Critical Function of PRDM2 in the Neoplastic Growth of Testicular Germ Cell Tumors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Plasmid Transient Transfection
2.3. RNA Isolation and Quantitative Reverse-Transcription PCR (qRT-PCR) Analysis
2.4. Western Blot and Immunoprecipitation
2.5. Cell Growth Analysis
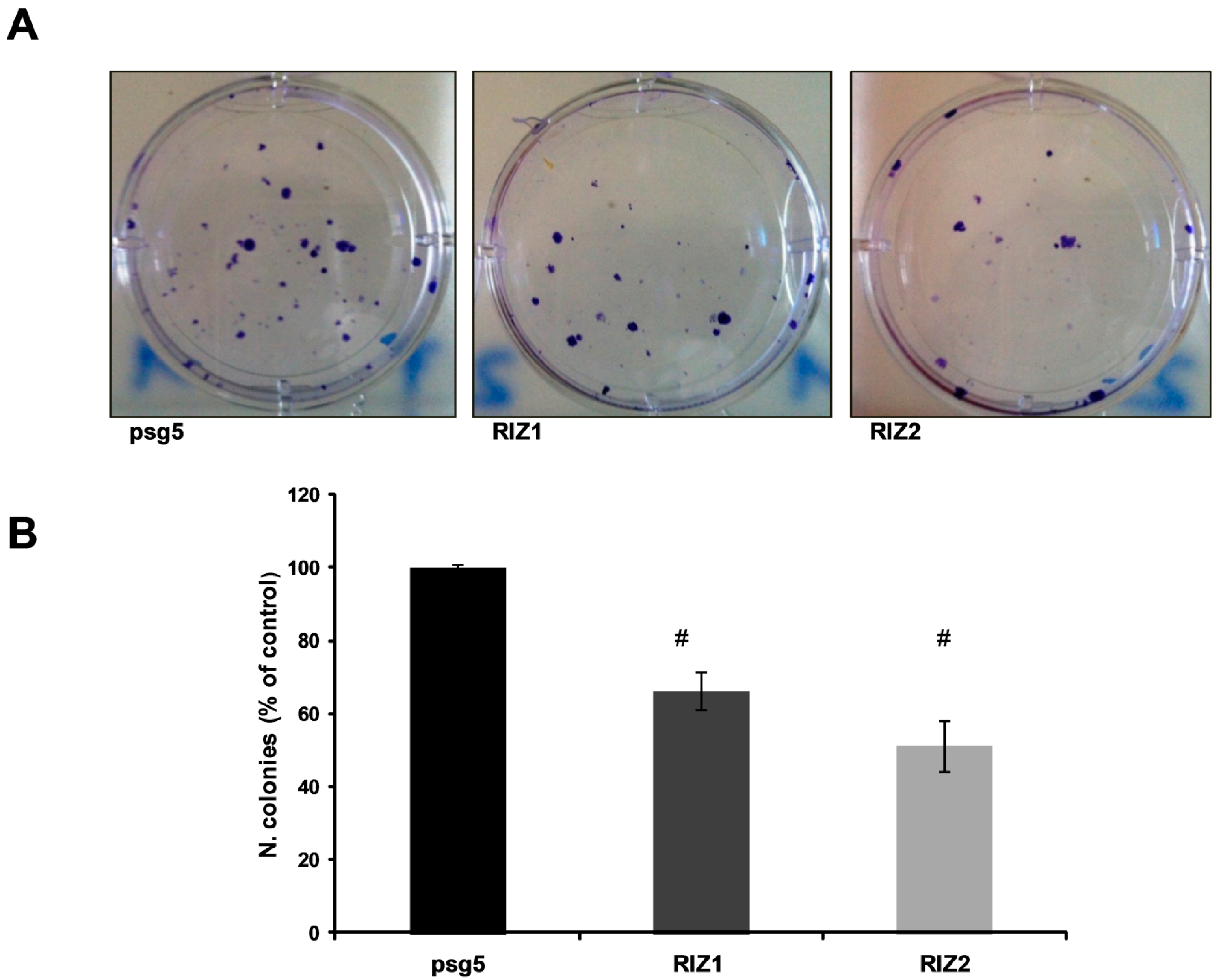
2.6. Clonogenic Assay
2.7. BrdU Incorporation Assay
2.8. TUNEL Assay
2.9. Statistical Analysis
3. Results
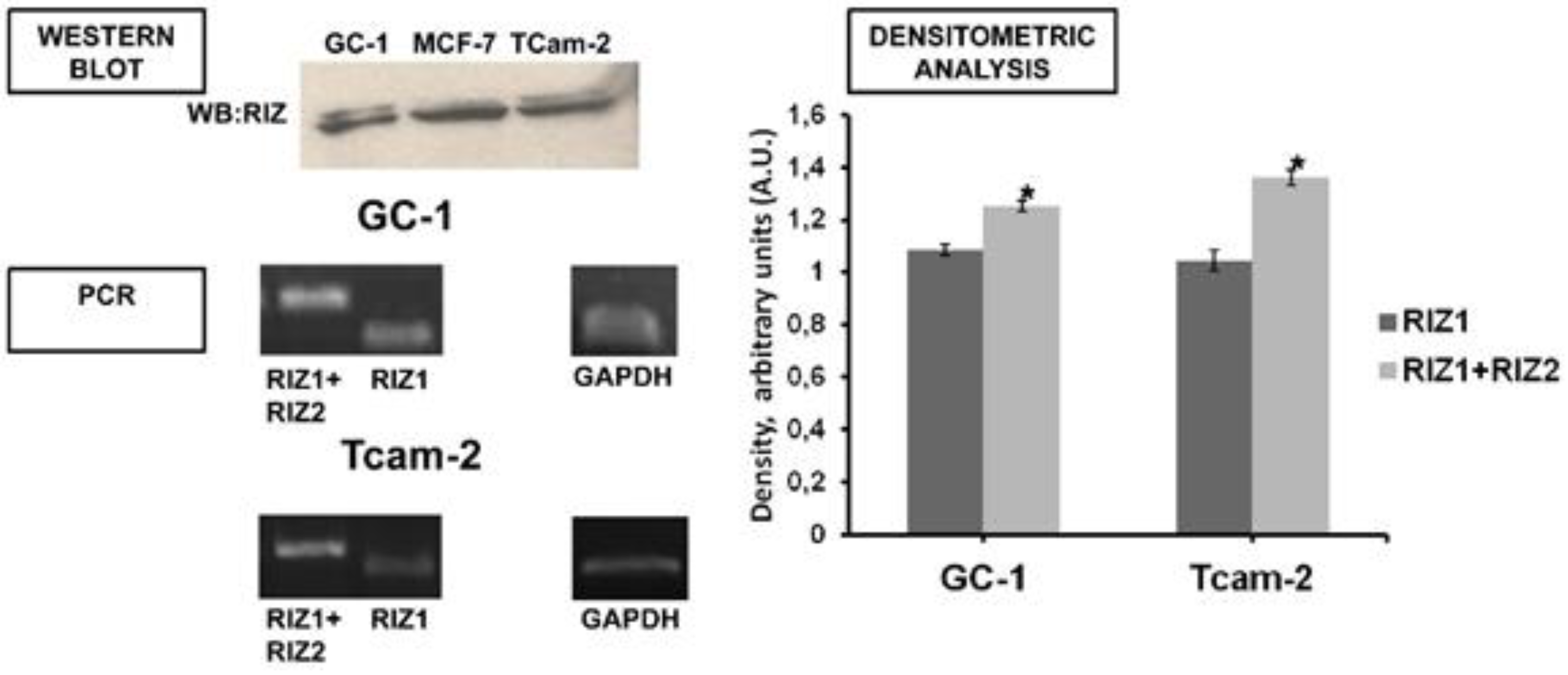
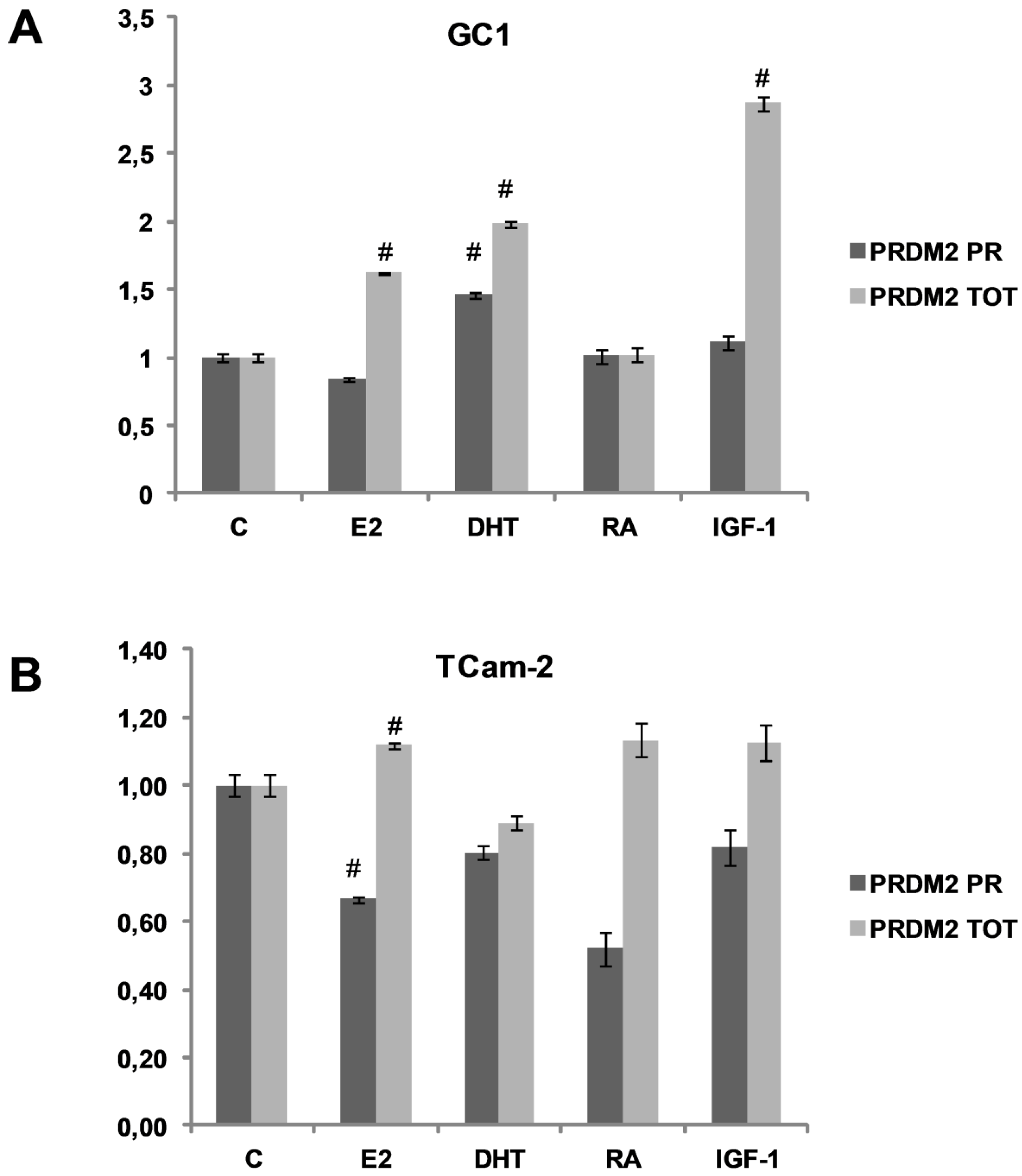
3.1. PRDM2 Expression Level is Modulated by Spermatogonial Proliferation and Differentiation Agents
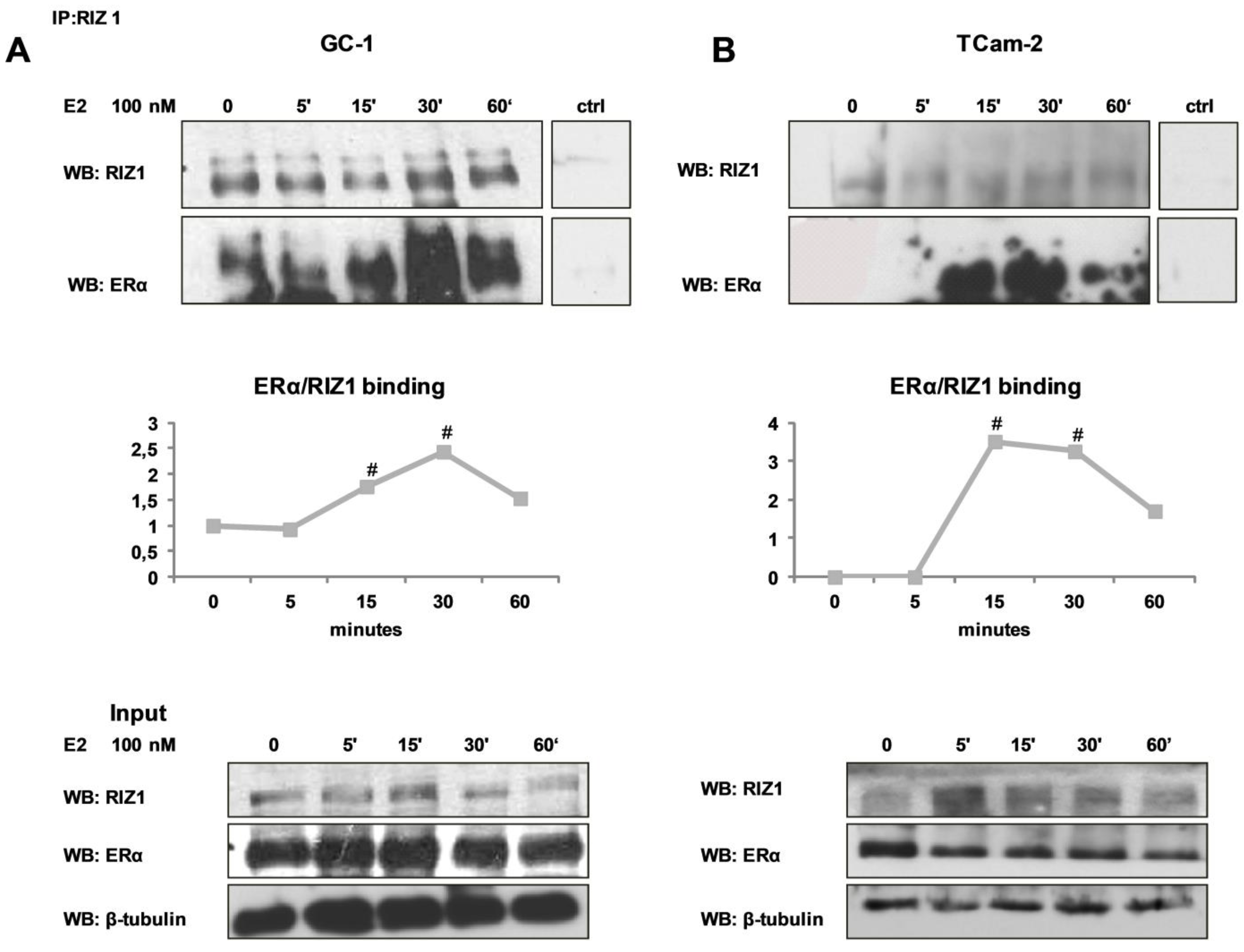
3.2. In GC-1 and TCam-2 Cell Lines RIZ1 Binds ERα and This Interaction is Modulated by Estradiol Treatment
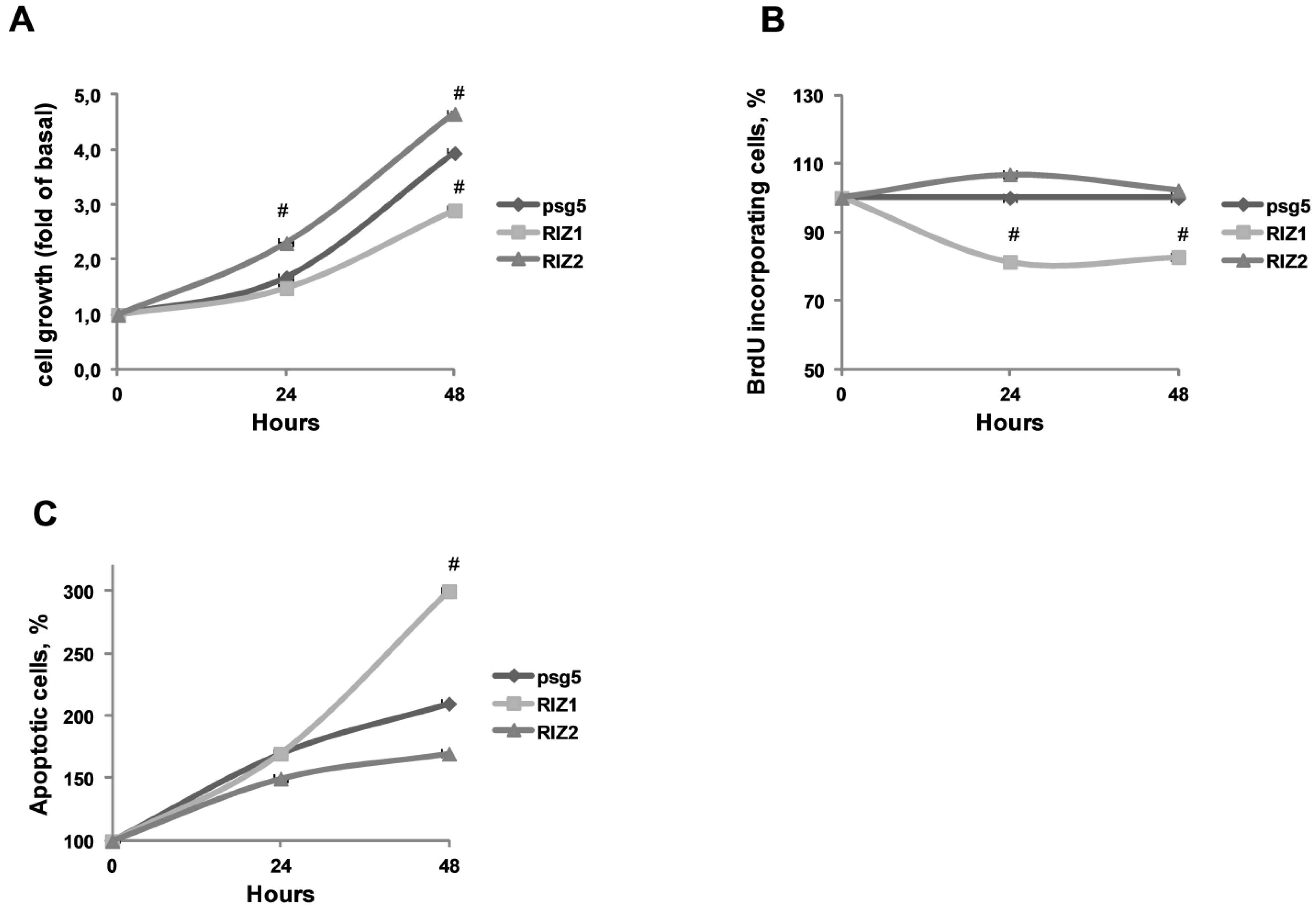
3.3. RIZ1 Over-Expression Inhibits GC-1 Cells Proliferation and Survival
3.4. RIZ1 Over-Expression Induces GC-1 Cells Apopotosis
3.5. PRDM2 Influences Tumor Growth
4. Discussion
5. Conclusions
Appendix A
Supplementary Materials
Acknowledgement
Author Contributions
Conflicts of Interest
Abbreviations
| TGCTs | Testicular germ cell tumors |
| NSGCTs | non-seminoma germ cell tumors |
| ERα | estrogen receptor α |
| ERβ | estrogen receptor β |
| E2 | 17β-Estradiol |
| MTT | 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide |
| BrdU | 5-bromo-2′-deoxy-uridine |
| DHT | 5α-dihydrotestosterone |
| SDS-PAGE | Sodium Dodecyl Sulphate-PolyAcrylamide Gel Electrophoresis |
References
- Chieffi, P. Molecular targets for the treatment of testicular germ cell tumors. Mini Rev. Med. Chem. 2007, 7, 755–759. [Google Scholar] [CrossRef] [PubMed]
- Chieffi, P. New prognostic markers and potential therapeutic targets in human testicular germ. Curr. Med. Chem. 2011, 18, 5033–5040. [Google Scholar] [CrossRef] [PubMed]
- Chieffi, P.; Franco, R.; Portella, G. Molecular and cell biology of testicular germ cell tumors. Int. Rev. Cell Mol. Biol. 2009, 278, 277–308. [Google Scholar] [PubMed]
- Oosterhuis, J.W.; Looijenga, L.H. Testicular germ-cell tumours in a broader perspective. Nat. Rev. Cancer 2005, 5, 210–222. [Google Scholar] [CrossRef] [PubMed]
- Houldsworth, J.; Reuter, V.; Bosl, G.J.; Chaganti, R.S. Aberrant expression of cyclin D2 is an early event in human male germ cell tumorigenesis. Cell Growth Differ. 1997, 8, 293–299. [Google Scholar] [PubMed]
- Lambard, S.; Carreau, S. Aromatase and oestrogens in human male germ cells. Int. J. Androl. 2005, 28, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Pais, V.; Leav, I.; Lau, K.M.; Jiang, Z.; Ho, S.M. Estrogen receptor-beta expression in human testicular germ cell tumors. Clin. Cancer Res. 2003, 9, 4475–4482. [Google Scholar] [PubMed]
- Rago, V.; Romeo, F.; Aquila, S.; Montanaro, D.; Ando, S.; Carpino, A. Cytochrome P450 aromatase expression in human seminoma. Reprod. Biol. Endocrinol. 2005. [Google Scholar] [CrossRef] [PubMed]
- Vicini, E.; Loiarro, M.; Di Agostino, S.; Corallini, S.; Capolunghi, F.; Carsetti, R.; Chieffi, P.; Geremia, R.; Stefanini, M.; Sette, C. 17-beta-estradiol elicits genomic and non-genomic responses in mouse male germ cells. J. Cell. Physiol. 2006, 206, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Henderson, B.E.; Benton, B.; Jing, J.; Yu, M.C.; Pike, M.C. Risk factors for cancer of the testis in young men. Int. J. Cancer 1979, 23, 598–602. [Google Scholar] [CrossRef] [PubMed]
- Wellejus, A.; Loft, S. Receptor-mediated ethinylestradiol-induced oxidative DNA damage in rat testicular. FASEB J. 2002, 16, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Katzenellenbogen, B.S.; Montano, M.M.; Ediger, T.R.; Sun, J.; Ekena, K.; Lazennec, G.; Martini, P.G.; McInerney, E.M.; Delage-Mourroux, R.; Weis, K.; et al. Estrogen receptors: Selective ligands, partners, and distinctive pharmacology. Recent Prog. Horm. Res. 2000, 55, 163–193. [Google Scholar] [PubMed]
- Pettersson, K.; Gustafsson, J.A. Role of estrogen receptor beta in estrogen action. Annu. Rev. Physiol. 2001, 63, 165–192. [Google Scholar] [CrossRef] [PubMed]
- Buyse, I.M.; Shao, G.; Huang, S. The retinoblastoma protein binds to RIZ, a zinc-finger protein that shares an epitope with the adenovirus E1A protein. Proc. Natl. Acad. Sci. USA 1995, 92, 4467–4471. [Google Scholar] [CrossRef] [PubMed]
- Huang, S. The retinoblastoma protein-interacting zinc finger gene RIZ in 1p36-linked cancers. Front. Biosci. 1999, 4, 528–532. [Google Scholar] [CrossRef]
- Abbondanza, C.; Medici, N.; Nigro, V.; Rossi, V.; Gallo, L.; Piluso, G.; Belsito, A.; Roscigno, A.; Bontempo, P.; Puca, A.A.; et al. The retinoblastoma-interacting zinc-finger protein RIZ is a downstream effector of estrogen action. Proc. Natl. Acad. Sci. USA 2000, 97, 3130–3135. [Google Scholar] [CrossRef] [PubMed]
- Carling, T.; Kim, K.C.; Yang, X.H.; Gu, J.; Zhang, X.K.; Huang, S. A Histone Methyltransferase Is Required for Maximal Response to Female Sex Hormones. Mol. Cell. Biol. 2004, 24, 7032–7042. [Google Scholar] [CrossRef] [PubMed]
- Medici, N.; Abbondanza, C.; Nigro, V.; Rossi, V.; Piluso, G.; Belsito, A.; Gallo, L.; Roscigno, A.; Bontempo, P.; Puca, A.A.; et al. Identification of a DNA binding protein cooperating with estrogen receptor as RIZ (retinoblastoma interacting zinc finger protein). Biochem. Biophys. Res. Commun. 1999, 264, 983–989. [Google Scholar] [CrossRef] [PubMed]
- Ancelin, K.; Lange, U.C.; Hajkova, P.; Schneider, R.; Bannister, A.J.; Kouzarides, T.; Surani, M.A. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat. Cell Biol. 2006, 8, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.A.; Haberland, M.; Arnold, M.A.; Sutherland, L.B.; McDonald, O.G.; Richardson, J.A.; Childs, G.; Harris, S.; Owens, G.K.; Olson, E.N. PRISM/PRDM6, a Transcriptional Repressor That Promotes the Proliferative Gene Program in Smooth Muscle Cells. Mol. Cell. Biol. 2006, 26, 2626–2636. [Google Scholar] [CrossRef] [PubMed]
- Di Zazzo, E.; De Rosa, C.; Abbondanza, C.; Moncharmont, B. PRDM Proteins: Molecular Mechanisms in Signal Transduction and Transcriptional Regulation. Biology 2013, 2, 107–141. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Yoshida, K.; Matsui, Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 2005, 438, 374–378. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Shao, G.; Steele-Perkins, G.; Huang, S. The retinoblastoma interacting zinc finger gene RIZ produces a PR domain-lacking product through an internal promoter. J. Biol. Chem. 1997, 272, 2984–2991. [Google Scholar] [CrossRef] [PubMed]
- Geli, J.; Kiss, N.; Kogner, P.; Larsson, C. Suppression of RIZ in biologically unfavourable neuroblastomas. Int. J. Oncol. 2010, 37, 1323–1330. [Google Scholar] [CrossRef] [PubMed]
- Geli, J.; Nord, B.; Frisk, T.; Edstrom Elder, E.; Ekstrom, T.J.; Carling, T.; Backdahl, M.; Larsson, C. Deletions and altered expression of the RIZ1 tumour suppressor gene in 1p36 in pheochromocytomas and abdominal paragangliomas. Int. J. Oncol. 2005, 26, 1385–1391. [Google Scholar] [CrossRef] [PubMed]
- Piao, Z.; Fang, W.; Malkhosyan, S.; Kim, H.; Horii, A.; Perucho, M.; Huang, S. Frequent frameshift mutations of RIZ in sporadic gastrointestinal and endometrial carcinomas with microsatellite instability. Cancer Res. 2000, 60, 4701–4704. [Google Scholar] [PubMed]
- Poetsch, M.; Dittberner, T.; Woenckhaus, C. Frameshift mutations of RIZ, but no point mutations in RIZ1 exons in malignant melanomas with deletions in 1p36. Oncogene 2002, 21, 3038–3042. [Google Scholar] [CrossRef] [PubMed]
- Sakurada, K.T.; Furukawa, T.; Kato, Y.; Kayama, T.; Huang, S.; Horii, A. RIZ, the retinoblastoma protein interacting zinc finger gene, is mutated in genetically unstable cancers of the pancreas, stomach, and colorectum. Genes Chromosomes Cancer 2001, 30, 207–211. [Google Scholar] [CrossRef]
- Sasaki, O.; Meguro, K.; Tohmiya, Y.; Funato, T.; Shibahara, S.; Sasaki, T. Altered expression of retinoblastoma protein-interacting zinc finger gene, RIZ, in human leukaemia. Br. J. Haematol. 2002, 119, 940–948. [Google Scholar] [CrossRef] [PubMed]
- Steele-Perkins, G.; Fang, W.; Yang, X.H.; van Gele, M.; Carling, T.; Gu, J.; Buyse, I.M.; Fletcher, J.A.; Liu, J.; Bronson, R.; et al. Tumor formation and inactivation of RIZ1, an Rb-binding member of a nuclear protein-methyltransferase superfamily. Genes Dev. 2001, 15, 2250–2262. [Google Scholar] [CrossRef] [PubMed]
- Mzoughi, S.; Tan, Y.X.; Low, D.; Guccione, E. The role of PRDMs in cancer: One family, two sides. Curr. Opin. Genet. Dev. 2016, 36, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Chadwick, R.B.; Jiang, G.L.; Bennington, G.A.; Yuan, B.; Johnson, C.K.; Stevens, M.W.; Niemann, T.H.; Peltomaki, P.; Huang, S.; de la Chapelle, A. Candidate tumor suppressor RIZ is frequently involved in colorectal carcinogenesis. Proc. Natl. Acad. Sci. USA 2000, 97, 2662–2667. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Piao, Z.; Buyse, I.M.; Simon, D.; Sheu, J.C.; Perucho, M.; Huang, S. Preferential loss of a polymorphic RIZ allele in human hepatocellular carcinoma. Br. J. Cancer 2001, 84, 743–747. [Google Scholar] [CrossRef] [PubMed]
- Rossi, V.; Staibano, S.; Pasquali, D.; de Rosa, C.; Mascolo, M.; Bellastella, G.; Visconti, D.; de Bellis, A.; Moncharmont, B.; de Rosa, G.; et al. Expression of RIZ1 protein (Retinoblastoma-interacting zinc-finger protein 1) in prostate cancer epithelial cells changes with cancer grade progression and is modulated in vitro by DHT and E2. J. Cell. Physiol. 2009, 221, 771–777. [Google Scholar] [CrossRef] [PubMed]
- Gazzerro, P.; Abbondanza, C.; D’Arcangelo, A.; Rossi, M.; Medici, N.; Moncharmont, B.; Puca, G.A. Modulation of RIZ gene expression is associated to estradiol control of MCF-7 breast cancer cell proliferation. Exp. Cell Res. 2006, 312, 340–349. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. Relative quantification. In Real Time PCR BIOS Advanced Methods; Dorak, T., Ed.; Taylor & Francis: Abingdon, UK, 2006; Volume 16, pp. 63–82. [Google Scholar]
- Abbondanza, C.; de Rosa, C.; D’Arcangelo, A.; Pacifico, M.; Spizuoco, C.; Piluso, G.; di Zazzo, E.; Gazzerro, P.; Medici, N.; Moncharmont, B.; et al. Identification of a functional estrogen-responsive enhancer element in the promoter 2 of PRDM2 gene in breast cancer cell lines. J. Cell. Physiol. 2012, 227, 964–975. [Google Scholar] [CrossRef] [PubMed]
- Porcile, C.; Di Zazzo, E.; Monaco, M.L.; D’Angelo, G.; Passarella, D.; Russo, C.; Di Costanzo, A.; Pattarozzi, A.; Gatti, M.; Bajetto, A.; et al. Adiponectin as novel regulator of cell proliferation in human glioblastoma. J. Cell. Physiol. 2014, 229, 1444–1454. [Google Scholar] [CrossRef] [PubMed]
- Di Zazzo, E.; Feola, A.; Zuchegna, C.; Romano, A.; Donini, C.F.; Bartollino, S.; Costagliola, C.; Frunzio, R.; Laccetti, P.; Di Domenico, M.; et al. The p85 regulatory subunit of PI3K mediates cAMP-PKA and insulin biological effects on MCF-7 cell growth and motility. Sci. World J. 2014. [Google Scholar] [CrossRef] [PubMed]
- Sirianni, R.; Chimento, A.; Ruggiero, C.; De Luca, A.; Lappano, R.; Andò, S.; Maggiolini, M.; Pezzi, V. The novel estrogen receptor, G protein-coupled receptor 30, mediates the proliferative effects induced by 17beta-estradiol on mouse spermatogonial GC-1 cell line. Endocrinology 2008, 149, 5043–5051. [Google Scholar] [CrossRef] [PubMed]
- Di Zazzo, E.; Department of Health Sciences, University of Molise, Campobasso, Italy. Unpublished results. 2014.
- Liu, Z.Y.; Wang, J.Y.; Liu, H.H.; Ma, X.M.; Wang, C.L.; Zhang, X.P.; Tao, Y.Q.; Lu, Y.C.; Liao, J.C.; Hu, G.H.; et al. Retinoblastoma protein-interacting zinc-finger gene 1 (RIZ1) dysregulation in human malignant meningiomas. Oncogene 2013, 32, 1216–1222. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Yu, J.X.; Liu, L.; Buyse, I.M.; Wang, M.S.; Yang, Q.C.; Nakagawara, A.; Brodeur, G.M.; Shi, Y.E.; Huang, S. RIZ1, but not the alternative RIZ2 product of the same gene, is underexpressed in breast cancer, and forced RIZ1 expression causes G2-M cell cycle arrest and/or apoptosis. Cancer Res. 1998, 58, 4238–4244. [Google Scholar] [PubMed]
- Jiang, G.; Liu, L.; Buyse, I.M.; Simon, D.; Huang, S. Decreased RIZ1 expression but not RIZ2 in hepatoma and suppression of hepatoma tumorigenicity by RIZ1. Int. J. Cancer 1999, 83, 541–546. [Google Scholar] [CrossRef]
- Jiang, G.L.; Huang, S. Adenovirus expressing RIZ1 in tumor suppressor gene therapy of microsatellite-unstable colorectal cancers. Cancer Res. 2001, 61, 1796–1798. [Google Scholar] [PubMed]
- Shadat, N.M.; Koide, N.; Khuda, I.I.; Dagvadorj, J.; Tumurkhuu, G.; Naiki, Y.; Komatsu, T.; Yoshida, T.; Yokochi, T. Retinoblastoma protein interacting zinc finger 1 (RIZ1) regulates the proliferation of monocytic leukemia cells via activation of p53. Cancer Investig. 2010, 28, 806–812. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Zazzo, E.; Porcile, C.; Bartollino, S.; Moncharmont, B. Critical Function of PRDM2 in the Neoplastic Growth of Testicular Germ Cell Tumors. Biology 2016, 5, 54. https://doi.org/10.3390/biology5040054
Di Zazzo E, Porcile C, Bartollino S, Moncharmont B. Critical Function of PRDM2 in the Neoplastic Growth of Testicular Germ Cell Tumors. Biology. 2016; 5(4):54. https://doi.org/10.3390/biology5040054
Chicago/Turabian StyleDi Zazzo, Erika, Carola Porcile, Silvia Bartollino, and Bruno Moncharmont. 2016. "Critical Function of PRDM2 in the Neoplastic Growth of Testicular Germ Cell Tumors" Biology 5, no. 4: 54. https://doi.org/10.3390/biology5040054
APA StyleDi Zazzo, E., Porcile, C., Bartollino, S., & Moncharmont, B. (2016). Critical Function of PRDM2 in the Neoplastic Growth of Testicular Germ Cell Tumors. Biology, 5(4), 54. https://doi.org/10.3390/biology5040054






