaling pathway. Cell Rep. 2012, 1, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Ikmi, A.; Gaertner, B.; Seidel, C.; Srivastava, M.; Zeitlinger, J.; Gibson, M.C. Molecular evolution of the Yap/Yorkie proto-oncogene and elucidation of its core transcriptional program. Mol. Biol. Evol. 2014, 31, 1375–1390. [Google Scholar] [CrossRef]
- Coste, A.; Jager, M.; Chambon, J.-P.; Manuel, M. Comparative study of Hippo pathway genes in cellular conveyor belts of a ctenophore and a cnidarian. EvoDevo 2016, 7, 4. [Google Scholar] [CrossRef]
- Chen, Y.; Han, H.; Seo, G.; Vargas, R.E.; Yang, B.; Chuc, K.; Zhao, H.; Wang, W. Systematic analysis of the Hippo pathway organization and oncogenic alteration in evolution. Sci. Rep. 2020, 10, 3173. [Google Scholar] [CrossRef]
- Elbediwy, A.; Thompson, B.J. Evolution of mechanotransduction via YAP/TAZ in animal epithelia. Curr. Opin. Cell Biol. 2018, 51, 117–123. [Google Scholar] [CrossRef]
- Hong, Y. aPKC: The Kinase that Phosphorylates Cell Polarity. F1000Research 2018, 7. [Google Scholar] [CrossRef]
- Stephens, R.; Lim, K.; Portela, M.; Kvansakul, M.; Humbert, P.O.; Richardson, H.E. The Scribble Cell Polarity Module in the Regulation of Cell Signaling in Tissue Development and Tumorigenesis. J. Mol. Biol. 2018, 430, 3585–3612. [Google Scholar] [CrossRef] [PubMed]
- Juanes, M.A.; Piatti, S. The final cut: Cell polarity meets cytokinesis at the bud neck in S. cerevisiae. Cell. Mol. Life Sci. 2016, 73, 3115–3136. [Google Scholar] [CrossRef] [PubMed]
- Piel, M.; Tran, P.T. Cell shape and cell division in fission yeast. Curr. Biol. 2009, 19, R823–R827. [Google Scholar] [CrossRef] [PubMed]
- Seybold, C.; Schiebel, E. Spindle pole bodies. Curr. Biol. 2013, 23, R858–R860. [Google Scholar] [CrossRef][Green Version]
- Komarnitsky, S.I.; Chiang, Y.C.; Luca, F.C.; Chen, J.; Toyn, J.H.; Winey, M.; Johnston, L.H.; Denis, C.L. DBF2 protein kinase binds to and acts through the cell cycle-regulated MOB1 protein. Mol. Cell. Biol. 1998, 18, 2100–2107. [Google Scholar] [CrossRef]
- McCollum, D.; Gould, K.L. Timing is everything: Regulation of mitotic exit and cytokinesis by the MEN and SIN. Trends Cell Biol. 2001, 11, 89–95. [Google Scholar] [CrossRef]
- Meitinger, F.; Palani, S.; Pereira, G. The power of MEN in cytokinesis. Cell Cycle 2012, 11, 219–228. [Google Scholar] [CrossRef]
- Luca, F.C.; Mody, M.; Kurischko, C.; Roof, D.M.; Giddings, T.H.; Winey, M. Saccharomyces cerevisiae Mob1p Is Required for Cytokinesis and Mitotic Exit. Mol. Cell. Biol. 2001, 21, 6972–6983. [Google Scholar] [CrossRef]
- Frenz, L.M.; Lee, S.E.; Fesquet, D.; Johnston, L.H. The budding yeast Dbf2 protein kinase localises to the centrosome and moves to the bud neck in late mitosis. J. Cell Sci. 2000, 113, 3399–3408. [Google Scholar]
- Mah, A.S.; Jang, J.; Deshaies, R.J. Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proc. Natl. Acad. Sci. USA 2001, 98, 7325–7330. [Google Scholar] [CrossRef]
- Hergovich, A. MOB control: Reviewing a conserved family of kinase regulators. Cell. Signal. 2011, 23, 1433–1440. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.E.; Frenz, L.M.; Wells, N.J.; Johnson, A.L.; Johnston, L.H. Order of function of the budding-yeast mitotic exit-network proteins Tem1, Cdc15, Mob1, Dbf2, and Cdc5. Curr. Biol. 2001, 11, 784–788. [Google Scholar] [CrossRef][Green Version]
- Mohl, D.A.; Huddleston, M.J.; Collingwood, T.S.; Annan, R.S.; Deshaies, R.J. Dbf2-Mob1 drives relocalization of protein phosphatase Cdc14 to the cytoplasm during exit from mitosis. J. Cell Biol. 2009, 184. [Google Scholar] [CrossRef] [PubMed]
- Simanis, V. Pombe’s thirteen-control of fission yeast cell division by the septation initiation network. J. Cell Sci. 2015, 128, 1465–1474. [Google Scholar] [CrossRef]
- Salimova, E.; Sohrmann, M.; Fournier, N.; Simanis, V. The S. pombe orthologue of the S. cerevisiae mob1 gene is essential and functions in signalling the onset of septum formation. J. Cell Sci. 2000, 113, 1695–1704. [Google Scholar]
- Sparks, C.A.; Morphew, M.; McCollum, D. Sid2p, a spindle pole body kinase that regulates the onset of cytokinesis. J. Cell Biol. 1999, 146, 777–790. [Google Scholar] [CrossRef]
- Hou, M.C.; Salek, J.; McCollum, D. Mob1p interacts with the Sid2p kinase and is required for cytokinesis in fission yeast. Curr. Biol. 2000, 10, 619–622. [Google Scholar] [CrossRef]
- Hou, M.-C.; Guertin, D.A.; McCollum, D. Initiation of cytokinesis is controlled through multiple modes of regulation of the Sid2p-Mob1p kinase complex. Mol. Cell. Biol. 2004, 24, 3262–3276. [Google Scholar] [CrossRef]
- Guertin, D.A.; Chang, L.; Irshad, F.; Gould, K.L.; McCollum, D. The role of the sid1p kinase and cdc14p in regulating the onset of cytokinesis in fission yeast. EMBO J. 2000, 19, 1803–1815. [Google Scholar] [CrossRef]
- Jaspersen, S.L.; Winey, M. The budding yeast spindle pole body: Structure, duplication, and function. Annu. Rev. Cell Dev. Biol. 2004, 20. [Google Scholar] [CrossRef]
- Schmidt, S.; Sohrmann, M.; Hofmann, K.; Woollard, A.; Simanis, V. The Spg1p GTPase is an essential, dosage-dependent inducer of septum formation in Schizosaccharomyces pombe. Genes Dev. 1997, 11, 1519–1534. [Google Scholar] [CrossRef] [PubMed]
- Sohrmann, M.; Schmidt, S.; Hagan, I.; Simanis, V. Asymmetric segregation on spindle poles of the Schizosaccharomyces pombe septum-inducing protein kinase Cdc7p. Genes Dev. 1998, 12, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Grallert, A.; Krapp, A.; Bagley, S.; Simanis, V.; Hagan, I.M. Recruitment of NIMA kinase shows that maturation of the S. pombe spindle-pole body occurs over consecutive cell cycles and reveals a role for NIMA in modulating SIN activity. Genes Dev. 2004, 18, 1007–1021. [Google Scholar] [CrossRef] [PubMed]
- García-Cortés, J.C.; McCollum, D. Proper timing of cytokinesis is regulated by Schizosaccharomyces pombe Etd1. J. Cell Biol. 2009, 186, 739–753. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.E.; McCollum, D.; Gould, K.L. Polar opposites: Fine-tuning cytokinesis through SIN asymmetry. Cytoskeleton 2012, 69, 686–699. [Google Scholar] [CrossRef] [PubMed]
- Pereira, G.; Tanaka, T.U.; Nasmyth, K.; Schiebel, E. Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. EMBO J. 2001, 20, 6359–6370. [Google Scholar] [CrossRef]
- Monje-Casas, F.; Amon, A. Cell polarity determinants establish asymmetry in MEN signaling. Dev. Cell 2009, 16, 132–145. [Google Scholar] [CrossRef][Green Version]
- Galagan, J.E.; Calvo, S.E.; Borkovich, K.A.; Selker, E.U.; Read, N.D.; Jaffe, D.; FitzHugh, W.; Ma, L.-J.; Smirnov, S.; Purcell, S.; et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature 2003, 422, 859–868. [Google Scholar] [CrossRef]
- Galagan, J.E.; Calvo, S.E.; Cuomo, C.; Ma, L.-J.; Wortman, J.R.; Batzoglou, S.; Lee, S.-I.; Baştürkmen, M.; Spevak, C.C.; Clutterbuck, J.; et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 2005, 438, 1105–1115. [Google Scholar] [CrossRef]
- Heilig, Y.; Schmitt, K.; Seiler, S. Phospho-regulation of the Neurospora crassa septation initiation network. PLoS ONE 2013, 8, e79464. [Google Scholar] [CrossRef]
- Colman-Lerner, A.; Chin, T.E.; Brent, R. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell 2001, 107, 739–750. [Google Scholar] [CrossRef]
- Weiss, E.L.; Kurischko, C.; Zhang, C.; Shokat, K.; Drubin, D.G.; Luca, F.C. The Saccharomyces cerevisiae Mob2p-Cbk1p kinase complex promotes polarized growth and acts with the mitotic exit network to facilitate daughter cell-specific localization of Ace2p transcription factor. J. Cell Biol. 2002, 158, 885–900. [Google Scholar] [CrossRef] [PubMed]
- Saputo, S.; Chabrier-Rosello, Y.; Luca, F.C.; Kumar, A.; Krysan, D.J. The RAM network in pathogenic fungi. Eukaryot. Cell 2012, 11, 708–717. [Google Scholar] [CrossRef]
- Kuravi, V.K.; Kurischko, C.; Puri, M.; Luca, F.C. Cbk1 kinase and Bck2 control MAP kinase activation and inactivation during heat shock. Mol. Biol. Cell 2011, 22, 4892–4907. [Google Scholar] [CrossRef] [PubMed]
- Du, L.-L.; Novick, P. Pag1p, a novel protein associated with protein kinase Cbk1p, is required for cell morphogenesis and proliferation in Saccharomyces cerevisiae. Mol. Biol. Cell 2002, 13, 503–514. [Google Scholar] [CrossRef]
- Bidlingmaier, S.; Weiss, E.L.; Seidel, C.; Drubin, D.G.; Snyder, M. The Cbk1p pathway is important for polarized cell growth and cell separation in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001, 21, 2449–2462. [Google Scholar] [CrossRef]
- Cavalier-Smith, T. Archamoebae: The ancestral eukaryotes? Biosystems 1991, 25, 25–38. [Google Scholar] [CrossRef]
- Wloga, D.; Frankel, J. From molecules to morphology: Cellular organization of Tetrahymena thermophila. Methods Cell Biol. 2012, 109, 83–140. [Google Scholar] [CrossRef]
- Iftode, F.; Cohen, J.; Ruiz, F.; Rueda, A.T.; Chen-Shan, L.; Adoutte, A.; Beisson, J. Development of surface pattern during division in Paramecium. I. Mapping of duplication and reorganization of cortical cytoskeletal structures in the wild type. Development. 1989, 105, 191–211. [Google Scholar]
- Frankel, J. Cell biology of Tetrahymena thermophila. Methods Cell Biol. 2000, 62, 27–125. [Google Scholar] [CrossRef]
- Ruehle, M.D.; Stemm-Wolf, A.J.; Pearson, C.G. Sas4 links basal bodies to cell division via Hippo signaling. J. Cell Biol. 2020, 219. [Google Scholar] [CrossRef]
- Jiang, Y.-Y.; Maier, W.; Baumeister, R.; Minevich, G.; Joachimiak, E.; Ruan, Z.; Kannan, N.; Clarke, D.; Frankel, J.; Gaertig, J. The Hippo Pathway Maintains the Equatorial Division Plane in the Ciliate Tetrahymena. Genetics 2017, 206, 873–888. [Google Scholar] [CrossRef]
- Frankel, J. What do genic mutations tell us about the structural patterning of a complex single-celled organism? Eukaryot. Cell 2008, 7, 1617–1639. [Google Scholar] [CrossRef]
- Slabodnick, M.M.; Ruby, J.G.; Dunn, J.G.; Feldman, J.L.; DeRisi, J.L.; Marshall, W.F. The kinase regulator mob1 acts as a patterning protein for stentor morphogenesis. PLoS Biol. 2014, 12, e1001861. [Google Scholar] [CrossRef]
- Morrison, D.A. Evolution of the Apicomplexa: Where are we now? Trends Parasitol. 2009, 25, 375–382. [Google Scholar] [CrossRef] [PubMed]
- McFadden, G.I.; Yeh, E. The apicoplast: Now you see it, now you don’t. Int. J. Parasitol. 2017, 47, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Frénal, K.; Dubremetz, J.F.; Lebrun, M.; Soldati-Favre, D. Gliding motility powers invasion and egress in Apicomplexa. Nat. Rev. Microbiol. 2017, 15, 645–660. [Google Scholar] [CrossRef] [PubMed]
- Seeber, F.; Steinfelder, S. Recent advances in understanding apicomplexan parasites. F1000Research 2016, 5, 1–10. [Google Scholar] [CrossRef]
- Hu, K.; Mann, T.; Striepen, B.; Beckers, C.J.M.; Roos, D.S.; Murray, J.M. Daughter cell assembly in the protozoan parasite Toxoplasma gondii. Mol. Biol. Cell 2002, 13, 593–606. [Google Scholar] [CrossRef] [PubMed]
- Morrissette, N.S.; Sibley, L.D. Disruption of microtubules uncouples budding and nuclear division in Toxoplasma gondii. J. Cell Sci. 2002, 115, 1017–1025. [Google Scholar]
- Fritz, H.M.; Buchholz, K.R.; Chen, X.; Durbin-Johnson, B.; Rocke, D.M.; Conrad, P.A.; Boothroyd, J.C. Transcriptomic analysis of toxoplasma development reveals many novel functions and structures specific to sporozoites and oocysts. PLoS ONE 2012, 7, e29998. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, C.; Maier, S.; Walker, R.A.; Rehrauer, H.; Joekel, D.E.; Winiger, R.R.; Basso, W.U.; Grigg, M.E.; Hehl, A.B.; Deplazes, P.; et al. An experimental genetically attenuated live vaccine to prevent transmission of Toxoplasma gondii by cats. Sci. Rep. 2019, 9, 1474. [Google Scholar] [CrossRef] [PubMed]
- Walker, R.A.; Sharman, P.A.; Miller, C.M.; Lippuner, C.; Okoniewski, M.; Eichenberger, R.M.; Ramakrishnan, C.; Brossier, F.; Deplazes, P.; Hehl, A.B.; et al. RNA Seq analysis of the Eimeria tenella gametocyte transcriptome reveals clues about the molecular basis for sexual reproduction and oocyst biogenesis. BMC Genom. 2015, 16, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Reid, A.J.; Blake, D.P.; Ansari, H.R.; Billington, K.; Browne, H.P.; Bryant, J.; Dunn, M.; Hung, S.S.; Kawahara, F.; Miranda-Saavedra, D.; et al. Genomic analysis of the causative agents of coccidiosis in domestic chickens. Genome Res. 2014, 24, 1676–1685. [Google Scholar] [CrossRef]
- Talevich, E.; Mirza, A.; Kannan, N. Structural and evolutionary divergence of eukaryotic protein kinases in Apicomplexa. BMC Evol. Biol. 2011, 11, 321. [Google Scholar] [CrossRef]
- Peixoto, L.; Chen, F.; Harb, O.S.; Davis, P.H.; Beiting, D.P.; Brownback, C.S.; Ouloguem, D.; Roos, D.S. Integrative genomic approaches highlight a family of parasite-specific kinases that regulate host responses. Cell Host Microbe 2010, 8, 208–218. [Google Scholar] [CrossRef]
- Yu, F.X.; Zhang, Y.; Park, H.W.; Jewell, J.L.; Chen, Q.; Deng, Y.; Pan, D.; Taylor, S.S.; Lai, Z.C.; Guan, K.L. Protein kinase A activates the Hippo pathway to modulate cell proliferation and differentiation. Genes Dev. 2013, 27, 1223–1232. [Google Scholar] [CrossRef]
- Jia, Y.; Marq, J.; Bisio, H.; Jacot, D.; Mueller, C.; Yu, L.; Choudhary, J.; Brochet, M.; Soldati-Favre, D. Crosstalk between PKA and PKG controls pH -dependent host cell egress of Toxoplasma gondii. EMBO J. 2017, 36, 3250–3267. [Google Scholar] [CrossRef]
- Hammarton, T.C.; Lillico, S.G.; Welburn, S.C.; Mottram, J.C. Trypanosoma brucei MOB1 is required for accurate and efficient cytokinesis but not for exit from mitosis. Mol. Microbiol. 2005, 56, 104–116. [Google Scholar] [CrossRef]
- Prestes, E.B.; Stoco, P.H.; de Moraes, M.H.; Moura, H.; Grisard, E.C. Messenger RNA levels of the Polo-like kinase gene (PLK) correlate with cytokinesis in the Trypanosoma rangeli cell cycle. Exp. Parasitol. 2019, 204, 107727. [Google Scholar] [CrossRef]
- De Oliveira, A.H.C.; Ruiz, J.C.; Cruz, A.K.; Greene, L.J.; Rosa, J.C.; Ward, R.J. Subproteomic analysis of soluble proteins of the microsomal fraction from two Leishmania species. Comp. Biochem. Physiol. Part D Genom. Proteom. 2006, 1, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Ankarklev, J.; Jerlström-Hultqvist, J.; Ringqvist, E.; Troell, K.; Svärd, S.G. Behind the smile: Cell biology and disease mechanisms of Giardia species. Nat. Rev. Microbiol. 2010, 8, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Markova, K.; Uzlikova, M.; Tumova, P.; Jirakova, K.; Hagen, G.; Kulda, J.; Nohynkova, E. Absence of a conventional spindle mitotic checkpoint in the binucleated single-celled parasite Giardia intestinalis. Eur. J. Cell Biol. 2016, 95, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Emery-Corbin, S.J.; Hamey, J.J.; Ansell, B.R.E.; Balan, B.; Tichkule, S.; Stroehlein, A.J.; Cooper, C.; McInerney, B.V.; Hediyeh-Zadeh, S.; Vuong, D.; et al. Eukaryote-Conserved Methylarginine Is Absent in Diplomonads and Functionally Compensated in Giardia. Mol. Biol. Evol. 2020, 1, 3–9. [Google Scholar] [CrossRef]
- Wang, S.E.; Amir, A.S.; Nguyen, T.; Poole, A.M.; Simoes-Barbosa, A. Spliceosomal introns in Trichomonas vaginalis revisited 06 Biological Sciences 0604 Genetics. Parasites Vectors 2018, 11, 1–9. [Google Scholar] [CrossRef]
- Grewal, J.S.; Lohia, A. Mechanism of Cell Division in Entamoeba histolytica. In Amebiasis; Springer: Tokyo, Japan, 2015; pp. 263–278. [Google Scholar] [CrossRef]
- Bulman, S.; Siemens, J.; Ridgway, H.J.; Eady, C.; Conner, A.J. Identification of genes from the obligate intracellular plant pathogen, Plasmodiophora brassicae. FEMS Microbiol. Lett. 2006, 264, 198–204. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. EggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- Stavridi, E.S.; Harris, K.G.; Huyen, Y.; Bothos, J.; Verwoerd, P.M.; Stayrook, S.E.; Pavletich, N.P.; Jeffrey, P.D.; Luca, F.C. Crystal structure of a human Mob1 protein: Toward understanding Mob-regulated cell cycle pathways. Structure 2003, 11, 1163–1170. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Le, S.Q.; Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Caydasi, A.K.; Kurtulmus, B.; Orrico, M.I.L.; Hofmann, A.; Ibrahim, B.; Pereira, G. Elm1 kinase activates the spindle position checkpoint kinase Kin4. J. Cell Biol. 2010, 190, 975–989. [Google Scholar] [CrossRef]
- Höfken, T.; Schiebel, E. Novel regulation of mitotic exit by the Cdc42 effectors Gic1 and Gic2. J. Cell Biol. 2004, 164, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, H.; Priest, C.; Lechner, J.; Pereira, G.; Schiebel, E. The yeast centrosome translates the positional information of the anaphase spindle into a cell cycle signal. J. Cell Biol. 2007, 179, 423–436. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, Y.M.; Mahowald, A.P.; Perlin, J.R.; Fuller, M.T. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science 2007, 315, 518–521. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, Y.M.; Fuller, M.T. Asymmetric centrosome behavior and the mechanisms of stem cell division. J. Cell Biol. 2008, 180, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Türkel, N.; Hemati, N.; Fuller, M.T.; Hunt, A.J.; Yamashita, Y.M. Centrosome misorientation reduces stem cell division during ageing. Nature 2008, 456, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Rebollo, E.; Sampaio, P.; Januschke, J.; Llamazares, S.; Varmark, H.; González, C. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev. Cell 2007, 12, 467–474. [Google Scholar] [CrossRef]
- Morlon-Guyot, J.; Francia, M.E.; Dubremetz, J.-F.; Daher, W. Towards a molecular architecture of the centrosome in Toxoplasma gondii. Cytoskeleton 2017, 74, 55–71. [Google Scholar] [CrossRef]
- King, N.; Westbrook, M.J.; Young, S.L.; Kuo, A.; Abedin, M.; Chapman, J.; Fairclough, S.; Hellsten, U.; Isogai, Y.; Letunic, I.; et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature 2008, 451, 783–788. [Google Scholar] [CrossRef]
- Abedin, M.; King, N. Diverse evolutionary paths to cell adhesion. Trends Cell Biol. 2010, 20, 734–742. [Google Scholar] [CrossRef] [PubMed]
- King, N.; Hittinger, C.T.; Carroll, S.B. Evolution of key cell signaling and adhesion protein families predates animal origins. Science 2003, 301, 361–363. [Google Scholar] [CrossRef] [PubMed]
- Meng, Z.; Moroishi, T.; Mottier-Pavie, V.; Plouffe, S.W.; Hansen, C.G.; Hong, A.W.; Park, H.W.; Mo, J.S.; Lu, W.; Lu, S.; et al. MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef]
- Plouffe, S.W.; Meng, Z.; Lin, K.C.; Lin, B.; Hong, A.W.; Chun, J.V.; Guan, K.L. Characterization of Hippo Pathway Components by Gene Inactivation. Mol. Cell 2016, 64, 993–1008. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, O.; Kukekova, A.V.; Aguirre, G.D.; Acland, G.M. Exonic SINE Insertion in STK38L Causes Canine Early Retinal Degeneration (erd). Genomics 2010, 96, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Berta, Á.I.; Boesze-Battaglia, K.; Genini, S.; Goldstein, O.; O’Brien, P.J.; Szél, Á.; Acland, G.M.; Beltran, W.A.; Aguirre, G.D. Photoreceptor cell death, proliferation and formation of hybrid rod/S-cone photoreceptors in the degenerating STK38L mutant retina. PLoS ONE 2011, 6, e24074. [Google Scholar] [CrossRef]
- Wheway, G.; Nazlamova, L.; Hancock, J.T. Signaling through the Primary Cilium. Front. Cell Dev. Biol. 2018, 6, 8. [Google Scholar] [CrossRef]
- Kim, M.; Kim, M.; Lee, M.-S.; Kim, C.-H.; Lim, D.-S. The MST1/2-SAV1 complex of the Hippo pathway promotes ciliogenesis. Nat. Commun. 2014, 5, 5370. [Google Scholar] [CrossRef]
- Frank, V.; Habbig, S.; Bartram, M.P.; Eisenberger, T.; Veenstra-Knol, H.E.; Decker, C.; Boorsma, R.A.C.; Göbel, H.; Nürnberg, G.; Griessmann, A.; et al. Mutations in NEK8 link multiple organ dysplasia with altered Hippo signalling and increased c-MYC expression. Hum. Mol. Genet. 2013, 22, 2177–2185. [Google Scholar] [CrossRef]
- Habbig, S.; Bartram, M.P.; Müller, R.U.; Schwarz, R.; Andriopoulos, N.; Chen, S.; Sägmüller, J.G.; Hoehne, M.; Burst, V.; Liebau, M.C.; et al. NPHP4, a cilia-associated protein, negatively regulates the Hippo pathway. J. Cell Biol. 2011, 193, 633–642. [Google Scholar] [CrossRef]
- Habbig, S.; Bartram, M.P.; Sägmüller, J.G.; Griessmann, A.; Franke, M.; Müller, R.-U.; Schwarz, R.; Hoehne, M.; Bergmann, C.; Tessmer, C.; et al. The ciliopathy disease protein NPHP9 promotes nuclear delivery and activation of the oncogenic transcriptional regulator TAZ. Hum. Mol. Genet. 2012, 21, 5528–5538. [Google Scholar] [CrossRef] [PubMed]
- Lobo, G.P.; Fulmer, D.; Guo, L.; Zuo, X.; Dang, Y.; Kim, S.H.; Su, Y.; George, K.; Obert, E.; Fogelgren, B.; et al. The exocyst is required for photoreceptor ciliogenesis and retinal development. J. Biol. Chem. 2017, 292, 14814–14826. [Google Scholar] [CrossRef] [PubMed]
| Protein | Described Functions | Functional Category | References | ||||
|---|---|---|---|---|---|---|---|
| TH | M | D | CC | ||||
| Mammals and birds | HsMOB1 | Tissue growth (suppressor); apoptotic signaling; mitotic exit; protein complex positioning to spindle midzone, mitotic spindle orientation; centrosome duplication and disjunction; cytokinesis | X | X | [13,14,27,28,29,30,34,48,55,56,57] | ||
| HsMOB2 | Tissue growth (suppressor); cortical development; mitotic spindle orientation; centrosome duplication; DNA damage response | X | X | X | [14,33,48,54,58,59,60,61] | ||
| HsMOB3 | Tissue growth (enhancer through MST1 inhibition); apoptotic signaling | X | X | [14,36] | |||
| HsMOB4 | Tissue growth (enhancer through MST1-MOB1 complex inhibition) | X | [34,35] | ||||
| MmMOB1 | Tissue growth (suppressor); lung and neuronal morphogenesis; stem cell differentiation and maintenance; centrosome duplication; actin cytoskeleton polarization | X | X | X | X | [10,37,38,39,40,53,62,63] | |
| MmMOB2 | Neuronal and cortical development; actin cytoskeleton organization. | X | [59,64] | ||||
| MmMOB4 | Dendritic arborization in neuronal development | X | [34] | ||||
| CfMOB1 | Photoreceptor growth; mitosis | X | X | [41] | |||
| GgMOB2 | Tissue growth (suppressor); follicle development | X | X | [42] | |||
| Insects | DmMOB1 | Tissue growth (suppressor); stem cell differentiation; chromosome segregation | X | X | X | [2,15,26,62,65] | |
| DmMOB2 | Wing hair, photoreceptor and neuromuscular junction morphogenesis | X | [66,67,68] | ||||
| DmMOB4 | Synapse morphogenesis and microtubule organization; spindle pole assembly; axonal transport of autophagosomes | X | X | [69,70,71] | |||
| Plants | AtMOB1 | Plant growth (enhancer); development of stem and root; sporogenesis and gametogenesis; stem cell differentiation and maintenance; apoptotic signaling; cytokinesis | X | X | X | X | [43,44,45,46] |
| MsMOB1 | Tissue growth (suppressor); stem cell association | X | X | [47,72] | |||
| Fungi | NcMOB1 | Tissue growth (enhancer); septum, aerial mycelium and fruiting body development; conidiation; ascosporogenesis; meiosis | X | X | X | X | [3] |
| NcMOB2 | Tissue growth (enhancer); hypha development; conidiation | X | X | X | [3] | ||
| NcMOB4 | Tissue growth (enhancer); fruiting body development; vegetative cell fusion | X | X | [3,24] | |||
| AnMOB4 | Tissue growth (suppressor); ascosporogenesis; meiosis. | X | X | X | [22] | ||
| SmMOB4 | Vegetative cell fusion; conidiation and ascosporogenesis; meiosis | X | X | X | [23] | ||
| ChMOB2 | Conidiation and ascosporogenesis; meiosis | X | X | [4] | |||
| P48 | D52 | W56 | N69 | M87 | A89 | A111 | Y114 | F132 | P133 | Y163 | F186 | F189 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identity in MOB proteins (multicellular) | 100% | 84% | 100% | 93% | 100% | 67% | 98% | 96% | 98% | 100% | 91% | 84% | 84% |
| Identity in MOB proteins (unicellular) | 97% | 67% | 94% | 64% | 92% | 78% | 100% | 92% | 92% | 89% | 92% | 89% | 86% |
| E33 | T35 | G37 | S38 | A44 | E51 | D63 | Q67 | M70 | T76 | Y92 | E93 | S110 | D126 | S182 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identity in MOB1-Like proteins (multicellular) | 50% | 94% | 88% | 100% | 100% | 94% | 100% | 94% | 69% | 81% | 81% | 94% | 94% | 94% | 75% |
| Identity in MOB1-Like proteins (unicellular) | 36% | 100% | 100% | 73% | 91% | 100% | 73% | 55% | 27% | 45% | 91% | 100% | 82% | 64% | 45% |
| E55 | A59 | Y72 | W97 | D128 | F140 | P141 | F144 | N180 | H185 | E192 | L207 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identity in non phocein-like MOB proteins (multicellular) | 74% | 97% | 85% | 94% | 74% | 100% | 94% | 100% | 94% | 91% | 82% | 74% |
| Identity in non phocein-like MOB proteins (unicellular) | 80% | 83% | 80% | 90% | 80% | 77% | 97% | 100% | 100% | 97% | 77% | 83% |
| C79 | C84 | H161 | H166 | |
|---|---|---|---|---|
| Identity in MOB proteins (multicellular) | 100% | 100% | 100% | 100% |
| Identity in MOB proteins (unicellular) | 97% | 94% | 100% | 100% |
| T12 | Y26 | T35 | S38 | S146 | |
|---|---|---|---|---|---|
| Identity in Mob1-like proteins (multicellular) | 75% | 63% | 94% | 100% | 44% |
| Identity in Mob1-like proteins (unicellular) | - | 45% | 100% | 73% | 27% |
| Y141 | S147 | |
|---|---|---|
| Identity in phocein-like proteins (multicellular) | 73% | 73% |
| Identity in phocein-like proteins (unicellular) | 0% | 40% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Delgado, I.L.S.; Carmona, B.; Nolasco, S.; Santos, D.; Leitão, A.; Soares, H. MOB: Pivotal Conserved Proteins in Cytokinesis, Cell Architecture and Tissue Homeostasis. Biology 2020, 9, 413. https://doi.org/10.3390/biology9120413
Delgado ILS, Carmona B, Nolasco S, Santos D, Leitão A, Soares H. MOB: Pivotal Conserved Proteins in Cytokinesis, Cell Architecture and Tissue Homeostasis. Biology. 2020; 9(12):413. https://doi.org/10.3390/biology9120413
Chicago/Turabian StyleDelgado, Inês L. S., Bruno Carmona, Sofia Nolasco, Dulce Santos, Alexandre Leitão, and Helena Soares. 2020. "MOB: Pivotal Conserved Proteins in Cytokinesis, Cell Architecture and Tissue Homeostasis" Biology 9, no. 12: 413. https://doi.org/10.3390/biology9120413
APA StyleDelgado, I. L. S., Carmona, B., Nolasco, S., Santos, D., Leitão, A., & Soares, H. (2020). MOB: Pivotal Conserved Proteins in Cytokinesis, Cell Architecture and Tissue Homeostasis. Biology, 9(12), 413. https://doi.org/10.3390/biology9120413






