Contrasting Responses to Nutrient Enrichment of Prokaryotic Communities Collected from Deep Sea Sites in the Southern Ocean
Abstract
:1. Introduction
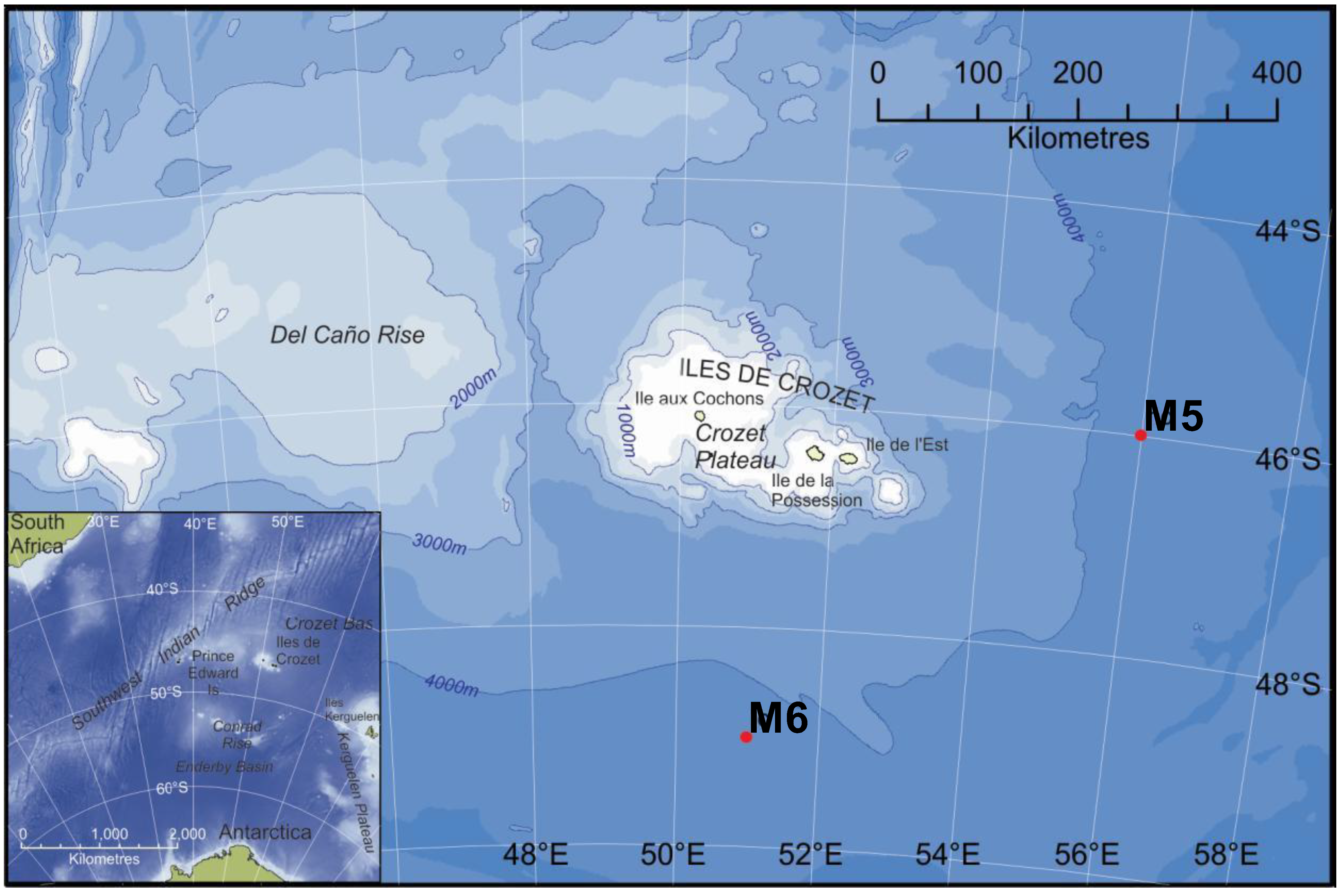
| M5 (HC) | M6 (LC) | |
|---|---|---|
| Depth (m) | 4,224 | 4,212 |
| Temperature (°C) | −0.22 | −0.21 |
| Salinity | 34.67 | 34.67 |
| Oxygen (μmol L−1) | 230.9 | 231.0 |
| Nitrate + Nitrite (μmol L−1) | 31.90 | 32.32 |
| Silicate (μmol L−1) | 154.4 | 155.3 |
| Phosphate (μmol L−1) | 2.19 | 2.19 |
| C flux to 100 m (g C m−2 y−1) | 40 | 10 |
| C flux to Seafloor (g C m−2 y−1) | 2 | 0.5 |
2. Results and Discussion
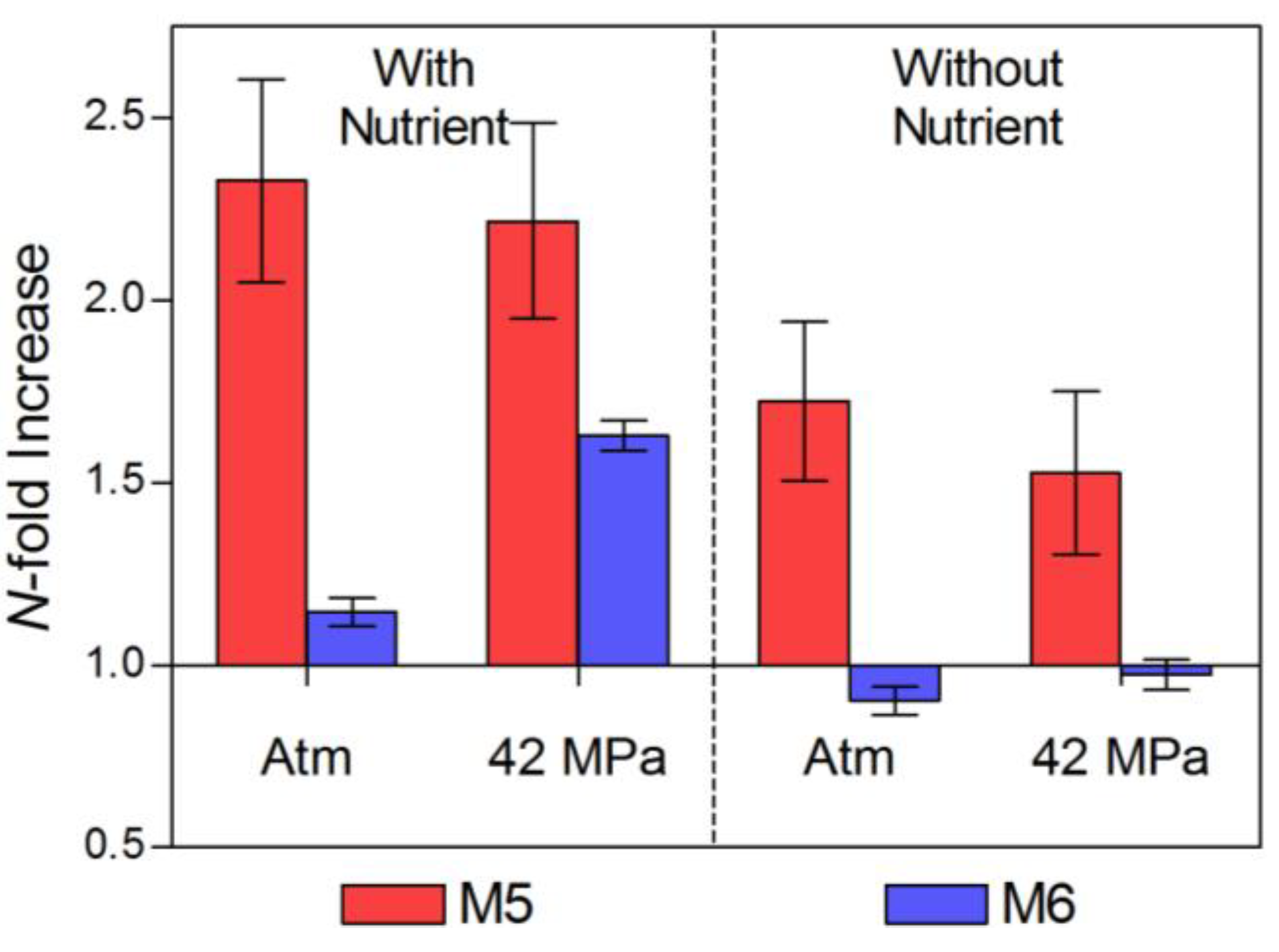
2.1. Pre-Incubation Prokaryotic Abundance
| M5 (High Chlorophyll site) | M6 (Low Chlorophyll site) | ||||||
|---|---|---|---|---|---|---|---|
| N | H | c | N | H | c | ||
| Time Zero | 1.19 (±0.14) | 2.593 | 0.104 | 3.95 (±0.08) | 3.021 | 0.064 | |
| Atm, N+ | 2.77 (±0 .08) | 2.789 | 0.08 | 4.52 (±0.13) | 1.307 | 0.368 | |
| 42 MPa, N+ | 2.64 (±0.10) | 2.71 | 0.088 | 6.44 (±0.10) | 2.118 | 0.182 | |
| Atm, N− | 2.05 (±0.10) | 2.845 | 0.082 | 3.56 (±0.14) | 2.674 | 0.092 | |
| 42 MPa, N− | 1.82 (±0.16) | 3.12 | 0.058 | 3.85 (±0.15) | 2.761 | 0.087 | |
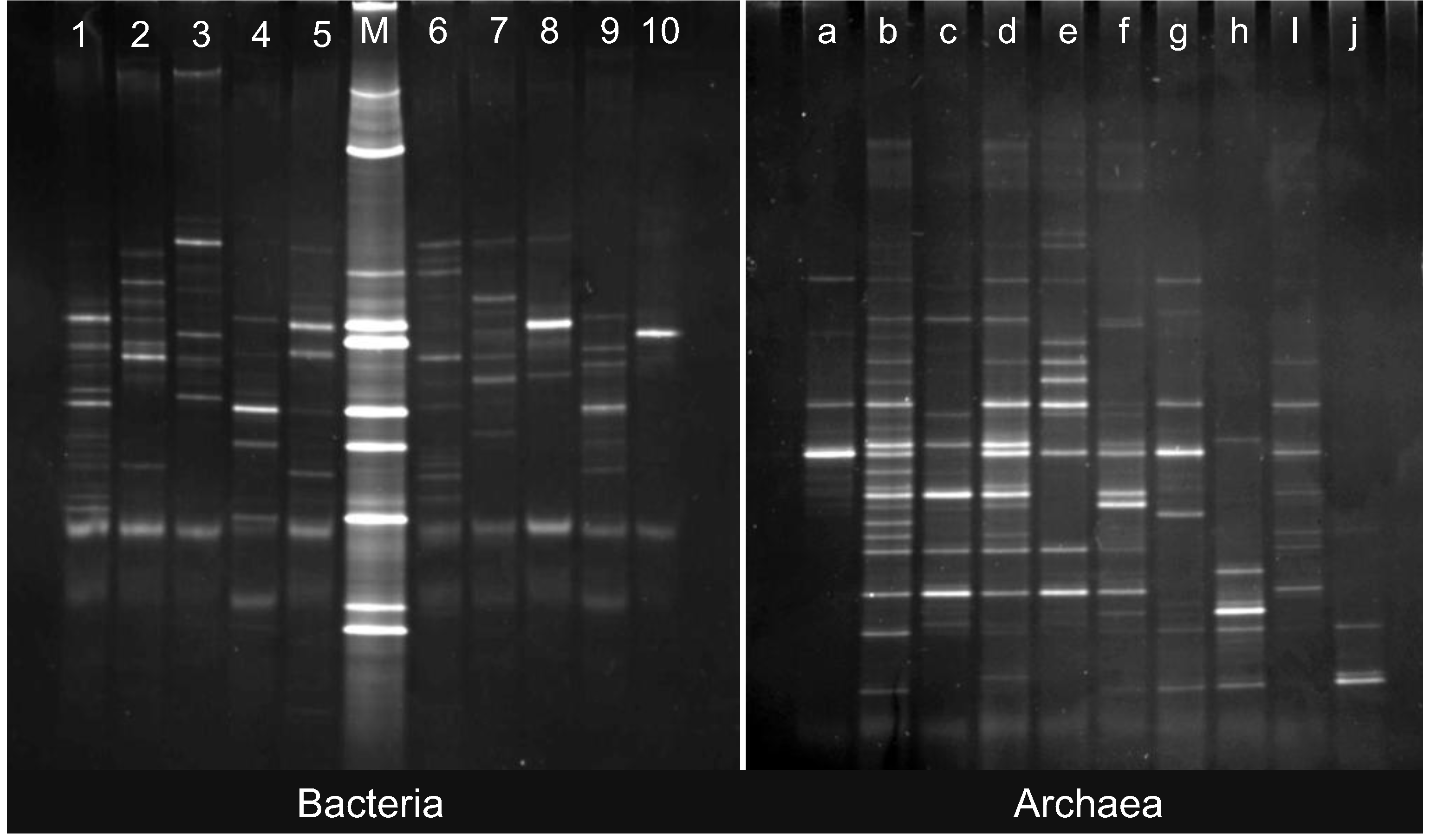
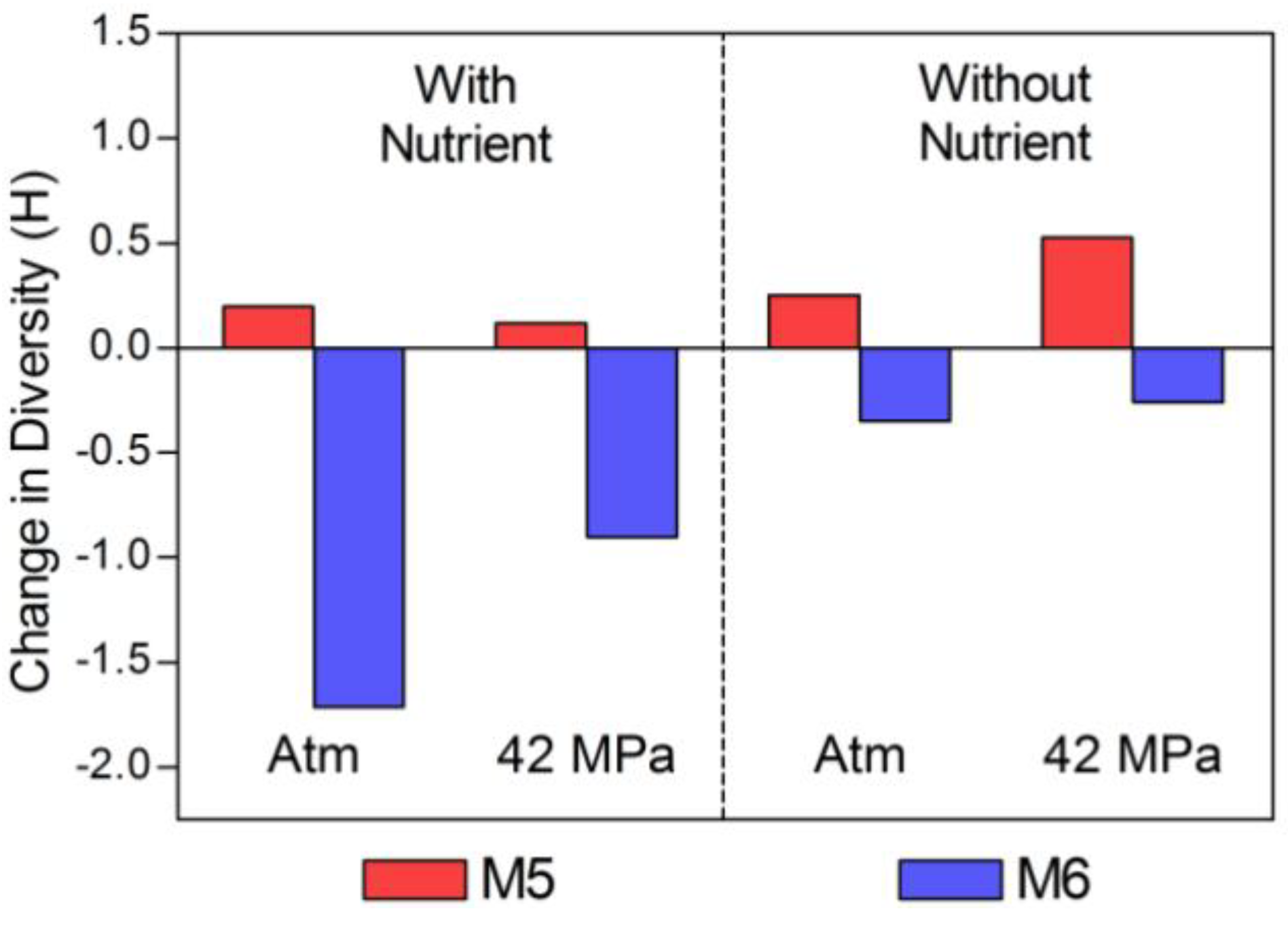
2.2. Changes in Abundance and Community Structure after Incubations
| HC (M5) | LC (M6) | |||
|---|---|---|---|---|
| Bact | Arch | Bact | Arch | |
| Time Zero | 2.34 | 1.122 | 2.459 | 2.203 |
| Atm, N+ | 1.963 | 2.224 | 0.446 | 1.127 |
| 42 MPa, N+ | 2.127 | 1.895 | 1.365 | 1.595 |
| Atm, N− | 1.852 | 2.449 | 1.99 | 2.161 |
| 42 MPa, N− | 1.84 | 2.851 | 2.276 | 1.819 |



3. Experimental Section
3.1. Study Area and Sampling Sites
3.2. Water Sampling
3.3. Enrichment Incubations
3.4. DNA Extraction
3.5. PCR and DGGE
3.6. Quantitative Analysis of DGGE Fingerprints


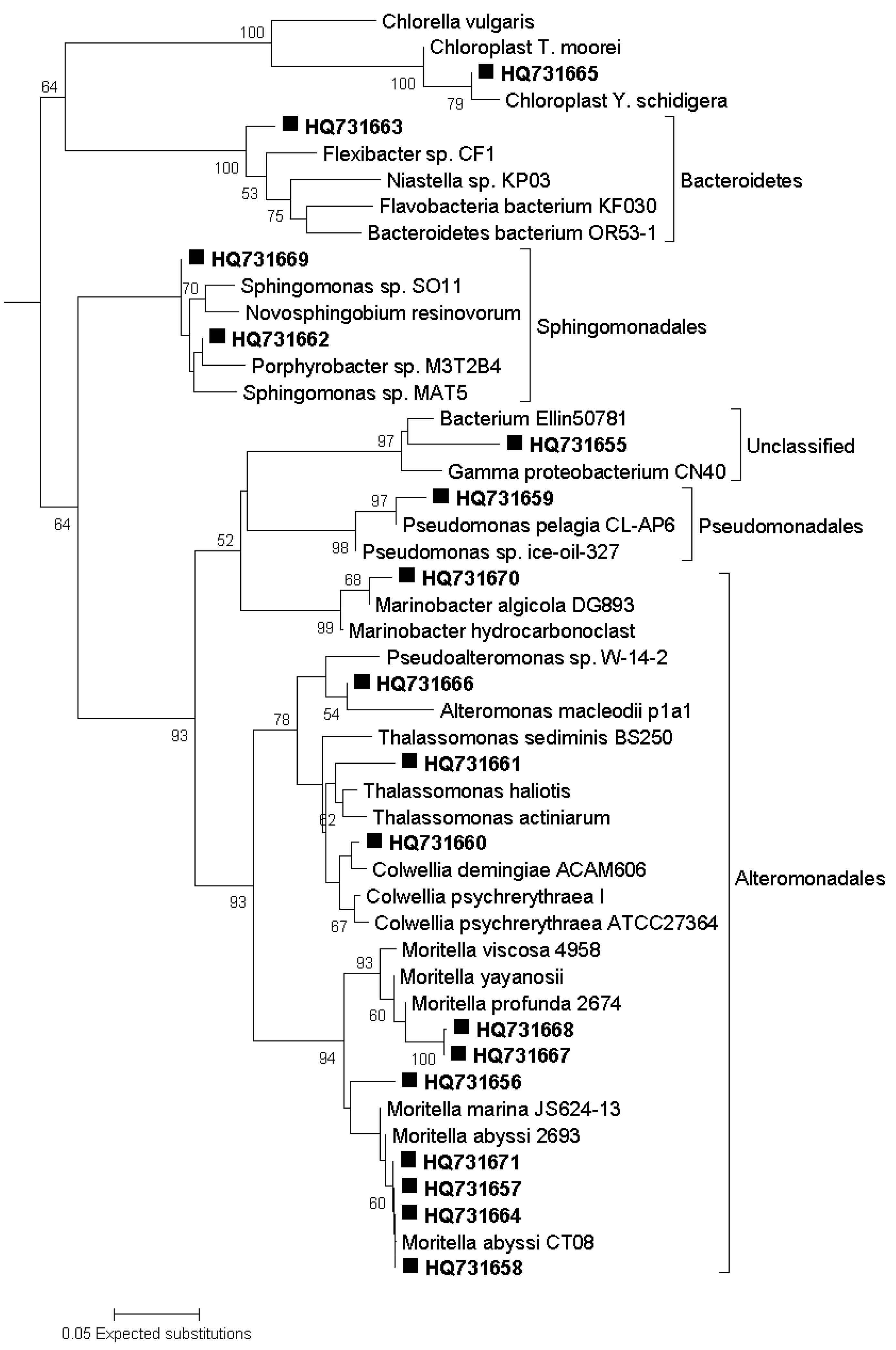
3.7. Sequencing of DGGE Bands
3.8. Phylogenetic Analysis of Sequence Data
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Yokokawa, T.; Nagata, T. Linking bacterial community structure to carbon fluxes in MARINE environments. J. Oceanogr. 2010, 66, 1–12. [Google Scholar] [CrossRef]
- Pedros-Alio, C. Ecology: Dipping into the rare biosphere. Science 2007, 315, 192–193. [Google Scholar] [CrossRef]
- Dumestre, J.F.; Casamayor, E.O.; Massana, R.; Pedros-Alio, C. Changes in bacterial and archaeal assemblages in an equatorial river induced by the water eutrophication of Petit Saut dam reservoir (French Guiana). Aquat. Microb. Ecol. 2002, 26, 209–221. [Google Scholar] [CrossRef]
- Lebaron, P.; Servais, P.; Troussellier, M.; Courties, C.; Vives-Rego, J.; Muyzer, G.; Bernard, L.; Guindulain, T.; Schafer, H.; Stackebrandt, E. Changes in bacterial community structure in seawater mesocosms differing in their nutrient status. Aquat. Microb. Ecol. 1999, 19, 255–267. [Google Scholar] [CrossRef]
- Leflaive, J.; Danger, M.; Lacroix, G.; Lyautey, E.; Oumarou, C.; Ten-Hage, L. Nutrient effects on the genetic and functional diversity of aquatic bacterial communities. FEMS Microbiol. Ecol. 2008, 66, 379–390. [Google Scholar] [CrossRef]
- Riemann, L.; Steward, G.F.; Azam, F. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl. Environ. Microbiol. 2000, 66, 578–587. [Google Scholar] [CrossRef]
- Egan, S.T.; McCarthy, D.M.; Patching, J.W.; Fleming, G.T.A. An investigation of the physiology and potential role of components of the deep ocean bacterial community (of the NE Atlantic) by enrichments carried out under minimal environmental change. Deep Sea Res. Part I Oceanogr. Res. Pap. 2012, 61, 11–20. [Google Scholar] [CrossRef]
- Jorgensen, B.B.; Boetius, A. Feast and famine: Microbial life in the deep-sea bed. Nat. Rev. Microbiol. 2007, 5, 770–781. [Google Scholar] [CrossRef]
- Tamburini, C.; Boutrif, M.; Garel, M.; Colwell, R.R.; Deming, J.W. Prokaryotic responses to hydrostatic pressure in the ocean—A review. Environ. Microbiol. 2013, 15, 1262–1274. [Google Scholar] [CrossRef]
- Pollard, R.T.; Venables, H.J.; Read, J.F.; Allen, J.T. Large-scale circulation around the Crozet Plateau controls an annual phytoplankton bloom in the Crozet Basin. DeepSea Res. Part II Top. Stud. Oceanogr. 2007, 54, 1915–1929. [Google Scholar] [CrossRef]
- Pollard, R.; Sanders, R.; Lucas, M.; Statham, P. The Crozet natural Iron Bloom and Export Experiment (CROZEX). DeepSea Res. Part II Top. Stud. Oceanogr. 2007, 54, 1905–1914. [Google Scholar] [CrossRef]
- Treguer, P.; Jacques, G. Dynamics of nutrients and phytoplankton, and fluxes of carbon, nitrogen and silicon in the Antarctic Ocean. Polar Biol. 1992, 12, 149–162. [Google Scholar]
- Pitchford, J.; Brindley, J. Iron limitation, grazing pressure and oceanic High Nutrient-Low Chlorophyll (HNLC) regions. J. Plankton Res. 1999, 21, 525–547. [Google Scholar] [CrossRef]
- Planquette, H.; Statham, P.J.; Fones, G.R.; Charette, M.A.; Moore, C.M.; Salter, I.; Nedelec, F.H.; Taylor, S.L.; French, M.; Baker, A.R.; et al. Dissolved iron in the vicinity of the Crozet Islands, Southern Ocean. DeepSea Res. Part II Top. Stud. Oceanogr. 2007, 54, 1999–2019. [Google Scholar] [CrossRef]
- Pollard, R.; Read, J. Circulation pathways and transports of the Southern Ocean in the vicinity of the Southwest Indian Ridge. J. Geophys. Res. 2001, 106, 2881–2898. [Google Scholar] [CrossRef]
- Young-Hyang, P.; Gamberoni, L.; Charriaud, E. Frontal structure, water masses, and circulation in the Crozet Basin. J. Geophys. Res. 1993, 98, 361–385. [Google Scholar]
- Pollard, R.T.; Lucas, M.I.; Read, J.F. Physical controls on biogeochemical zonation in the Southern Ocean. DeepSea Res. Part II Top. Stud. Oceanogr. 2002, 49, 3289–3305. [Google Scholar] [CrossRef]
- Hughes, J.A.; Smith, T.; Chaillan, F.; Bett, B.J.; Billett, D.S.M.; Boorman, B.; Fischer, E.H.; Frenz, M.; Wolff, G.A. Two abyssal sites in the Southern Ocean influenced by different organic matter inputs: Environmental characterisation and preliminary observations on the benthic foraminifera. DeepSea Res. Part II Top. Stud. Oceanogr. 2007, 54, 2275–2290. [Google Scholar] [CrossRef]
- Martin, J.H.; Knauer, G.A.; Karl, D.M.; Broenkow, W.W. VERTEX: Carbon cycling in the northeast Pacific. DeepSea Res. Part I Oceanogr. Res. Pap. 1987, 34, 267–285. [Google Scholar] [CrossRef]
- Sanders, R.; Morris, P.J.; Stinchcombe, M.; Seeyave, S.; Venables, H.; Lucas, M. New production and the f ratio around the Crozet Plateau in austral summer 2004–2005 diagnosed from seasonal changes in inorganic nutrient levels. DeepSea Res. Part II Top. Stud. Oceanogr. 2007, 54, 2191–2207. [Google Scholar] [CrossRef]
- Turley, C. Bacteria in the cold deep-sea benthic boundary layer and sediment-water interface of the NE Atlantic. FEMS Microbiol. Ecol. 2000, 33, 89–99. [Google Scholar]
- Jamieson, R.E.; Rogers, A.D.; Billett, D.S.M.; Pearce, D.A. Bacterial biodiversity in deep-sea sediments from two regions of contrasting surface water productivity near the Crozet Islands, Southern Ocean. Deep Sea Res. Part A Oceanogr. Res. Pap. 2013, in press. [Google Scholar]
- Danovaro, R.; Corinaldesi, C.; Luna, G.M.; Magagnini, M.; Manini, E.; Pusceddu, A. Prokaryote diversity and viral production in deep-sea sediments and seamounts. DeepSea Res. Part II Top. Stud. Oceanogr. 2009, 56, 738–747. [Google Scholar] [CrossRef]
- Bendtsen, J.; Lundsgaard, C.; Middelboe, M.; Archer, D. Influence of bacterial uptake on deep-ocean dissolved organic carbon. Glob. Biogeochem. Cycle 2002, 16, 74:1–74:12. [Google Scholar]
- Danovaro, R.; della Croce, N.; Dell’Anno, A.; Pusceddu, A. A depocenter of organic matter at 7800 m depth in the SE Pacific Ocean. DeepSea Res. Part I Oceanogr. Res. Pap. 2003, 50, 1411–1420. [Google Scholar] [CrossRef]
- Fang, J.; Zhang, L.; Bazylinski, D.A. Deep-sea piezosphere and piezophiles: Geomicrobiology and biogeochemistry. Trends Microbiol. 2010, 18, 413–422. [Google Scholar] [CrossRef]
- Forney, L.J.; Zhou, X.; Brown, C.J. Molecular microbial ecology: Land of the one-eyed king. Curr. Opin. Microbiol. 2004, 7, 210–220. [Google Scholar] [CrossRef]
- Grossart, H.P.; Gust, G. Hydrostatic pressure affects physiology and community structure of marine bacteria during settling to 4000 m: An experimental approach. Mar. Ecol. Prog. Ser. 2009, 390, 97–104. [Google Scholar] [CrossRef]
- Muyzer, G.; de Waal, E.C.; Uitterlinden, A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar]
- Fromin, N.; Hamelin, J.; Tarnawski, S.; Roesti, D.; Jourdain-Miserez, K.; Forestier, N.; Teyssier-Cuvelle, S.; Gillet, F.; Aragno, M.; Rossi, P. Statistical analysis of denaturing gel electrophoresis (DGE) fingerprinting patterns. Environ. Microbiol. 2002, 4, 634–643. [Google Scholar] [CrossRef]
- Rink, B.; Seeberger, S.; Martens, T.; Duerselen, C.D.; Simon, M.; Brinkhoff, T. Effects of phytoplankton bloom in a coastal ecosystem on the composition of bacterial communities. Aquat. Microb. Ecol. 2007, 48, 47–60. [Google Scholar] [CrossRef]
- Yoshida, A.; Nishimura, M.; Kogure, K. Bacterial community structure in the Sulu Sea and adjacent areas. DeepSea Res. Part II Top. Stud. Oceanogr. 2007, 54, 103–113. [Google Scholar] [CrossRef]
- Wu, L.; Yu, Y.; Zhang, T.; Feng, W.; Zhang, X.; Li, W. PCR-DGGE fingerprinting analysis of plankton communities and its relationship to lake trophic status. Int. Rev. Hydrobiol. 2009, 94, 528–541. [Google Scholar] [CrossRef]
- Bonin, P.C.; Michotey, V.D.; Mouzdahir, A.; Rontani, J.-F. Anaerobic biodegradation of squalene: Using DGGE to monitor the isolation of denitrifying Bacteria taken from enrichment cultures. FEMS Microbiol. Ecol. 2002, 42, 37–49. [Google Scholar] [CrossRef]
- Nakagawa, T.; Fukui, M. Phylogenetic characterization of microbial mats and streamers from a Japanese alkaline hot spring with a thermal gradient. J. Gen. Appl. Microbiol. 2002, 48, 211–222. [Google Scholar] [CrossRef]
- Stephen, J.R.; Chang, Y.-J.; Gan, Y.D.; Peacock, A.; Pfiffner, S.M.; Barcelona, M.J.; White, D.C.; Macnaughton, S.J. Microbial characterization of a JP-4 fuel-contaminated site using a combined lipid biomarker/polymerase chain reaction-denaturing gradient gel electrophoresis (PCR-DGGE)-based approach. Environ. Microbiol. 1999, 1, 231–241. [Google Scholar] [CrossRef]
- Murray, A.E.; Preston, C.M.; Massana, R.; Taylor, L.T.; Blakis, A.; Wu, K.; DeLong, E.F. Seasonal and spatial variability of bacterial and archaeal assemblages in the coastal waters near Anvers Island, Antarctica. Appl. Environ. Microbiol. 1998, 64, 2585–2595. [Google Scholar]
- Spiegelman, D.; Whissell, G.; Greer, C.W. A survey of the methods for the characterization of microbial consortia and communities. Can. J. Microbiol. 2005, 51, 355–386. [Google Scholar] [CrossRef]
- Grossi, V.; Yakimov, M.M.; Al Ali, B.; Tapilatu, Y.; Cuny, P.; Goutx, M.; La Cono, V.; Giuliano, L.; Tamburini, C. Hydrostatic pressure affects membrane and storage lipid compositions of the piezotolerant hydrocarbon-degrading Marinobacter hydrocarbonoclasticus strain #5. Environ. Microbiol. 2010, 12, 2020–2033. [Google Scholar] [CrossRef]
- Yayanos, A.A.; Dietz, A.S. Death of a hadal deep-sea bacterium after decompression. Science 1983, 220, 497–498. [Google Scholar]
- Chastain, R.A.; Yayanos, A.A. Ultrastructural changes in an obligately barophilic marine bacterium after decompression. Appl. Environ. Microbiol. 1991, 57, 1489–1497. [Google Scholar]
- Park, C.B.; Clark, D.S. Rupture of the cell envelope by decompression of the deep-sea methanogen methanococcus jannaschii. Appl. Environ. Microbiol. 2002, 68, 1458–1463. [Google Scholar] [CrossRef]
- Yayanos, A.A.; Dietz, A.S. Thermal inactivation of a deep-sea barophilic bacterium, isolate CNPT-3. Appl. Environ. Microbiol. 1982, 43, 1481–1489. [Google Scholar]
- Patching, J.W.; Eardly, D. Bacterial biomass and activity in the deep waters of the eastern Atlantic—Evidence of a barophilic community. Deep Sea Res. Part I Oceanogr. Res. Pap. 1997, 44, 1655–1670. [Google Scholar] [CrossRef]
- Eardly, D.F.; Carton, M.W.; Gallagher, J.M.; Patching, J.W. Bacterial abundance and activity in deep-sea sediments from the eastern North Atlantic. Prog. Oceanog. 2001, 50, 245–259. [Google Scholar] [CrossRef]
- Nagata, T.; Fukuda, H.; Fukuda, R.; Koike, I. Bacterioplankton distribution and production in deep Pacific waters: Large-Scale geographic variations and possible coupling with sinking particle fluxes. Limnol. Oceanogr. 2000, 45, 426–435. [Google Scholar] [CrossRef]
- Thingstad, T.F. Elements of a theory for the mechanisms controlling abundance, diversity and biogeochemical role of lytic viruses in aquatic systems. Limnol. Oceanogr. 2000, 45, 1320–1328. [Google Scholar] [CrossRef]
- Pernthaler, J. Predation on prokaryotes in the water column and its ecological implications. Nat. Rev. Microbiol. 2005, 3, 537–536. [Google Scholar] [CrossRef]
- Hyun, J.H.; Kim, K.H. Bacterial abundance and production during the unique spring phytoplankton bloom in the central Yellow Sea. Mar. Ecol. Prog. Ser. 2003, 252, 77–88. [Google Scholar] [CrossRef]
- Guixa-Boixereu, N.; Lysnes, K.; Pedros-Alio, C. Viral lysis and bacterivory during a phytoplankton bloom in a coastal water microcosm. Appl. Environ. Microbiol. 1999, 65, 1949–1958. [Google Scholar]
- Pinhassi, J.; Sala, M.M.; Havskum, H.; Peters, F.; Guadayol, O.; Malits, A.; Marrase, C. Changes in bacterioplankton composition under different phytoplankton regimens. Appl. Environ. Microbiol. 2004, 70, 6753–6766. [Google Scholar] [CrossRef]
- Jurgens, K.; Matz, C. Predation as a shaping force for the phenotypic and genotypic composition of planktonic bacteria. Antonie Leeuwenhoek 2002, 81, 413–434. [Google Scholar] [CrossRef]
- ZoBell, C.E.; Anderson, D.Q. Observations on the multiplication of bacteria in different volumes of sea water and the influence of oxygen tension and solid surfaces. Biol. Bull. (Woods Hole) 1936, 71, 324–342. [Google Scholar] [CrossRef]
- Zobell, C.E. The effect of solid surfaces upon bacterial activity. J. Bacteriol. 1943, 46, 39–56. [Google Scholar]
- Kato, C.; Bartlett, D.H. The molecular biology of barophilic bacteria. Extremophiles 1997, 1, 111–116. [Google Scholar] [CrossRef]
- Jannasch, H.W.; Taylor, C.D. Deep-sea microbiology. Annu. Rev. Microbiol. 1984, 38, 487–514. [Google Scholar] [CrossRef]
- Bent, S.J.; Forney, L.J. The tragedy of the uncommon: Understanding limitations in the analysis of microbial diversity. ISME J. 2008, 2, 689–695. [Google Scholar] [CrossRef]
- Sogin, M.L.; Morrison, H.G.; Huber, J.A.; Welch, D.M.; Huse, S.M.; Neal, P.R.; Arrieta, J.M.; Herndl, G.J. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. USA 2006, 103, 12115–12120. [Google Scholar] [CrossRef]
- Fuhrman, J.A. Microbial community structure and its functional implications. Nature 2009, 459, 193–199. [Google Scholar] [CrossRef]
- Muylaert, K.; VanDerGucht, K.; Vloemans, N.; Meester, L.; Gillis, M.; Vyverman, W. Relationship between bacterial community composition and bottom-up versus top-down variables in four eutrophic shallow lakes. Appl. Environ. Microbiol. 2002, 68, 4740–4750. [Google Scholar] [CrossRef]
- Zwart, G.; Kamst-van Agterveld, M.P.; van der Werff-Staverman, I.; Hagen, F.; Hoogveld, H.L.; Gons, H.J. Molecular characterization of cyanobacterial diversity in a shallow eutrophic lake. Environ. Microbiol. 2005, 7, 365–377. [Google Scholar] [CrossRef]
- Park, J.W.; Crowley, D.E. Nested PCR bias: A case study of Pseudomonas spp in soil microcosms. J. Environ. Monit. 1039, 12, 985–988. [Google Scholar] [CrossRef]
- Touzet, N.; McCarthy, D.; Fleming, G.T. Molecular fingerprinting of lacustrian cyanobacterial communities: Regional patterns in summer diversity. FEMS Microbiol. Ecol. 2013. [Google Scholar] [CrossRef]
- Bianchi, A.; Garcin, J. Bacterial response to hydrostatic-pressure in seawater samples collected in mixed-water and stratified-water conditions. Mar. Ecol. Prog. Ser. 1994, 111, 137–141. [Google Scholar] [CrossRef]
- Jannasch, H.W.; Wirsen, C.O.; Taylor, C.D. Deep-Sea Bacteria: Isolation in the Absence of Decompression. Science 1982, 216, 1315–1317. [Google Scholar]
- Herndl, G.J.; Reinthaler, T.; Teira, E.; van Aken, H.; Veth, C.; Pernthaler, A.; Pernthaler, J. Contribution of archaea to total prokaryotic production in the deep Atlantic Ocean. Appl. Environ. Microbiol. 2005, 71, 2303–2309. [Google Scholar] [CrossRef]
- Ingalls, A.E.; Shah, S.R.; Hansman, R.L.; Aluwihere, L.I.; Santos, G.R.; Druffel, E.R.M.; Pearson, A. Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc. Natl. Acad. Sci. USA 2006, 103, 6442–6447. [Google Scholar] [CrossRef]
- Oger, P.M.; Jebbar, M. The many ways of coping with pressure. Res. Microbiol. 2010, 161, 799–809. [Google Scholar] [CrossRef]
- Parkes, R.J.; Cragg, B.A.; Getliff, J.M.; Harvey, S.M.; Fry, J.C.; Lewis, C.A.; Rowland, S.J. A quantitative study of microbial decomposition of biopolymers in Recent sediments from the Peru Margin. Mar. Geol. 1993, 113, 55–66. [Google Scholar] [CrossRef]
- Don, R.H.; Cox, P.T.; Wainwright, B.J.; Baker, K.; Mattick, J.S. “Touchdown” PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991. [Google Scholar] [CrossRef]
- Achenbach, L.; Woese, C.R. 16S and 23S rRNA-like Primers. In Archaea—A Laboratory Manual; Sower, K.R., Schreier, H.J., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1995; pp. 521–523. [Google Scholar]
- DeLong, E.F. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 1992, 89, 5685–5689. [Google Scholar] [CrossRef]
- Ovreas, L.; Forney, L.; Daae, F.; Torsvik, V. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl. Environ. Microbiol. 1997, 63, 3367–3373. [Google Scholar]
- Sheffield, V.C.; Cox, D.R.; Lerman, L.S.; Myers, R.M. Attachment of a 40 base-pair G+C-rich sequence to genomic DNA fragments by the polymerase chain reaction results in improved detection of single base changes. Proc. Natl. Acad. Sci. USA 1989, 86, 232–236. [Google Scholar] [CrossRef]
- Kovach, W. MVSP-A Multivariate Statistical Package for Windows, ver. 3.13q; Kovach Computing Services: Pentraeth, Wales, UK, 1998. [Google Scholar]
- Simpson, E.H. Measurement of diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Shannon, C.E.; Weaver, W. The Mathematical Theory of Communication; University of Illinois Press: Urbana, IL, USA, 1963; p. 29. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Chenna, R.; Sugawara, H.; Koike, T.; Lopez, R.; Gibson, T.J.; Higgins, D.G.; Thompson, J.D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003, 31, 3497–3500. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, N. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
McCarthy, D.M.; Pearce, D.A.; Patching, J.W.; Fleming, G.T.A. Contrasting Responses to Nutrient Enrichment of Prokaryotic Communities Collected from Deep Sea Sites in the Southern Ocean. Biology 2013, 2, 1165-1188. https://doi.org/10.3390/biology2031165
McCarthy DM, Pearce DA, Patching JW, Fleming GTA. Contrasting Responses to Nutrient Enrichment of Prokaryotic Communities Collected from Deep Sea Sites in the Southern Ocean. Biology. 2013; 2(3):1165-1188. https://doi.org/10.3390/biology2031165
Chicago/Turabian StyleMcCarthy, David M., David A. Pearce, John W. Patching, and Gerard T. A. Fleming. 2013. "Contrasting Responses to Nutrient Enrichment of Prokaryotic Communities Collected from Deep Sea Sites in the Southern Ocean" Biology 2, no. 3: 1165-1188. https://doi.org/10.3390/biology2031165
APA StyleMcCarthy, D. M., Pearce, D. A., Patching, J. W., & Fleming, G. T. A. (2013). Contrasting Responses to Nutrient Enrichment of Prokaryotic Communities Collected from Deep Sea Sites in the Southern Ocean. Biology, 2(3), 1165-1188. https://doi.org/10.3390/biology2031165




