First De Novo Transcriptome of the Copepod Rhincalanus gigas from Antarctic Waters
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Zooplankton Sampling
2.3. DNA Extraction and Genotyping
2.4. RNA Extraction
2.5. RNA Sequencing
2.6. De Novo Assembly and Functional Annotation
3. Results
3.1. Genotyping
3.2. De Novo Assembled Reference Transcriptome
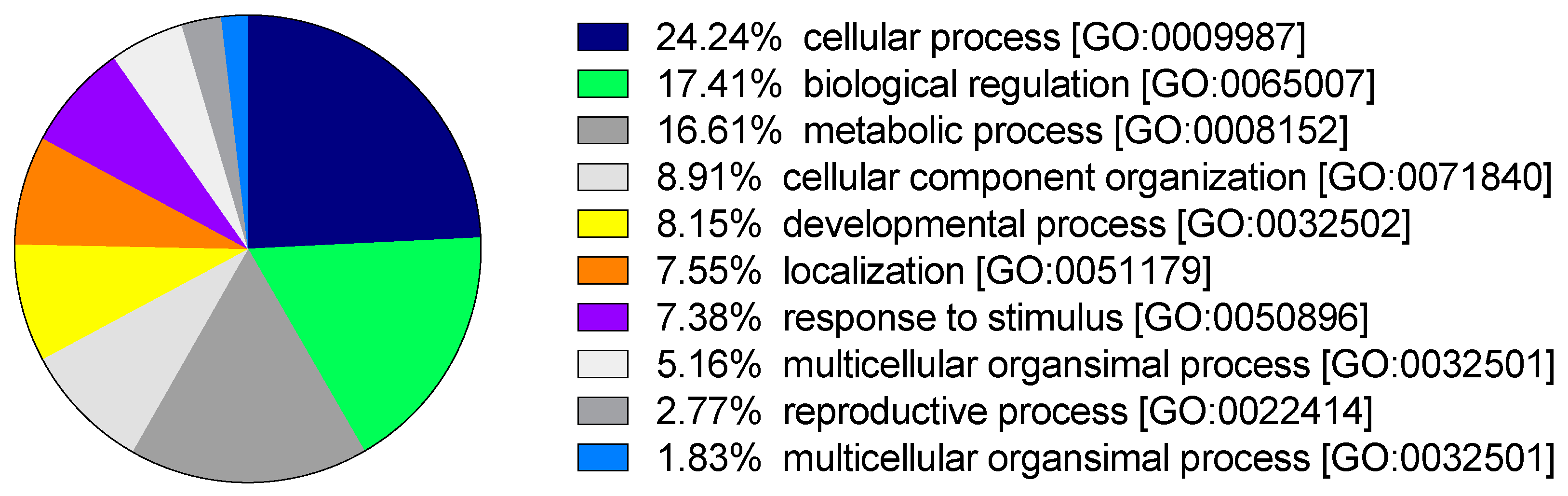
3.3. Functional Annotation
3.4. Adaptation to Cold: Glutathione Metabolism and Antifreeze Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Convey, P.; Peck, L.S. Antarctic environmental change and biological responses. Sci. Adv. 2019, 5, eaaz0888. [Google Scholar] [CrossRef] [PubMed]
- Voronina, N.M. Comparative abundance and distribution of major filter-feeders in the Antarctic pelagic zone. J. Mar. Syst. 1998, 17, 375–390. [Google Scholar] [CrossRef]
- Kang, S.; Ahn, D.-H.; Lee, J.H.; Lee, S.G.; Shin, S.C.; Lee, J.; Min, G.-S.; Lee, H.; Kim, H.-W.; Kim, S.; et al. The genome of the Antarctic-endemic copepod, Tigriopus kingsejongensis. GigaScience 2017, 6. [Google Scholar] [CrossRef]
- Sales, G.; Deagle, B.E.; Calura, E.; Martini, P.; Biscontin, A.; Pittà, C.D.; Kawaguchi, S.; Romualdi, C.; Meyer, B.; Costa, R.; et al. KrillDB: A de novo transcriptome database for the Antarctic krill (Euphausia superba). PLoS ONE 2017, 12, e0171908. [Google Scholar] [CrossRef]
- Johnson, K.M.; Hofmann, G.E. A transcriptome resource for the Antarctic pteropod Limacina helicina antarctica. Mar. Genom. 2016, 28, 25–28. [Google Scholar] [CrossRef]
- Atkinson, A. Life cycles of Calanoides acutus, Calanus simillimus and Rhincalanus gigas (Copepoda: Calanoida) within the Scotia Sea. Mar. Biol. 1991, 109, 79–91. [Google Scholar] [CrossRef]
- Goetze, E. Cryptic speciation on the high seas; global phylogenetics of the copepod family Eucalanidae. Proc. R. Soc. Lond. B 2003, 270, 2321–2331. [Google Scholar] [CrossRef]
- Ward, P.; Atkinson, A.; Schnack-Schiel, S.B.; Murray, A.W.A. Regional variation in the life cycle of Rhincalanus gigas (Copepoda: Calanoida) in the Atlantic Sector of the Southern Ocean—Re-examination of existing data (1928 to 1993). Mar. Ecol. Prog. Ser. 1997, 157, 261–275. [Google Scholar]
- Schaafsma, F.L.; Cherel, Y.; Flores, H.; van Franeker, J.A.; Lea, M.-A.; Raymond, B.; van de Putte, A.P. Review: The energetic value of zooplankton and nekton species of the Southern Ocean. Mar. Biol. 2018, 165, 129. [Google Scholar] [CrossRef]
- Michels, J.; Appel, E.; Gorb, S.N. Functional diversity of resilin in Arthropoda. Beilstein J. Nanotechnol. 2016, 7, 1241–1259. [Google Scholar] [CrossRef]
- Schründer, S.; Schnack-Schiel, S.B.; Auel, H.; Sartoris, F.J. Control of Diapause by Acidic pH and Ammonium Accumulation in the Hemolymph of Antarctic Copepods. PLoS ONE 2013, 8, e77498. [Google Scholar] [CrossRef][Green Version]
- Atkinson, A. Life cycle strategies of epipelagic copepods in the Southern Ocean. J. Mar. Syst. 1998, 15, 289–311. [Google Scholar] [CrossRef]
- Garcia, M.D.; Dutto, M.S.; Chazarreta, C.J.; Berasategui, A.A.; Schloss, I.R.; Hoffmeyer, M.S. Micro- and mesozooplankton successions in an Antarctic coastal environment during a warm year. PLoS ONE 2020, 15, e0232614. [Google Scholar] [CrossRef]
- Abdel-Mageed, W.M.; Lehri, B.; Jarmusch, S.A.; Miranda, K.; Al-Wahaibi, L.H.; Stewart, H.A.; Jamieson, A.J.; Jaspars, M.; Karlyshev, A.V. Whole genome sequencing of four bacterial strains from South Shetland Trench revealing biosynthetic and environmental adaptation gene clusters. Mar. Genom. 2020, 100782. [Google Scholar] [CrossRef]
- Lauritano, C.; Carotenuto, Y.; Miralto, A.; Procaccini, G.; Ianora, A. Copepod Population-Specific Response to a Toxic Diatom Diet. PLoS ONE 2012, 7, e47262. [Google Scholar] [CrossRef]
- Lauritano, C.; Carotenuto, Y.; Procaccini, G.; Turner, J.T.; Ianora, A. Changes in expression of stress genes in copepods feeding upon a non-brevetoxin-producing strain of the dinoflagellate Karenia brevis. Harmful Algae 2013, 28, 23–30. [Google Scholar] [CrossRef]
- Asai, S.; Ianora, A.; Lauritano, C.; Lindeque, P.K.; Carotenuto, Y. High-quality RNA extraction from copepods for Next Generation Sequencing: A comparative study. Mar. Genom. 2015, 24, 115–118. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644. [Google Scholar]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Boutet, E.; Lieberherr, D.; Tognolli, M.; Schneider, M.; Bansal, P.; Bridge, A.J.; Poux, S.; Bougueleret, L.; Xenarios, I. UniProtKB/Swiss-Prot, the Manually Annotated Section of the UniProt KnowledgeBase: How to Use the Entry View. Methods Mol. Biol. 2016, 1374, 23–54. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Lenz, P.H.; Roncalli, V.; Hassett, R.P.; Wu, L.-S.; Cieslak, M.C.; Hartline, D.K.; Christie, A.E. De Novo Assembly of a Transcriptome for Calanus finmarchicus (Crustacea, Copepoda)—The Dominant Zooplankter of the North Atlantic Ocean. PLoS ONE 2014, 9, e88589. [Google Scholar] [CrossRef]
- Roncalli, V.; Cieslak, M.C.; Sommer, S.A.; Hopcroft, R.R.; Lenz, P.H. De novo transcriptome assembly of the calanoid copepod Neocalanus flemingeri: A new resource for emergence from diapause. Mar. Genom. 2018, 37, 114–119. [Google Scholar] [CrossRef]
- Roncalli, V.; Christie, A.E.; Sommer, S.A.; Cieslak, M.C.; Hartline, D.K.; Lenz, P.H. A deep transcriptomic resource for the copepod crustacean Labidocera madurae: A potential indicator species for assessing near shore ecosystem health. PLoS ONE 2017, 12, e0186794. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- Schultz, J.; Copley, R.R.; Doerks, T.; Ponting, C.P.; Bork, P. SMART: A web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000, 28, 231–234. [Google Scholar] [CrossRef]
- Kiko, R. Acquisition of freeze protection in a sea-ice crustacean through horizontal gene transfer? Polar Biol. 2010, 33, 543–556. [Google Scholar] [CrossRef]
- Bucklin, A.; DiVito, K.R.; Smolina, I.; Choquet, M.; Questel, J.M.; Hoarau, G.; O’Neill, R.J. Population Genomics of Marine Zooplankton. In Population Genomics: Marine Organisms; Oleksiak, M.F., Rajora, O.P., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 61–102. ISBN 978-3-030-37936-0. [Google Scholar]
- Tarrant, A.M.; Nilsson, B.; Hansen, B.W. Molecular physiology of copepods—From biomarkers to transcriptomes and back again. Comp. Biochem. Physiol. Part D Genom. Proteom. 2019, 30, 230–247. [Google Scholar] [CrossRef]
- Asai, S.; Sanges, R.; Lauritano, C.; Lindeque, P.K.; Esposito, F.; Ianora, A.; Carotenuto, Y. De Novo Transcriptome Assembly and Gene Expression Profiling of the Copepod Calanus helgolandicus Feeding on the PUA-Producing Diatom Skeletonema marinoi. Mar. Drugs 2020, 18, 392. [Google Scholar] [CrossRef]
- Russo, E.; Lauritano, C.; d’Ippolito, G.; Fontana, A.; Sarno, D.; von Elert, E.; Ianora, A.; Carotenuto, Y. RNA-Seq and differential gene expression analysis in Temora stylifera copepod females with contrasting non-feeding nauplii survival rates: An environmental transcriptomics study. BMC Genom. 2020, 21, 693. [Google Scholar] [CrossRef]
- Li, N.; Flanagan, B.A.; Partridge, M.; Huang, E.J.; Edmands, S. Sex differences in early transcriptomic responses to oxidative stress in the copepod Tigriopus californicus. BMC Genom. 2020, 21, 759. [Google Scholar] [CrossRef]
- Boyen, J.; Fink, P.; Mensens, C.; Hablützel, P.I.; De Troch, M. Fatty acid bioconversion in harpacticoid copepods in a changing environment: A transcriptomic approach. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020, 375, 20190645. [Google Scholar] [CrossRef] [PubMed]
- Semmouri, I.; De Schamphelaere, K.A.C.; Van Nieuwerburgh, F.; Deforce, D.; Janssen, C.R.; Asselman, J. Spatio-temporal patterns in the gene expression of the calanoid copepod Temora longicornis in the Belgian part of the North Sea. Mar. Environ. Res. 2020, 160, 105037. [Google Scholar] [CrossRef] [PubMed]
- Fürtauer, L.; Weiszmann, J.; Weckwerth, W.; Nägele, T. Dynamics of Plant Metabolism during Cold Acclimation. Int. J. Mol. Sci. 2019, 20, 5411. [Google Scholar] [CrossRef]
- Li, X.C.; Peris, D.; Hittinger, C.T.; Sia, E.A.; Fay, J.C. Mitochondria-encoded genes contribute to evolution of heat and cold tolerance in yeast. Sci Adv. 2019, 5, eaav1848. [Google Scholar] [CrossRef]
- Paget, C.M.; Schwartz, J.-M.; Delneri, D. Environmental systems biology of cold-tolerant phenotype in Saccharomyces species adapted to grow at different temperatures. Mol. Ecol. 2014, 23, 5241–5257. [Google Scholar] [CrossRef]
- Enriquez, T.; Colinet, H. Cold acclimation triggers lipidomic and metabolic adjustments in the spotted wing drosophila Drosophila suzukii (Matsumara). Am. J. Physiol. Regul. Integr. Comp. Physiol. 2019, 316, R751–R763. [Google Scholar] [CrossRef]
- Lauritano, C.; Procaccini, G.; Ianora, A. Gene expression patterns and stress response in marine copepods. Mar. Environ. Res. 2012, 76, 22–31. [Google Scholar] [CrossRef]
- Han, J.; Puthumana, J.; Lee, M.-C.; Kim, S.; Lee, J.-S. Different susceptibilities of the Antarctic and temperate copepods Tigriopus kingsejongensis and T. japonicus to ultraviolet (UV) radiation. Mar. Ecol. Prog. Ser. 2016, 561, 99–107. [Google Scholar] [CrossRef][Green Version]
- Roncalli, V.; Cieslak, M.C.; Passamaneck, Y.; Christie, A.E.; Lenz, P.H. Glutathione S-Transferase (GST) Gene Diversity in the Crustacean Calanus finmarchicus—Contributors to Cellular Detoxification. PLoS ONE 2015, 10, e0123322. [Google Scholar] [CrossRef]
- Patarnello, T.; Verde, C.; di Prisco, G.; Bargelloni, L.; Zane, L. How will fish that evolved at constant sub-zero temperatures cope with global warming? Notothenioids as a case study. Bioessays 2011, 33, 260–268. [Google Scholar] [CrossRef] [PubMed]
- Verde, C.; Giordano, D.; di Prisco, G. The adaptation of polar fishes to climatic changes: Structure, function and phylogeny of haemoglobin. IUBMB Life 2008, 60, 29–40. [Google Scholar] [CrossRef]
- Arai, T.; Fukami, D.; Hoshino, T.; Kondo, H.; Tsuda, S. Ice-binding proteins from the fungus Antarctomyces psychrotrophicus possibly originate from two different bacteria through horizontal gene transfer. FEBS J. 2019, 286, 946–962. [Google Scholar] [CrossRef]
- Peck, L.S. Chapter 3 Antarctic Marine Biodiversity: Adaptations, Environments and Responses to Change. In Oceanography and Marine Biology: An Annual Review; Hawkins, S.J., Evans, A.J., Dale, A.C., Firth, L.B., Smith, I.P., Eds.; CRC Press: Boca Raton, FL, USA; Taylor & Francis Group: London, UK; New York, NY, USA, 2018; Volume 56, pp. 2–133. [Google Scholar] [CrossRef]

| R. gigas Reference Transcriptome | |
|---|---|
| Trinity Transcripts (n) | 78,285 |
| Trinity predicted genes (n) | 31,851 |
| Minimum length (bp) | 301 |
| Maximum length (bp) | 10,033 |
| Average contig length (bp) | 877 |
| GC content (%) | 40.52 |
| N50 (bp) | 1143 |
| N25 (bp) | 2089 |
| N75 (bp) | 612 |
| Mapping | |
| Overall mapping (%) | 81.8 |
| Mapping >1 time (%) | 44 |
| BUSCO Eukaryotic Genes | |
| Complete (%) | 70 |
| Fragmented (%) | 12 |
| Missing (%) | 22 |
| R. gigas | C. finmarchicus | N. flemingeri | L. madurae | |
|---|---|---|---|---|
| Sequencing | Illumina NextSeq | Illumina HiSeq | Illumina NextSeq | Illumina NextSeq |
| NCBI BioProject | PRJNA639356 | PRJNA236528 | PRJNA324453 | PRJNA324849 |
| De novo assembly | ||||
| Transcripts (n) | 78,285 | 206,041 | 140,841 | 211,002 |
| Minimum length (bp) | 301 | 301 | 301 | 301 |
| Maximum length (bp) | 10,033 | 23,068 | 24,981 | 23,836 |
| N50 | 1143 | 1418 | 1452 | 1184 |
| Overall self-mapping (%) | 82 | 89 | 92 | 90.8 |
| Functional annotation | ||||
| Transcripts with coding region (n) | 61,983 (79.1%) | np | 108,092 (76.7%) | 72,391 (32%) |
| Transcripts with BLAST hits (n) | 34,238 | 28,616 | 62,126 | 62,980 |
| Transcripts with GO terms (n) | 24,426 | 10,334 | 59,544 | 60,097 |
| BUSCO | ||||
| Complete (%) | 70 | 79 | 79 | 76 |
| Fragmented (%) | 12 | 8 | 6 | 11 |
| Missing (%) | 22 | 12 | 15 | 12 |
| Pathway Name | Entry | Class | Transcripts |
|---|---|---|---|
| Purine metabolism | 00230 | Nucleotide metabolism (metabolism) | 210 |
| Cysteine and methionine metabolism | 00270 | Amino-acid metabolism (metabolism) | 128 |
| Pyrimidine metabolism | 00240 | Nucleotide metabolism (metabolism) | 121 |
| Glutathione metabolism | 00480 | Other amino-acid metabolism (metabolism) | 116 |
| Starch and sucrose metabolism | 00500 | Carbohydrate metabolism (metabolism) | 115 |
| mTOR signaling pathway | 04150 | Signal transduction (environmental information processing) | 115 |
| Glycolysis/Gluconeogenesis | 00010 | Carbohydrate metabolism (metabolism) | 106 |
| Aminoacyl transfer RNA (tRNA) biosynthesis | 00970 | Translation (genetic information processing) | 100 |
| Amino sugar and nucleotide sugar metabolism | 00520 | Carbohydrate metabolism (metabolism) | 99 |
| PI3K/Akt signaling pathway | 04151 | Signal transduction (environmental information processing) | 97 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lauritano, C.; Roncalli, V.; Ambrosino, L.; Cieslak, M.C.; Ianora, A. First De Novo Transcriptome of the Copepod Rhincalanus gigas from Antarctic Waters. Biology 2020, 9, 410. https://doi.org/10.3390/biology9110410
Lauritano C, Roncalli V, Ambrosino L, Cieslak MC, Ianora A. First De Novo Transcriptome of the Copepod Rhincalanus gigas from Antarctic Waters. Biology. 2020; 9(11):410. https://doi.org/10.3390/biology9110410
Chicago/Turabian StyleLauritano, Chiara, Vittoria Roncalli, Luca Ambrosino, Matthew C. Cieslak, and Adrianna Ianora. 2020. "First De Novo Transcriptome of the Copepod Rhincalanus gigas from Antarctic Waters" Biology 9, no. 11: 410. https://doi.org/10.3390/biology9110410
APA StyleLauritano, C., Roncalli, V., Ambrosino, L., Cieslak, M. C., & Ianora, A. (2020). First De Novo Transcriptome of the Copepod Rhincalanus gigas from Antarctic Waters. Biology, 9(11), 410. https://doi.org/10.3390/biology9110410








