Physiological and Phylogenetic Characterization of Rhodotorula diobovata DSBCA06, a Nitrophilous Yeast
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Strain Isolation
2.3. Molecular Characterization
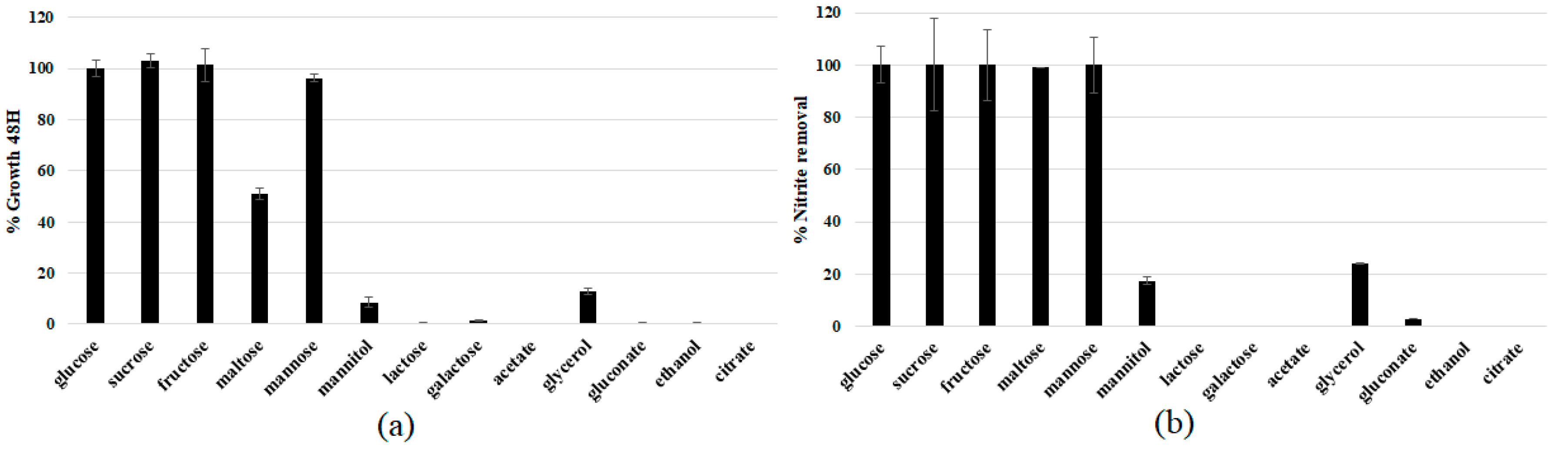
2.4. Characterization and Optimization of Catabolic Features
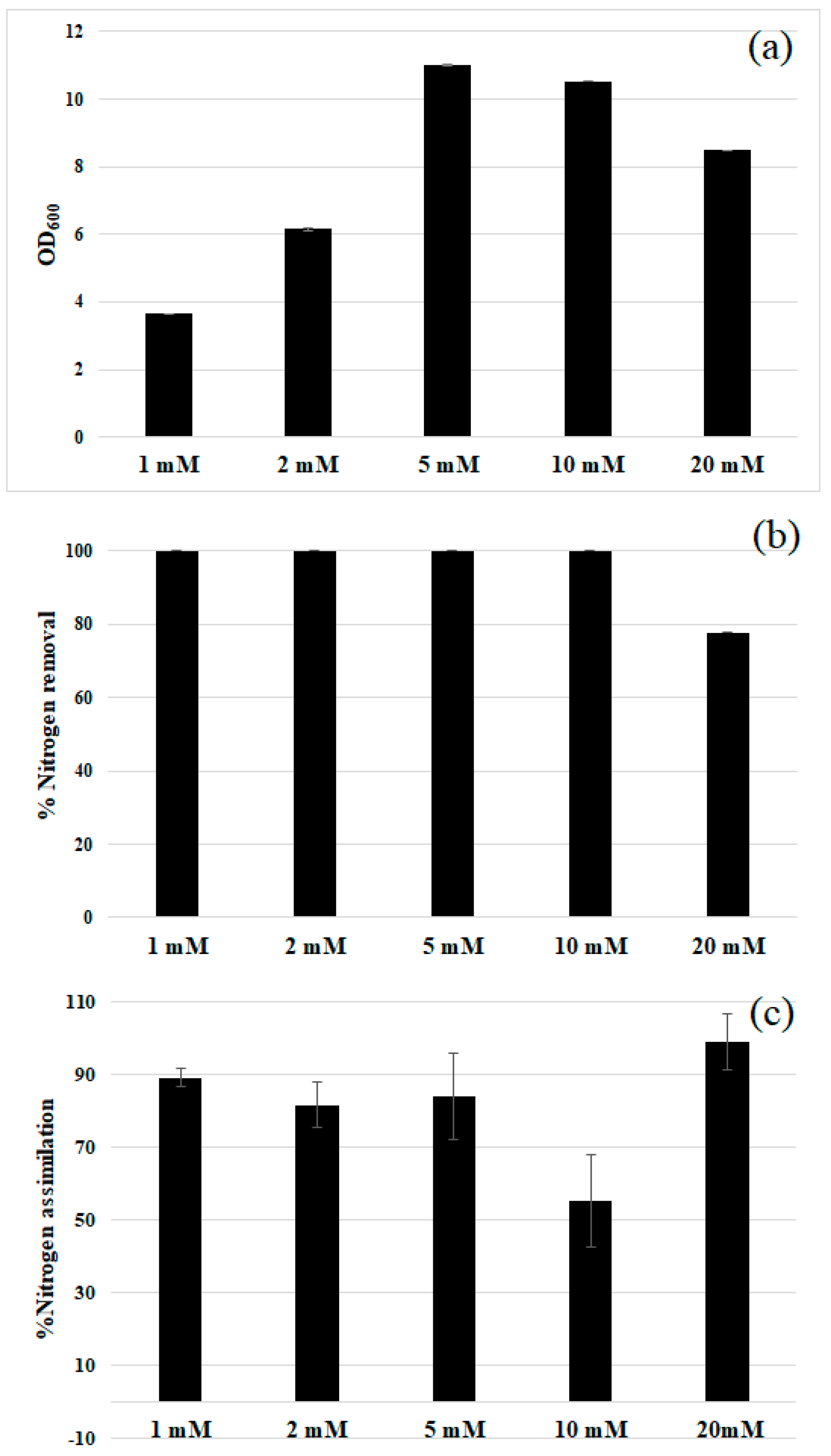
2.5. Nitrogen Assimilation
2.6. Analytic Determinations
2.7. Process Scale-Up
3. Results and Discussion
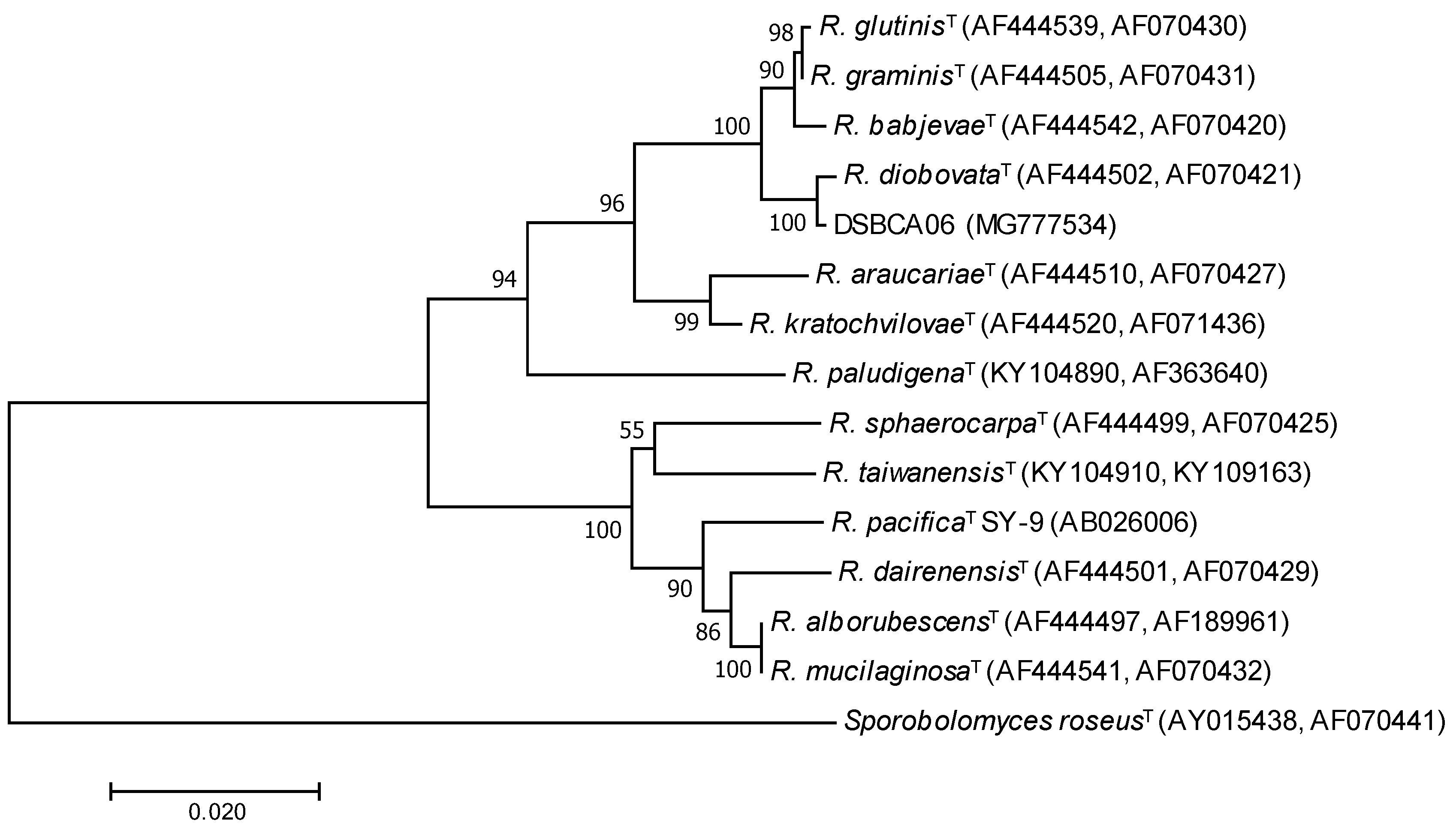
3.1. Phylogenetic Characterization
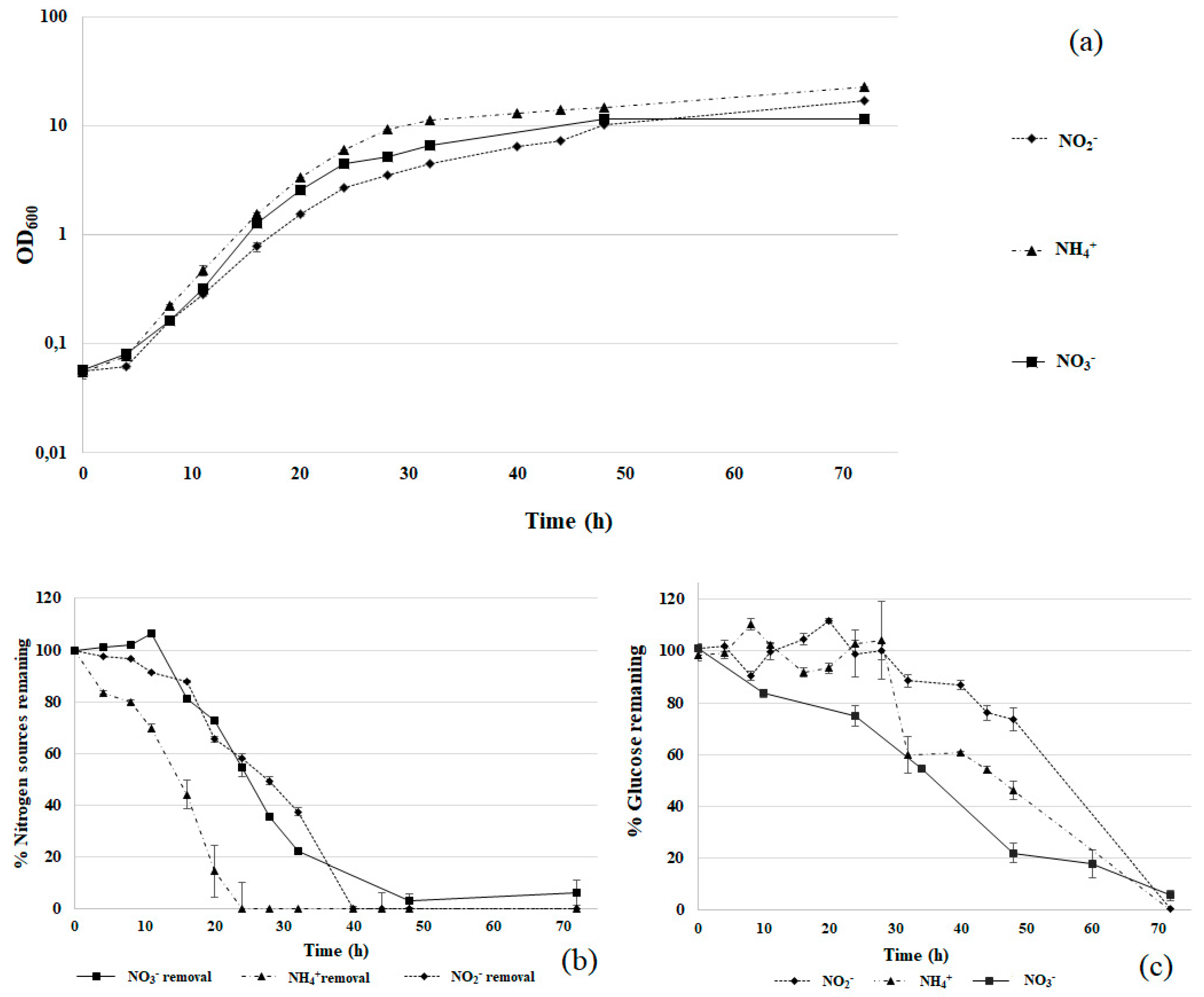
3.2. Comparison of Nitrogen Sources
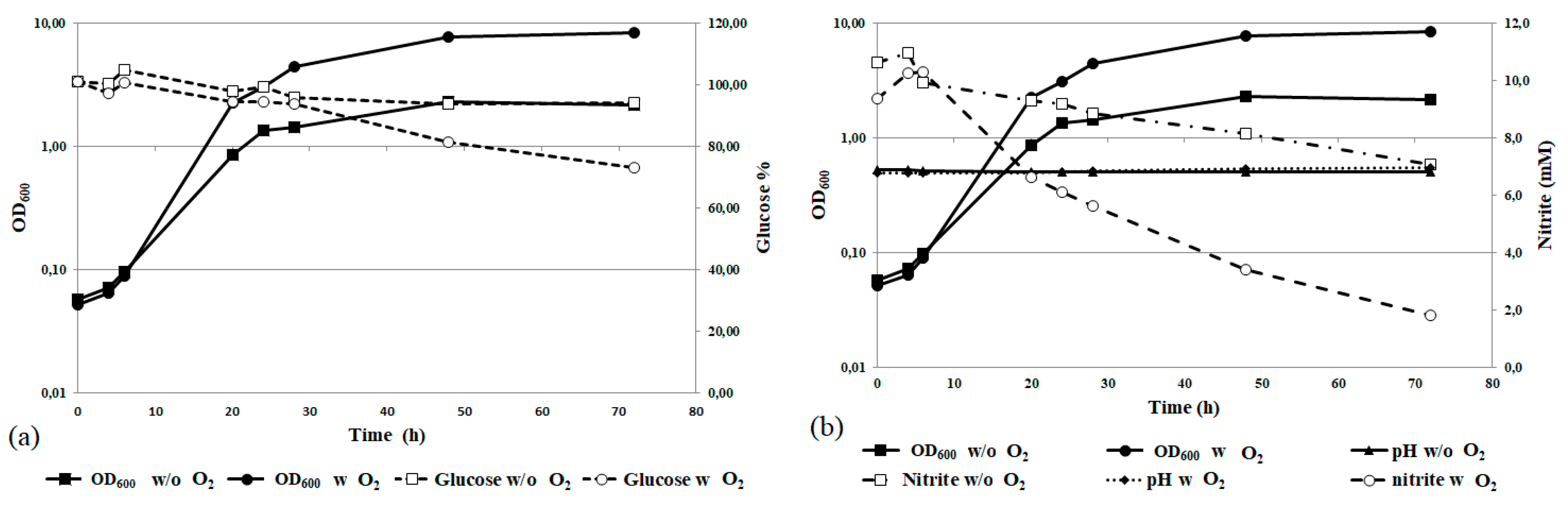
3.3. Physiological Characterization and Optimization of Growth Conditions
3.4. Nitrite Tolerance
3.5. Heavy Metal Tolerance
3.6. Nitrite Removal at Bioreactor Scale
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lewis, W.M.; Morris, D.P. Toxicity of Nitrite to Fish: A Review. Trans. Am. Fish. Soc. 1986, 115, 183–195. [Google Scholar] [CrossRef]
- Howarth, R.W.; Marino, R. Nitrogen as the limiting nutrient for eutrophication in coastal marine ecosystems: Evolving views over three decades. Limnol. Oceanogr. 2006, 51, 364–376. [Google Scholar] [CrossRef]
- Camargo, J.A.; Alonso, Á. Ecological and toxicological effects of inorganic nitrogen pollution in aquatic ecosystems: A global assessment. Environ. Int. 2006, 32, 831–849. [Google Scholar] [CrossRef] [PubMed]
- Blasco, R.; Martínez-Luque, M.; Madrid, M.P.; Castillo, F.; Moreno-Vivián, C. Rhodococcus sp. RB1 grows in the presence of high nitrate and nitrite concentrations and assimilates nitrate in moderately saline environments. Arch. Microbiol. 2001, 175, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Giles, J. Nitrogen study fertilizes fears of pollution. Nature 2005, 433, 791. [Google Scholar] [CrossRef] [PubMed]
- Ye, R.W.; Thomas, S.M. Microbial nitrogen cycles: Physiology, genomics and applications. Curr. Opin. Microbiol. 2001, 4, 307–312. [Google Scholar] [CrossRef]
- Ahn, Y.H. Sustainable nitrogen elimination biotechnologies: A review. Process Biochem. 2006, 41, 1709–1721. [Google Scholar] [CrossRef]
- Cabrera, E.; González-Montelongo, R.; Giraldez, T.; de la Rosa, D.A.; Siverio, J.M. Molecular components of nitrate and nitrite efflux in yeast. Eukaryot. Cell 2014, 13, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Siverio, J.M. Assimilation of nitrate by yeasts. FEMS Microbiol. Rev. 2002, 26, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Johnson, E.A. Biotechnology of non-Saccharomyces yeasts-the basidiomycetes. Appl. Microbiol. Biotechnol. 2013, 97, 7563–7577. [Google Scholar] [CrossRef] [PubMed]
- Navarro, F.J.; Perdomo, G.; Tejera, P.; Medina, B.; Machín, F.; Guillén, R.M.; Lancha, A.; Siverio, J.M. The role of nitrate reductase in the regulation of the nitrate assimilation pathway in the yeast Hansenula polymorpha. FEMS Yeast Res. 2003, 4, 149–155. [Google Scholar] [CrossRef]
- Kubisi, A.; Ali, A.H.; Hipkin, C.R. Nitrite assimilation by the yeast Candida nitratophila. New Phytol. 1996, 132, 313–316. [Google Scholar] [CrossRef]
- Vigliotta, G.; Di Giacomo, M.; Carata, E.; Massardo, D.R.; Tredici, S.M.; Silvestro, D.; Paolino, M.; Pontieri, P.; Del Giudice, L.; Parente, D.; et al. Nitrite metabolism in Debaryomyces hansenii TOB-Y7, a yeast strain involved in tobacco fermentation. Appl. Microbiol. Biotechnol. 2007, 75, 633–645. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Almanza, A.; Cesar Montanez, J.; Aguilar-González, M.A.; Martínez-Ávila, C.; Rodríguez-Herrera, R.; Aguilar, C.N. Rhodotorula glutinis as source of pigments and metabolites for food industry. Food Biosci. 2014, 5, 64–72. [Google Scholar] [CrossRef]
- Frengova, G.; Beshkova, D. Carotenoids from Rhodotorula and Phaffia: Yeasts of biotechnological importance. J. Ind. Microbiol. Biotechnol. 2009, 36, 163–180. [Google Scholar] [CrossRef] [PubMed]
- Csutak, O.; Stoica, I.; Vassu, T. Evaluation of production, stability and activity of biosurfactants from yeasts with application in bioremediation of oil-polluted environment. Rev. Chim. 2012, 63, 973–977. [Google Scholar]
- Bai, J.; Li, Z.; Fan, F.; Wu, X.; Tian, W.; Yin, X.; Zhao, L.; Fan, F.; Tian, L.; Wang, Y.; et al. Biosorption of uranium by immobilized cells of Rhodotorula glutinis. J. Radioanal. Nucl. Chem. 2014, 299, 1517–1524. [Google Scholar] [CrossRef]
- Nasirian, N.; Mirzaie, M.; Cicek, N.; Levin, D.B. Lipid and carotenoid synthesis by Rhodosporidium diobovatum, grown on glucose versus glycerol, and its biodiesel properties. Can. J. Microbiol. 2018, 64, 277–289. [Google Scholar] [CrossRef] [PubMed]
- Rescigno, A.; Casañola-Martin, G.M.M.; Sanjust, E.; Zucca, P.; Marrero-Ponce, Y. Vanilloid Derivatives as Tyrosinase Inhibitors Driven by Virtual Screening-Based QSAR Models. Drug Test. Anal. 2011, 3, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Zucca, P.; Littarru, M.; Rescigno, A.; Sanjust, E. Cofactor recycling for selective enzymatic biotransformation of cinnamaldehyde to cinnamyl alcohol. Biosci. Biotechnol. Biochem. 2009, 73, 1224–1226. [Google Scholar] [CrossRef] [PubMed]
- Wickerham, L.J. Taxonomy of Yeasts; United States Department of Agriculture: Washington, DC, USA, 1951.
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Consortium, F.B.; Bolchacova, E.; Voigt, K.; et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef] [PubMed]
- Seifert, K.A. Progress towards DNA barcoding of fungi. Mol. Ecol. Resour. 2009, 9, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.G. Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 2013, 12, 1229–1235. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, D.J.D.; Nason, A. Determination of nitrate and nitrite. Methods Enzymol. 1957, 3, 982–984. [Google Scholar]
- Miranda, K.M.; Espey, M.G.; Wink, D.A. A rapid, simple spectrophotometric method for simultaneous detection of nitrate and nitrite. Nitric Oxide Biol. Chem. 2001, 5, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Usai, A.; Peddio, D.; Cincotti, A. Kinetics of nitrate-and nitrite-removal by Rhodotorula glutinis: Determination of a reaction mechanism. Chem. Eng. Trans. 2016, 49, 457–462. [Google Scholar]
- Rau, R.; Molitoris, H.P. Determination and significance of nitrate reductase in marine fungi. Kiel. Meeresforsh. Sonderh. 1991, 8, 369–375. [Google Scholar]
- Munch, G.; Sestric, R.; Sparling, R.; Levin, D.B.; Cicek, N. Lipid production in the under-characterized oleaginous yeasts, Rhodosporidium babjevae and Rhodosporidium diobovatum, from biodiesel-derived waste glycerol. Bioresour. Technol. 2015, 185, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Kolouchová, I.; Ma͗átková, O.; Sigler, K.; Masák, J.; Řezanka, T. Production of palmitoleic and linoleic acid in oleaginous and nonoleaginous yeast biomass. Int. J. Anal. Chem. 2016, 2016, 7583684. [Google Scholar] [CrossRef] [PubMed]
- Barman, P.; Kati, A.; Mandal, A.K.; Bandyopadhyay, P.; Mohapatra, P.K.D. Biopotentiality of Bacillus cereus PB45 for nitrogenous waste detoxification in ex situ model. Aquacult. Int. 2017, 25, 1167–1183. [Google Scholar] [CrossRef]
- Sollai, F.A.; Zucca, P.; Rescigno, A.; Dumitriu, E.; Sanjust, E. Sporobolomyces salmonicolor as a tool for nitrate removal from wastewaters. Environ. Eng. Manag. J. 2012, 11, 1455–1460. [Google Scholar]
- Gonzalez, C.; Gonzalez, G.; Avila, J.; Perez, M.D.; Brito, N.; Siverio, J.M. Nitrite causes reversible inactivation of nitrate reductase in the yeast Hansenula anomala. Microbiology 1994, 140, 2633–2637. [Google Scholar] [CrossRef] [PubMed]
- Easterling, E.R.; French, W.T.; Hernandez, R.; Licha, M. The effect of glycerol as a sole and secondary substrate on the growth and fatty acid composition of Rhodotorula glutinis. Bioresour. Technol. 2009, 100, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Saenge, C.; Cheirsilp, B.; Suksaroge, T.T.; Bourtoom, T. Potential use of oleaginous red yeast Rhodotorula glutinis for the bioconversion of crude glycerol from biodiesel plant to lipids and carotenoids. Process Biochem. 2011, 46, 210–218. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Fell, J.W.; Boekhout, T. The Yeasts; Elsevier Science: New York, NY, USA, 2010; Volumes 1–3. [Google Scholar]
- Wang, Q.M.; Yurkov, A.M.; Göker, M.; Lumbsch, H.T.; Leavitt, S.D.; Groenewald, M.; Theelen, B.; Liu, X.Z.; Boekhout, T.; Bai, F.Y. Phylogenetic classification of yeasts and related taxa within Pucciniomycotina. Stud. Mycol. 2015, 81, 149–189. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Das, P.; Sen, R. Towards commercial production of microbial surfactants. Trends Biotechnol. 2006, 24, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Gorchev, H.G.; Ozolins, G. WHO guidelines for drinking-water quality. WHO Chron. 2011, 38, 104–108. [Google Scholar] [CrossRef]
- Bollag, J.-M.; Henninger, N.M. Effects of nitrite toxicity on soil bacteria under aerobic and anaerobic conditions. Soil Biol. Biochem. 1978, 10, 377–381. [Google Scholar] [CrossRef]
- Siddarame Gowda, T.K.; Siddaramappa, R.; Sethunathan, N. Heterotrophic nitrification and nitrite tolerance by Aspergillus carneus (van tiegh) blochwitz, a predominant fungus isolated from benomyl-amended soil. Soil Biol. Biochem. 1976, 8, 435–437. [Google Scholar] [CrossRef]
- Li, X.; Poon, C.S.; Liu, P.S. Heavy metal contamination of urban soils and street dusts in Hong Kong. Appl. Geochem. 2001, 16, 1361–1368. [Google Scholar] [CrossRef]
- Gadd, G.M.; Griffiths, A.J. Microorganisms and heavy metal toxicity. Microb. Ecol. 1977, 4, 303–317. [Google Scholar] [CrossRef] [PubMed]
- Goyer, R.A.; Clarkson, T.W. Toxic effects of metals. In Casarett & Doull’s Toxicology. The Basic Science of Poisons, 5th ed.; Klaassen, C.D., Ed.; McGraw-Hill Health Professions Division: Columbus, OH, USA, 1996; ISBN 71054766. [Google Scholar]
- Blackwell, K.J.; Singleton, I.; Tobin, J.M. Metal cation uptake by yeast: A review. Appl. Microbiol. Biotechnol. 1995, 43, 579–584. [Google Scholar] [CrossRef] [PubMed]
- Salinas, E.; Elorza De Orellano, M.; Rezza, I.; Martinez, L.; Marchesvky, E.; Sanz De Tosetti, M. Removal of cadmium and lead from dilute aqueous solutions by Rhodotorula rubra. Bioresour. Technol. 2000, 72, 107–112. [Google Scholar] [CrossRef]
- Rapta, P.; Polovka, M.; Zalibera, M.; Breierova, E.; Zitnanova, I.; Marova, I.; Certik, M. Scavenging and antioxidant properties of compounds synthesized by carotenogenic yeasts stressed by heavy metals—EPR spin trapping study. Biophys. Chem. 2005, 116, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Vadkertiová, R.; Sláviková, E.; Vadkertiova, R.; Slavikova, E. Metal tolerance of yeasts isolated from water, soil and plant environments. J. Basic Microbiol. 2006, 46, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yuan, H.; Hu, X. Cadmium-resistance in growing Rhodotorula sp. Y11. Bioresour. Technol. 2008, 99, 1339–1344. [Google Scholar] [CrossRef] [PubMed]
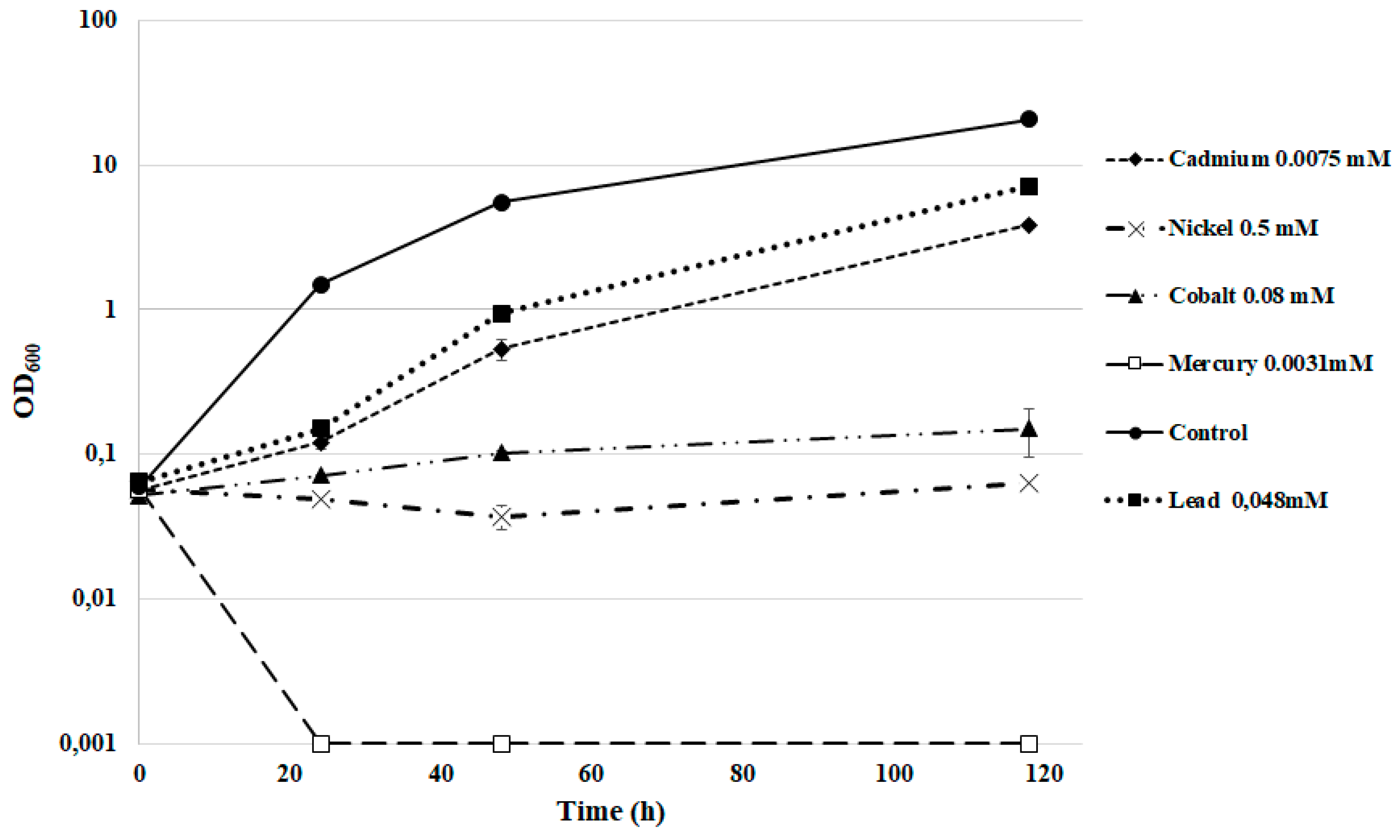
| Metal | mg/kg | mM |
|---|---|---|
| Cadmium acetate | 2 | 0.0075 |
| Cobalt acetate | 20 | 0.08 |
| Mercury acetate | 1 | 0.0031 |
| Nickel chloride | 120 | 0.5 |
| Lead acetate | 100 | 0.264 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Civiero, E.; Pintus, M.; Ruggeri, C.; Tamburini, E.; Sollai, F.; Sanjust, E.; Zucca, P. Physiological and Phylogenetic Characterization of Rhodotorula diobovata DSBCA06, a Nitrophilous Yeast. Biology 2018, 7, 39. https://doi.org/10.3390/biology7030039
Civiero E, Pintus M, Ruggeri C, Tamburini E, Sollai F, Sanjust E, Zucca P. Physiological and Phylogenetic Characterization of Rhodotorula diobovata DSBCA06, a Nitrophilous Yeast. Biology. 2018; 7(3):39. https://doi.org/10.3390/biology7030039
Chicago/Turabian StyleCiviero, Enrico, Manuela Pintus, Claudio Ruggeri, Elena Tamburini, Francesca Sollai, Enrico Sanjust, and Paolo Zucca. 2018. "Physiological and Phylogenetic Characterization of Rhodotorula diobovata DSBCA06, a Nitrophilous Yeast" Biology 7, no. 3: 39. https://doi.org/10.3390/biology7030039
APA StyleCiviero, E., Pintus, M., Ruggeri, C., Tamburini, E., Sollai, F., Sanjust, E., & Zucca, P. (2018). Physiological and Phylogenetic Characterization of Rhodotorula diobovata DSBCA06, a Nitrophilous Yeast. Biology, 7(3), 39. https://doi.org/10.3390/biology7030039