Sequence Determinants of Substrate Ambiguity in a HAD Phosphosugar Phosphatase of Arabidopsis Thaliana
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Computational Biology
2.3. Site-Directed Mutagenesis
2.4. Purification of Recombinant Proteins
2.5. Activity Assays
3. Results
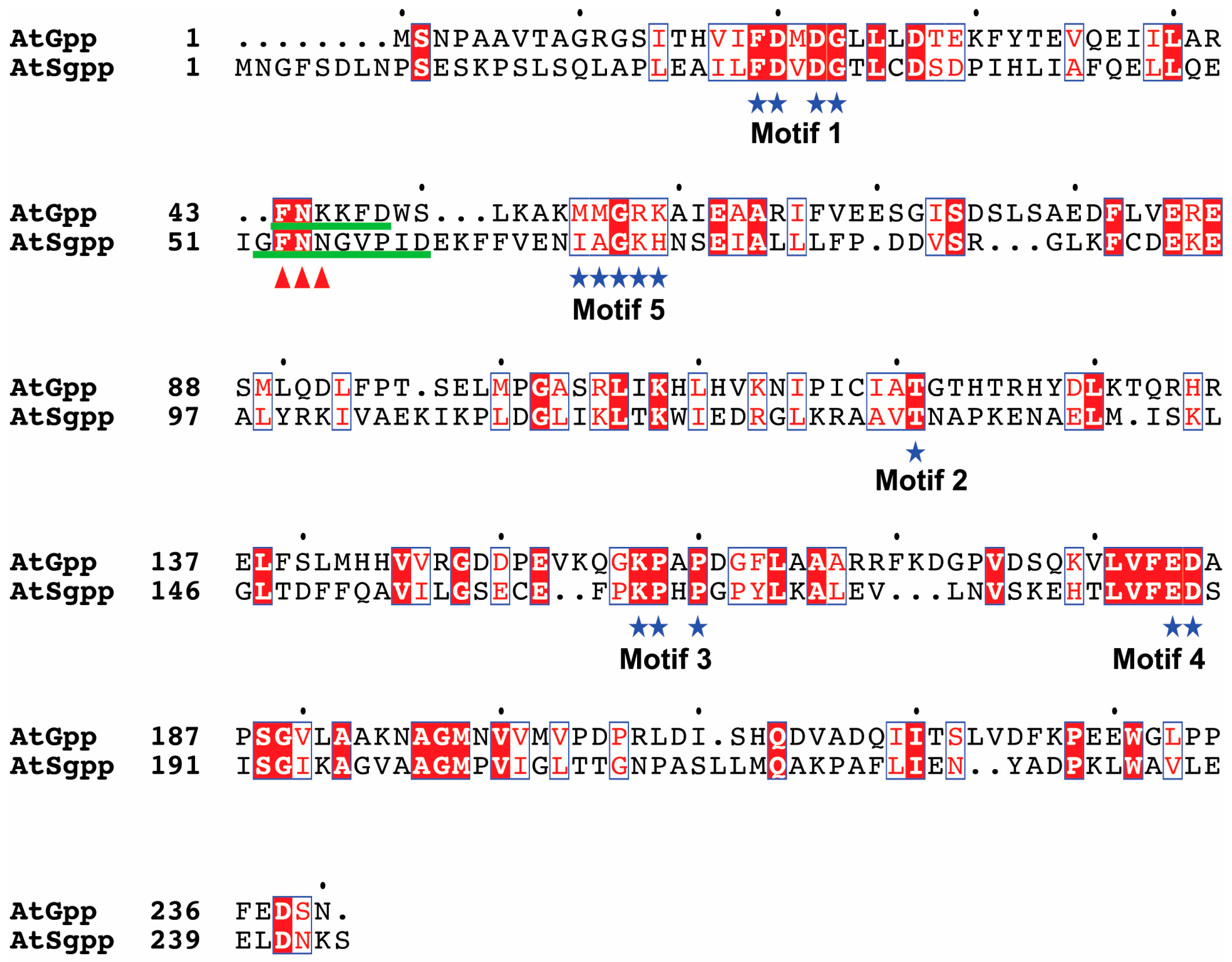
3.1. Sequence and Structure Comparisons
3.2. Comparison of Kinetic Analysis
3.3. pH Rate Profile Comparison
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Crick, F.H. On protein synthesis. Symp. Soc. Exp. Biol. 1958, 12, 138–163. [Google Scholar] [PubMed]
- Crick, F. Central dogma of molecular biology. Nature 1970, 227, 561–563. [Google Scholar] [CrossRef] [PubMed]
- Fischer, E. Einfluss der Configuration auf die Wirkung der Enzyme. Eur. J. Inorg. Chem. 1894, 27, 2985–2993. [Google Scholar]
- Hult, K.; Berglund, P. Enzyme promiscuity: Mechanism and applications. Trends Biotechnol. 2007, 25, 231–238. [Google Scholar] [CrossRef]
- Bornscheuer, U.T.; Kazlauskas, R.J. Catalytic promiscuity in biocatalysis: Using old enzymes to form new bonds and follow new pathways. Angew. Chem. 2004, 43, 6032–6040. [Google Scholar] [CrossRef]
- Jensen, R.A. Enzyme recruitment in evolution of new function. Annu. Rev. Microbiol. 1976, 30, 409–425. [Google Scholar] [CrossRef] [PubMed]
- Khersonsky, O.; Malitsky, S.; Rogachev, I.; Tawfik, D.S. Role of chemistry versus substrate binding in recruiting promiscuous enzyme functions. Biochemistry 2011, 50, 2683–2690. [Google Scholar] [CrossRef]
- Ma, W.; Tang, C.; Lai, L. Specificity of trypsin and chymotrypsin: Loop-motion-controlled dynamic correlation as a determinant. Biophys. J. 2005, 89, 1183–1193. [Google Scholar] [CrossRef]
- Carbonell, P.; Faulon, J.L. Molecular signatures-based prediction of enzyme promiscuity. Bioinformatics 2010, 26, 2012–2019. [Google Scholar] [CrossRef]
- Babbitt, P.C.; Hasson, M.S.; Wedekind, J.E.; Palmer, D.R.; Barrett, W.C.; Reed, G.H.; Rayment, I.; Ringe, D.; Kenyon, G.L.; Gerlt, J.A. The enolase superfamily: A general strategy for enzyme-catalyzed abstraction of the alpha-protons of carboxylic acids. Biochemistry 1996, 35, 16489–16501. [Google Scholar] [CrossRef]
- O’Brien, P.J.; Herschlag, D. Catalytic promiscuity and the evolution of new enzymatic activities. Chem. Biol. 1999, 6, R91–R105. [Google Scholar] [CrossRef]
- Romero, P.A.; Arnold, F.H. Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell Biol. 2009, 10, 866–876. [Google Scholar] [CrossRef] [PubMed]
- Yoshikuni, Y.; Ferrin, T.E.; Keasling, J.D. Designed divergent evolution of enzyme function. Nature 2006, 440, 1078–1082. [Google Scholar] [CrossRef] [PubMed]
- Pandya, C.; Farelli, J.D.; Dunaway-Mariano, D.; Allen, K.N. Enzyme promiscuity: Engine of evolutionary innovation. J. Biol. Chem. 2014, 289, 30229–30236. [Google Scholar] [CrossRef] [PubMed]
- Caparros-Martin, J.A.; McCarthy-Suarez, I.; Culianez-Macia, F.A. The kinetic analysis of the substrate specificity of motif 5 in a HAD hydrolase-type phosphosugar phosphatase of Arabidopsis thaliana. Planta 2014, 240, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Burroughs, A.M.; Allen, K.N.; Dunaway-Mariano, D.; Aravind, L. Evolutionary genomics of the HAD superfamily: Understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J. Mol. Biol. 2006, 361, 1003–1034. [Google Scholar] [CrossRef] [PubMed]
- Rao, K.N.; Kumaran, D.; Seetharaman, J.; Bonanno, J.B.; Burley, S.K.; Swaminathan, S. Crystal structure of trehalose-6-phosphate phosphatase-related protein: Biochemical and biological implications. Protein Sci. 2006, 15, 1735–1744. [Google Scholar] [CrossRef] [PubMed]
- Caparros-Martin, J.A.; Reiland, S.; Kochert, K.; Cutanda, M.C.; Culianez-Macia, F.A. Arabidopsis thaliana AtGppl and AtGpp2: Two novel low molecular weight phosphatases involved in plant glycerol metabolism. Plant Mol. Biol. 2007, 63, 505–517. [Google Scholar] [CrossRef]
- Caparros-Martin, J.A.; McCarthy-Suarez, I.; Culianez-Macia, F.A. HAD hydrolase function unveiled by substrate screening: Enzymatic characterization of Arabidopsis thaliana subclass I phosphosugar phosphatase AtSgpp. Planta 2013, 237, 943–954. [Google Scholar] [CrossRef]
- Lahiri, S.D.; Zhang, G.; Dai, J.; Dunaway-Mariano, D.; Allen, K.N. Analysis of the substrate specificity loop of the HAD superfamily cap domain. Biochemistry 2004, 43, 2812–2820. [Google Scholar] [CrossRef]
- Allen, K.N.; Dunaway-Mariano, D. Phosphoryl group transfer: Evolution of a catalytic scaffold. Trends Biochem. Sci. 2004, 29, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Schenk, P.M.; Baumann, S.; Mattes, R.; Steinbiss, H.H. Improved high-level expression system for eukaryotic genes in Escherichia coli using T7 RNA polymerase and rare ArgtRNAs. BioTechniques 1995, 19, 196–198. [Google Scholar] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Corpet, F.; Servant, F.; Gouzy, J.; Kahn, D. ProDom and ProDom-CG: Tools for protein domain analysis and whole genome comparisons. Nucleic Acids Res. 2000, 28, 267–269. [Google Scholar] [CrossRef] [PubMed]
- Bairoch, A.; Apweiler, R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000, 28, 45–48. [Google Scholar] [CrossRef] [PubMed]
- Higgins, D.G. CLUSTAL V: Multiple alignment of DNA and protein sequences. Methods Mol. Biol. 1994, 25, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Lambert, C.; Leonard, N.; De Bolle, X.; Depiereux, E. ESyPred3D: Prediction of proteins 3D structures. Bioinformatics 2002, 18, 1250–1256. [Google Scholar] [CrossRef] [PubMed]
- Sayle, R.A.; Milner-White, E.J. RASMOL: Biomolecular graphics for all. Trends Biochem. Sci. 1995, 20, 374–376. [Google Scholar] [CrossRef]
- Rostkowski, M.; Olsson, M.H.; Sondergaard, C.R.; Jensen, J.H. Graphical analysis of pH-dependent properties of proteins predicted using PROPKA. BMC Struct. Biol. 2011, 11, 6. [Google Scholar] [CrossRef]
- Cormack, B. Directed mutagenesis using the polymerase chain reaction. Curr. Protoc. Mol. Biol. 2001, 3, 4–11. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Schagger, H.; von Jagow, G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987, 166, 368–379. [Google Scholar] [CrossRef]
- Sussman, I.; Avron, M. Characterization and partial purification of dl-glycerol-1-phosphatase from Dunaliella salina. Biochim. Biophys. Acta 1981, 661, 199–204. [Google Scholar] [CrossRef]
- Ames, B.N. Methods in Enzimology; Academic Press: Cambridge, MA, USA, 1966; Volume 8. [Google Scholar]
- Zhang, G.; Morais, M.C.; Dai, J.; Zhang, W.; Dunaway-Mariano, D.; Allen, K.N. Investigation of metal ion binding in phosphonoacetaldehyde hydrolase identifies sequence markers for metal-activated enzymes of the HAD enzyme superfamily. Biochemistry 2004, 43, 4990–4997. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Dunaway-Mariano, D.; Allen, K.N. HAD superfamily phosphotransferase substrate diversification: Structure and function analysis of HAD subclass IIB sugar phosphatase BT4131. Biochemistry 2005, 44, 8684–8696. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Khersonsky, O.; Tawfik, D.S. Enzyme promiscuity: A mechanistic and evolutionary perspective. Annu. Rev. Biochem. 2010, 79, 471–505. [Google Scholar] [CrossRef] [PubMed]
- Copley, S.D. An evolutionary biochemist’s perspective on promiscuity. Trends Biochem. Sci. 2015, 40, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Risso, V.A.; Gavira, J.A.; Mejia-Carmona, D.F.; Gaucher, E.A.; Sanchez-Ruiz, J.M. Hyperstability and substrate promiscuity in laboratory resurrections of Precambrian beta-lactamases. J. Am. Chem. Soc. 2013, 135, 2899–2902. [Google Scholar] [CrossRef]
- Chan, K.K.; Fedorov, A.A.; Fedorov, E.V.; Almo, S.C.; Gerlt, J.A. Structural basis for substrate specificity in phosphate binding (beta/alpha)8-barrels: D-allulose 6-phosphate 3-epimerase from Escherichia coli K-12. Biochemistry 2008, 47, 9608–9617. [Google Scholar] [CrossRef] [PubMed]
- Daughtry, K.D.; Huang, H.; Malashkevich, V.; Patskovsky, Y.; Liu, W.; Ramagopal, U.; Sauder, J.M.; Burley, S.K.; Almo, S.C.; Dunaway-Mariano, D.; et al. Structural basis for the divergence of substrate specificity and biological function within HAD phosphatases in lipopolysaccharide and sialic acid biosynthesis. Biochemistry 2013, 52, 5372–5386. [Google Scholar] [CrossRef] [PubMed]
- Lahiri, S.D.; Zhang, G.; Dunaway-Mariano, D.; Allen, K.N. Diversification of function in the haloacid dehalogenase enzyme superfamily: The role of the cap domain in hydrolytic phosphoruscarbon bond cleavage. Bioorganic Chem. 2006, 34, 394–409. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.E. Analysing the pH-dependent properties of proteins using pKa calculations. J. Mol. Graph. Model. 2007, 25, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Pandya, C.; Liu, C.; Al-Obaidi, N.F.; Wang, M.; Zheng, L.; Toews Keating, S.; Aono, M.; Love, J.D.; Evans, B.; et al. Panoramic view of a superfamily of phosphatases through substrate profiling. Proc. Natl. Acad. Sci. USA 2015, 112, E1974–E1983. [Google Scholar] [CrossRef] [PubMed]
- Dellus-Gur, E.; Toth-Petroczy, A.; Elias, M.; Tawfik, D.S. What makes a protein fold amenable to functional innovation? Fold polarity and stability trade-offs. J. Mol. Biol. 2013, 425, 2609–2621. [Google Scholar] [CrossRef]
- Yasutake, Y.; Yao, M.; Sakai, N.; Kirita, T.; Tanaka, I. Crystal structure of the Pyrococcus horikoshii isopropylmalate isomerase small subunit provides insight into the dual substrate specificity of the enzyme. J. Mol. Biol. 2004, 344, 325–333. [Google Scholar] [CrossRef]
- Waz, S.; Nakamura, T.; Hirata, K.; Koga-Ogawa, Y.; Chirifu, M.; Arimori, T.; Tamada, T.; Ikemizu, S.; Nakabeppu, Y.; Yamagata, Y. Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity. J. Biol. Chem. 2017, 292, 2785–2794. [Google Scholar] [CrossRef]
| Substrate | AtSgpp | AtSgpp3Δ |
|---|---|---|
| kcat (s−1) | ||
| D-ribose-5-phosphate | 3.9 (3.6–4.2) × 10 [0.3] | 3.1 (2.7–3.5) × 10 [0.4] |
| 2-deoxy-D-glucose-6-phosphate | 3.3 (3.2–3.4) × 10 [0.1] | 2.3 (2.2–2.4) × 10 [0.1] |
| D-mannose-6-phosphate | 3.2 (2.5–3.9) × 10 [0.6] | 1.9 (1.7–2.1) × 10 [0.2] |
| D-glucose-6-phosphate | 2.4 (1.8–3) × 10 [0.5] | 1.2 (1.1–1.3) × 10 [0.1] |
| D-fructose-6-phosphate | 1.9 (1.7–2.1) × 10 [0.2] | 1.1 (0.8–1.4) × 10 [0.3] |
| DL-glycerol-3-phosphate | 2.3 (2–2.6) × 10 [0.3] | 0.7 (0.5–0.9) × 10 [0.2] |
| Km (M) | ||
| D-ribose-5-phosphate | 3.6 (3.5–3.7) × 10−3 [0.1] | 3.7 (3.5–3.9) × 10−3 [0.2] |
| 2-deoxy-D-glucose-6-phosphate | 4.6 (4–5.2) × 10−3 [0.5] | 4.7 (4.5–4.9) × 10−3 [0.2] |
| D-mannose-6-phosphate | 4.9 (4.2–5.6) × 10−3 [0.6] | 6.7 (6.6–6.8) × 10−3 [0.1] |
| D-glucose-6-phosphate | 7.1 (6.8–7.4) × 10−3 [0.3] | 8.3 (8–8.6) × 10−3 [0.3] |
| D-fructose-6-phosphate | 7.7 (7.3–8.1) × 10−3 [0.4] | 10 (9.5–10.5) × 10−3 [0.4] |
| DL-glycerol-3-phosphate | 7.3 (7.1–7.5) × 10−3 [0.2] | 14 (13.7–14.3) × 10−3 [0.3] |
| kcat/Km (M−1s−1) | ||
| D-ribose-5-phosphate | 1.07 × 104 | 8.3 × 103 |
| 2-deoxy-D-glucose-6-phosphate | 7.2 × 103 | 4.9 × 103 |
| D-mannose-6-phosphate | 6.7 × 103 | 2.8 × 103 |
| D-glucose-6-phosphate | 3.4 × 103 | 1.5 × 103 |
| D-fructose-6-phosphate | 2.5 × 103 | 1.2 × 103 |
| DL-glycerol-3-phosphate | 3.1 × 103 | 0.5 × 103 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caparrós-Martín, J.A.; McCarthy-Suárez, I.; Culiáñez-Macià, F.A. Sequence Determinants of Substrate Ambiguity in a HAD Phosphosugar Phosphatase of Arabidopsis Thaliana. Biology 2019, 8, 77. https://doi.org/10.3390/biology8040077
Caparrós-Martín JA, McCarthy-Suárez I, Culiáñez-Macià FA. Sequence Determinants of Substrate Ambiguity in a HAD Phosphosugar Phosphatase of Arabidopsis Thaliana. Biology. 2019; 8(4):77. https://doi.org/10.3390/biology8040077
Chicago/Turabian StyleCaparrós-Martín, José A., Iva McCarthy-Suárez, and Francisco A. Culiáñez-Macià. 2019. "Sequence Determinants of Substrate Ambiguity in a HAD Phosphosugar Phosphatase of Arabidopsis Thaliana" Biology 8, no. 4: 77. https://doi.org/10.3390/biology8040077
APA StyleCaparrós-Martín, J. A., McCarthy-Suárez, I., & Culiáñez-Macià, F. A. (2019). Sequence Determinants of Substrate Ambiguity in a HAD Phosphosugar Phosphatase of Arabidopsis Thaliana. Biology, 8(4), 77. https://doi.org/10.3390/biology8040077






