Prospective Mapping of Transcriptional Changes Associated with Lipid and Carotenoid Production in Rhodotorula glutinis Using Different Feeding Approaches
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Fed-Batch Fermentation of R. glutinis
2.2. Detection of Lipid Bodies, Dry Cell Weight (DCW) and Reducing Sugar in the Culture Media
2.3. Total Lipids (TL) Detection and GC Analysis of Fatty Acids Methyl Esters
2.4. Extraction, Quantification and Identification of Total Carotenoids
2.5. Transcriptomic Sequencing
2.6. Statistical Analysis
3. Result and Discussion
3.1. Biomass, Lipid, and Carotenoid Production by R. glutinis Under Different Fed-Batch Fermentation Strategies
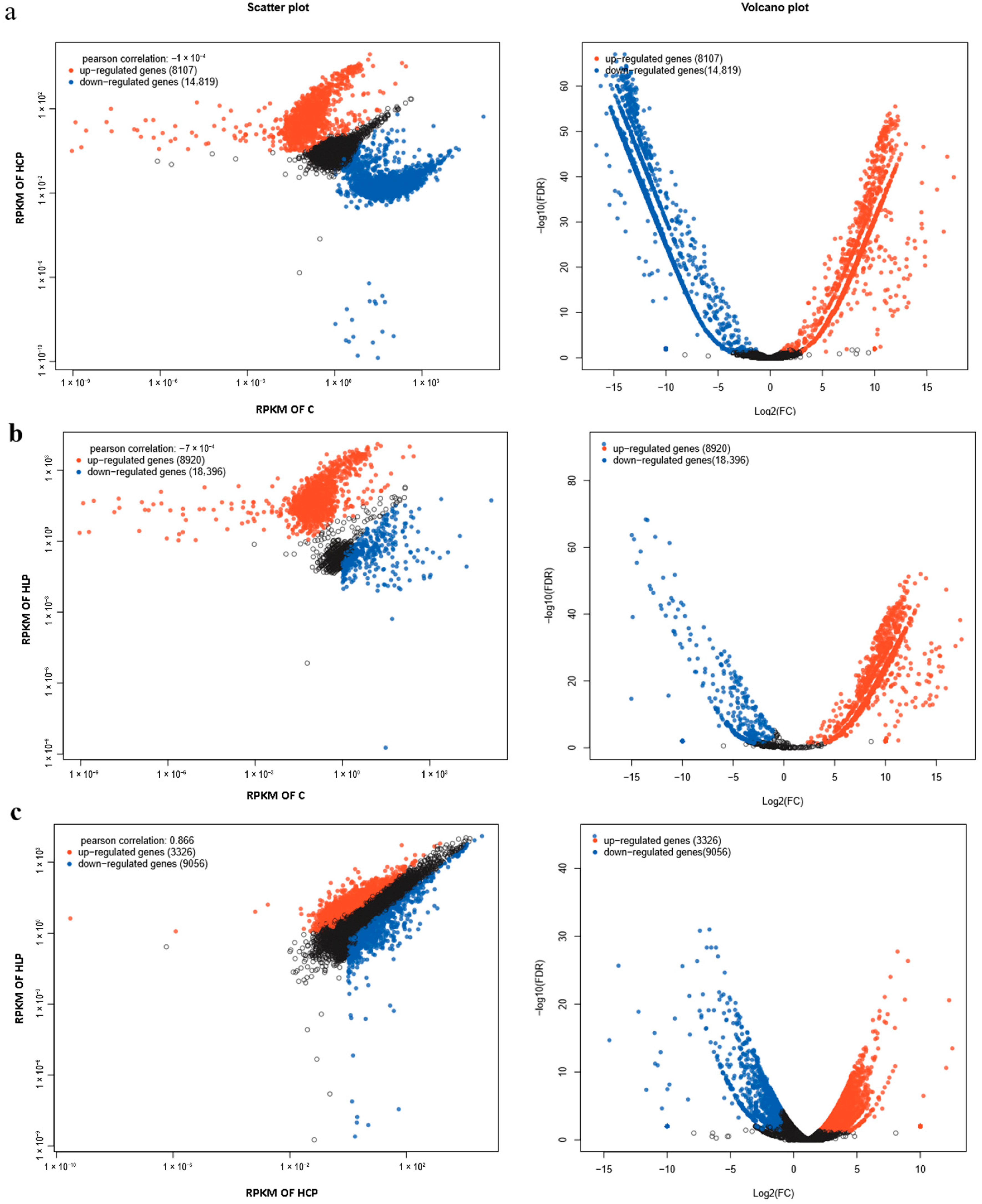
3.2. Differential Gene Expression (DEG)
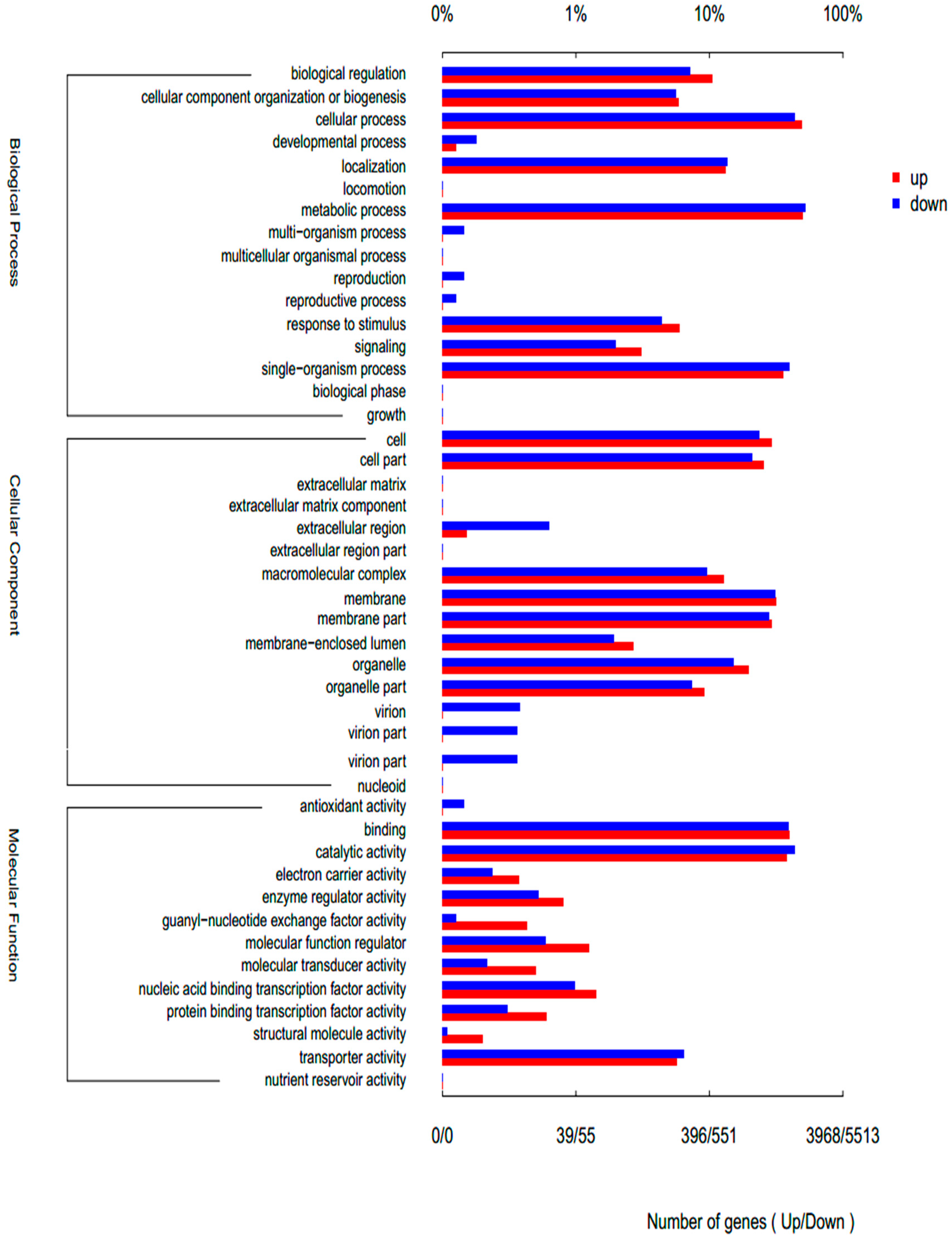
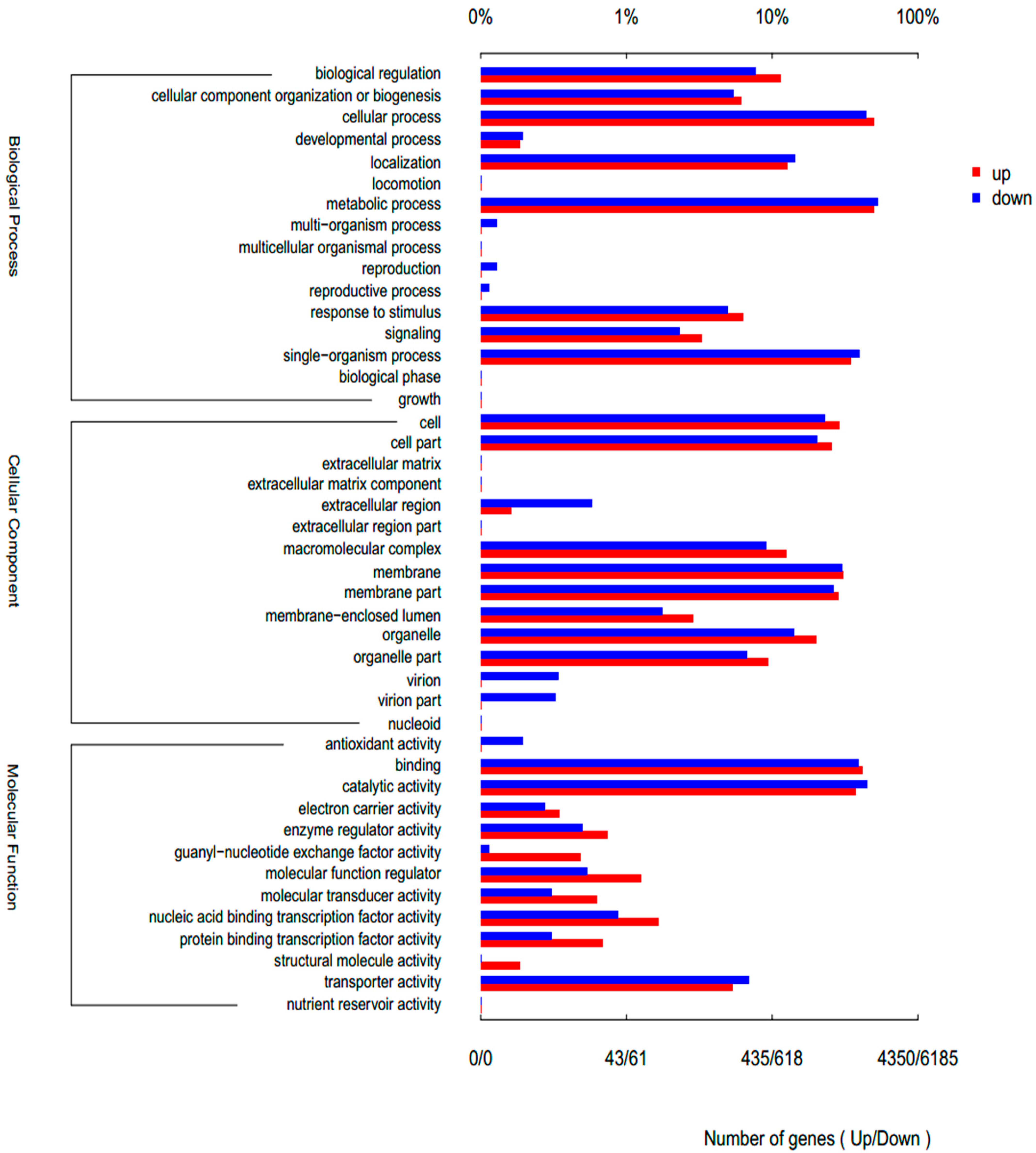
3.3. Gene Ontology (GO) Enrichment Analysis
3.4. Central Carbon Metabolic Pathways
3.5. Lipid Metabolism
3.6. Carotenoid Biosynthetic Pathway
4. Conclusions
5. Limitations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| R. glutinis | Rhodotorula glutinis |
| C/N | Carbon-to-nitrogen ratios |
| C | Control |
| HLP | High lipid production |
| HCP | High carotenoid production |
| FAS1/2 | Fatty acid synthase complex |
| PPP | Pentose phosphate pathway |
| CrtZ | Putative beta-carotene hydroxylase |
| gapN | NADP+-dependent glyceraldehyde-3-phosphate dehydrogenase |
| C/S | Carbon to sulfur |
| DCW | Dry Cell Weight |
| TL | Total lipids |
| GC | Gas Chromatography |
| RSEM | Expectation-Maximization |
| DEG | Differentially expressed genes |
| SRA | Sequence Read Archive |
| Clipid | Cellular lipid |
| Ccar | Cellular Carotenoid |
| TP | Total Pigment |
References
- Elfeky, N.; Rizk, A.; Gharieb, M.M. Exploring the lipids, carotenoids, and vitamins content of Rhodotorula glutinis with selenium supplementation under lipid accumulating and growth proliferation conditions. BMC Microbiol. 2024, 24, 451. [Google Scholar] [CrossRef] [PubMed]
- Elfeky, N.; Elmahmoudy, M.; Bao, Y. Manipulation of Culture Conditions: Tool for Correlating/Improving Lipid and Carotenoid Production by Rhodotorula glutinis. Processes 2020, 8, 140. [Google Scholar] [CrossRef]
- Kot, A.M.; Błażejak, S.; Kurcz, A.; Gientka, I.; Kieliszek, M. Rhodotorula glutinis-potential source of lipids, carotenoids, and enzymes for use in industries. Appl. Microbiol. Biotechnol. 2016, 100, 6103–6117. [Google Scholar] [CrossRef]
- Thancharoen, K.; Malasri, A.; Leamsingkorn, W.; Boonyalit, P. Selection of Oleaginous Yeasts with Lipid Accumulation by the Measurement of Sudan Black B for Benefits of Biodiesel. Int. J. Pharm. Med. Biol. Sci. 2017, 6, 53–57. [Google Scholar] [CrossRef]
- Zoz, L.; Carvalho, J.C.; Soccol, V.T.; Casagrande, T.C.; Cardoso, L. Torularhodin and torulene: Bioproduction, properties and prospective applications in food and cosmetics—A review. Braz. Arch. Biol. Technol. 2015, 58, 278–288. [Google Scholar] [CrossRef]
- Tkáčová, J.; Čaplová, J.; Klempová, T.; Čertík, M. Correlation between lipid and carotenoid synthesis in torularhodin-producing Rhodotorula glutinis. Ann. Microbiol. 2017, 67, 541–551. [Google Scholar] [CrossRef]
- Gong, G.; Liu, L.; Zhang, X.; Tan, T. Comparative evaluation of different carbon sources supply on simultaneous production of lipid and carotene of Rhodotorula glutinis with irradiation and the assessment of key gene transcription. Bioresour. Technol. 2019, 288, 121559. [Google Scholar] [CrossRef]
- Braunwald, T.; Schwemmlein, L.; Graeff-Hönninger, S.; French, W.T.; Hernandez, R.; Holmes, W.E.; Claupein, W. Effect of different C/N ratios on carotenoid and lipid production by Rhodotorula glutinis. Appl. Microbiol. Biotechnol. 2013, 97, 6581–6588. [Google Scholar] [CrossRef] [PubMed]
- Nunes, D.D.; Pillay, V.L.; Van Rensburg, E.; Pott, R.W.M. Oleaginous microorganisms as a sustainable oil source with a focus on downstream processing and cost-lowering production strategies: A review. Bioresour. Technol. Rep. 2024, 26, 101871. [Google Scholar] [CrossRef]
- Tang, W.; Wang, Y.; Zhang, J.; Cai, Y.; He, Z. Biosynthetic pathway of carotenoids in Rhodotorula and strategies for enhanced their production. J. Microbiol. Biotechnol. 2019, 29, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Elfeky, N.; Elmahmoudy, M.; Zhang, Y.; Guo, J.L.; Bao, Y. Lipid and carotenoid production by Rhodotorula glutinis with a combined cultivation mode of nitrogen, sulfur, and aluminium stress. Appl. Sci. 2019, 9, 2444. [Google Scholar] [CrossRef]
- Gong, G.; Liu, L.; Zhang, X.; Tan, T. Multi-omics metabolism analysis on irradiation-induced oxidative stress to Rhodotorula glutinis. Appl. Microbiol. Biotechnol. 2019, 103, 361–374. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhang, S.; Liu, H.; Shen, H.; Lin, X.; Yang, F.; Zhou, Y.J.; Jin, G.; Ye, M.; Zou, H.; et al. A multi-omic map of the lipid-producing yeast Rhodosporidium toruloides. Nat. Commun. 2012, 3, 1111. [Google Scholar] [CrossRef]
- Kimura, K.; Yamaoka, M.; Kamisaka, Y. Rapid estimation of lipids in oleaginous fungi and yeasts using Nile red fluorescence. J. Microbiol. Methods 2004, 56, 331–338. [Google Scholar] [CrossRef]
- Sitepu, R.; Sestric, R.; Ignatia, L.; Levin, D.; German, J.; Gillies, L.A.; Almada, L.A.; Boundy-Mills, K.L. Manipulation of culture conditions alters lipid content and fatty acid profiles of a wide variety of known and new oleaginous yeast species. Bioresour. Technol. 2013, 144, 360–369. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Mishra, S.; Suh, W.; Farooq, W.; Moon, M.; Shrivastav, A.; Park, M.S.; Yang, J. Rapid quantification of microalgal lipids in aqueous medium by a simple colorimetric method. Bioresour. Technol. 2014, 155, 330–333. [Google Scholar] [CrossRef] [PubMed]
- Van Wychen, S.; Ramirez, K.; Laurens, L.M. Determination of Total Lipids as Fatty Acid Methyl Esters (FAME) by In Situ Transesterification; Technical Report; NREL: Golden, CO, USA, 2013. [Google Scholar]
- Frengova, G.; Sirnova, E.; Pavlova, K.; Beshkova, D. Formation of Carotenoids by Rhodotorula glutinis in whey ultrafiltrate. Biotechnol. Bioeng. 1994, 44, 888–894. [Google Scholar] [CrossRef]
- Weber, R.W.; Anke, H.; Davoli, P. Simple method for the extraction and reversed-phase high-performance liquid chromatographic analysis of carotenoid pigments from red yeasts (Basidiomycota, Fungi). J. Chromatogr. A 2007, 1145, 118–122. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Saenge, C.; Cheirsilp, B.; Suksaroge, T.T.; Bourtoom, T. Potential use of oleaginous red yeast Rhodotorula glutinis for the bioconversion of crude glycerol from biodiesel plant to lipids and carotenoids. Process Biochem. 2011, 46, 210–218. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, S.; Zhu, Z.; Shen, H.; Lin, X.; Jin, X.; Jiao, X.; Zhao, Z.K. Systems analysis of phosphate limitation-induced lipid accumulation by the oleaginous yeast Rhodosporidium toruloides. Biotechnol. Biofuels 2018, 11, 148. [Google Scholar] [CrossRef]
- Mondala, A.H.; Hernandez, R.; French, T.; McFarland, L.; Domingo, J.W.S.; Meckes, M.; Ryu, H.; Iker, B. Enhanced Lipid and Biodiesel Production from Glucose-Fed Activated Sludge: Kinetics and Microbial Community Analysis. AIChE J. 2012, 58, 1279–1290. [Google Scholar] [CrossRef]
- Dias, C.; Sousa, S.; Caldeira, J.; Reis, A.T. New dual-stage pH control fed-batch cultivation strategy for the improvement of lipids and carotenoids production by the red yeast Rhodosporidium toruloides NCYC 921. Bioresour. Technol. 2015, 189, 309–318. [Google Scholar] [CrossRef]
- El-Banna, A.A.; Abd El-Razek, A.M.; El-Mahdy, A.R. Some Factors Affecting the Production of Carotenoids by Rhodotorula glutinis var. glutinis. Food Nutr. Sci. 2012, 3, 64–71. [Google Scholar] [CrossRef]
- Bhosale, P.B.; Gadre, R.V. Production of β-carotene by a mutant of Rhodotorula glutinis. Appl. Microbiol. Biotechnol. 2001, 55, 423–427. [Google Scholar] [CrossRef]
- Han, M.; Xu, Z.-Y.; Du, C.; Qian, H.; Zhang, W.-G. Effects of nitrogen on the lipid and carotenoid accumulation of oleaginous yeast Sporidiobolus pararoseus. Bioprocess Biosyst. Eng. 2016, 39, 1425–1433. [Google Scholar] [CrossRef]
- Ratledge, C. The role of malic enzyme as the provider of NADPH in oleaginous microorganisms: A reappraisal and unsolved problems. Biotechnol. Lett. 2014, 36, 1557–1568. [Google Scholar] [CrossRef]
- Rashida, Z.; Srinivasan, R.; Cyanam, M.; Laxman, S. Kog1/Raptor mediates metabolic rewiring during nutrient limitation by controlling SNF1/AMPK activity. Sci. Adv. 2021, 7, eabe5544. [Google Scholar] [CrossRef]
- Fraser, P.D.; Bramley, P.M. The biosynthesis and nutritional uses of carotenoids. Prog. Lipid Res. 2004, 43, 228–265. [Google Scholar] [CrossRef] [PubMed]
- Conrad, M.; Schothorst, J.; Kankipati, H.N.; Van Zeebroeck, G.; Rubio-Texeira, M.; Thevelein, J.M. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2014, 38, 254–299. [Google Scholar] [CrossRef]
- Metur, S.P.; Klionsky, D.J. Nutrient-dependent signaling pathways that control autophagy in yeast. FEBS Lett. 2024, 598, 32–47. [Google Scholar] [CrossRef]
- Hariri, H.; Rogers, S.; Ugrankar, R.; Liu, Y.L.; Feathers, J.R.; Henne, W.M. Lipid droplet biogenesis is spatially coordinated at ER–vacuole contacts under nutritional stress. EMBO Rep. 2018, 19, 57–72. [Google Scholar] [CrossRef]
- Koh, H.-J.; Lee, S.-M.; Son, B.-G.; Lee, S.-H.; Ryoo, Z.Y.; Chang, K.-T.; Park, J.-W.; Park, D.-C.; Song, B.J.; Veech, R.L.; et al. Cytosolic NADP+-dependent isocitrate dehydrogenase plays a key role in lipid metabolism. J. Biol. Chem. 2004, 279, 39968–39974. [Google Scholar] [CrossRef]
- Lin, Z.; Ni, C.; Jiang, H.; Yang, H.; Deng, L.; Liu, P.; Li, X.; Yu, Y.; Li, W.; Wang, R.; et al. Guanine Nucleotide Exchange Factors and Small GTPases: Their Regulation and Functions, Diseases, and Therapeutic Targets. MedComm 2025, 6, e70362. [Google Scholar] [CrossRef]
- Yang, Y.; Ye, Z.; Guo, M.; Chen, G. Transcriptomic analysis of the effects of nutritional conditions on Rhodosporidium toruloides lipid production. Biochem. Eng. J. 2026, 225, 109945. [Google Scholar] [CrossRef]
- Sasaki, Y.; Yoshikuni, Y. Metabolic engineering for valorization of macroalgae biomass. Metab. Eng. 2022, 71, 42–61. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Yan, P.; Li, Y.; Liu, X.; Wang, Z.; Chen, T.; Zhao, X. Enhancing β-carotene production in Escherichia coli by perturbing central carbon metabolism and improving the NADPH supply. Front. Bioeng. Biotechnol. 2020, 8, 585. [Google Scholar] [CrossRef] [PubMed]
- Hynes, M.J.; Murray, S.L. ATP-citrate lyase is required for production of cytosolic acetyl coenzyme A and development in Aspergillus nidulans. Eukaryot. Cell 2010, 9, 1039–1048. [Google Scholar] [CrossRef]
- Partow, S.; Hyland, P.B.; Mahadevan, R. Synthetic rescue couples NADPH generation to metabolite overproduction in Saccharomyces cerevisiae. Metab. Eng. 2017, 43, 64–70. [Google Scholar] [CrossRef]
- Ratledge, C. Regulation of lipid accumulation in oleaginous microorganisms. Biochem. Soc. Trans. 2002, 30, 1047–1050. [Google Scholar] [CrossRef] [PubMed]
- Ratledge, C.; Wynn, J.P. The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Adv. Appl. Microbiol. 2002, 51, 1–51. [Google Scholar]
- Dulermo, T.; Nicaud, J.M. Involvement of the G3P shuttle and β-oxidation pathway in the control of TAG synthesis and lipid accumulation in Yarrowia lipolytica. Metab. Eng. 2011, 13, 482–491. [Google Scholar] [CrossRef]
- Zhang, S.; Skerker, J.M.; Rutter, C.D.; Maurer, M.J.; Arkin, A.P.; Rao, C.V. Engineering Rhodosporidium toruloides for increased lipid production. Biotechnol. Bioeng. 2016, 113, 1056–1066. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Shi, F.; Zhan, W. Overexpression of ZWF1 and POS5 improves carotenoid biosynthesis in recombinant Saccharomyces cerevisiae. Lett. Appl. Microbiol. 2015, 61, 354–360. [Google Scholar] [CrossRef]
- Lee, P.C.; Momen, A.Z.R.; Mijts, B.N.; Schmidt-Dannert, C. Biosynthesis of Structurally Novel Carotenoids in Escherichia coli. Chem. Biol. 2003, 10, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.K.; Matsuda, S.; Hoshino, T.; Peng, X.; Misawa, N. Characterization of bacterial β-carotene 3,3′-hydroxylases, CrtZ, and P450 in astaxanthin biosynthetic pathway and adonirubin production by gene combination in Escherichia coli. Appl. Microbiol. Biotechnol. 2006, 72, 1238–1246. [Google Scholar] [CrossRef]
- Linden, H. Carotenoid Hydroxylase from Haematococcus pluvialis: CDNA Sequence, Regulation and Functional Complementation. Biochim. Biophys. Acta 1999, 1446, 203–212. [Google Scholar] [CrossRef]
- Mata-Gómez, L.C.; Montañez, J.C.A.; Méndez-Zavala, A.; Aguilar, C.N. Biotechnological production of carotenoids by yeasts: An overview. Microb. Cell Fact. 2014, 13, 12. [Google Scholar] [CrossRef] [PubMed]
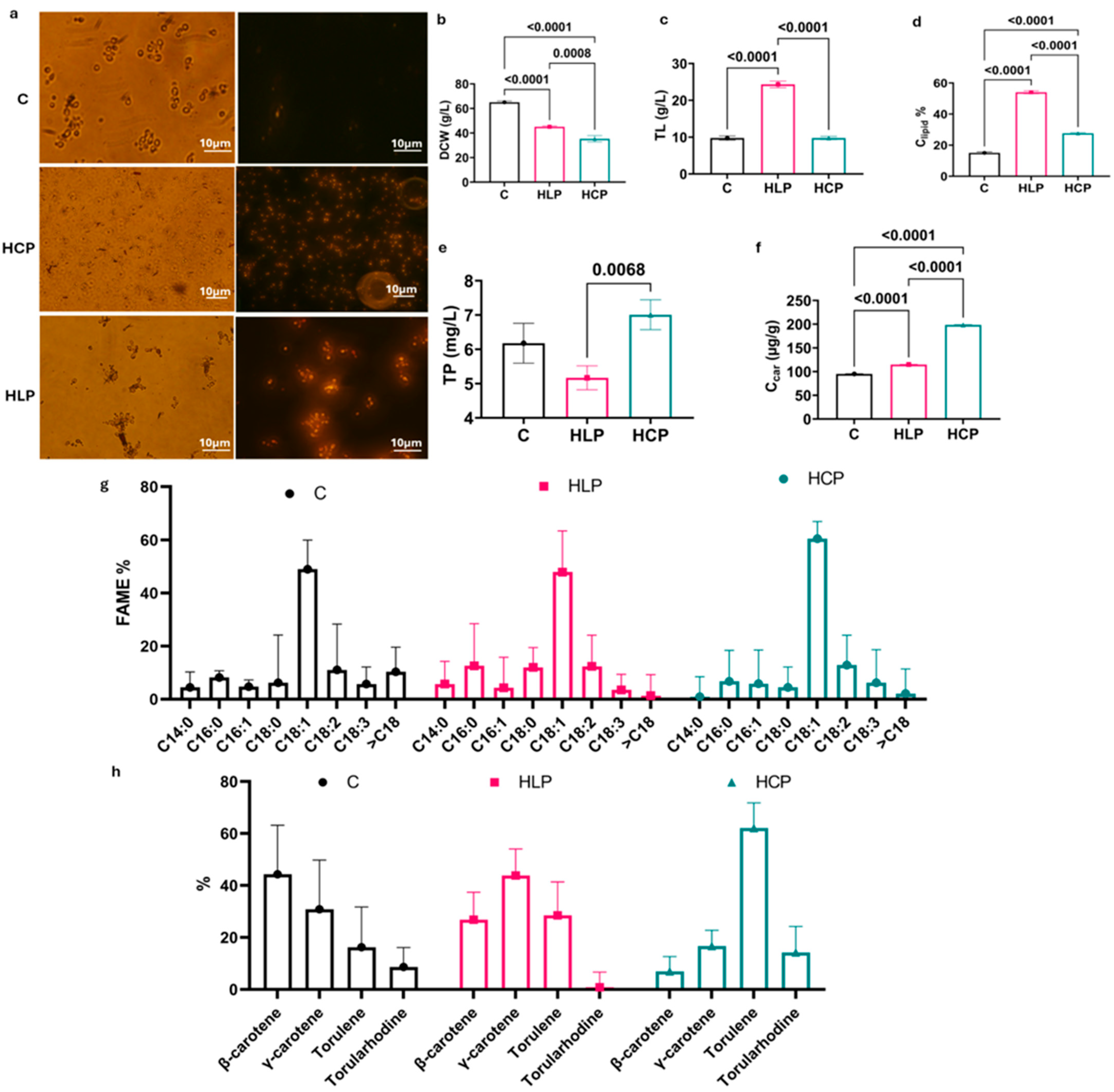

| Group | Medium Composition (g/L) | Metal | Feeding Solution (g/100 mL Distilled Water) | Feeding Rate (mL/h) | Residual Glucose (g/L) |
|---|---|---|---|---|---|
| C | Glucose (20), peptone (10), yeast extract (10) | None | Glucose: Yeast Extract (50:50) | 5 | >5.00 |
| HLP | Glucose (60), yeast extract (5), (NH4)2SO4 (1), KH2PO4 (1.5), MgSO4 (1) | BaCl2 and MnCl2 | Glucose (100) | 4 | >25.0 |
| HCP | Glucose (60), yeast extract (5), (NH4)2SO4 (2), KH2PO4 (2), MgSO4 (3) | Al2SO4 | Glucose: MgSO4 (100:0.20) | 4 | >25.0 |
| Accession | Bioproject Accession | Biosample Accession | Group |
|---|---|---|---|
| SRR10738942 | PRJNA596366 | SAMN13620008 | C |
| SRR10738940 | PRJNA596366 | SAMN13620010 | HCP |
| SRR10738939 | PRJNA596366 | SAMN13620011 | HLP |
| Symbol | Definition | HCP/C | HLP/C | HLP/HCP | Log2 FC HCP/C (p Value) | Log2 FC HLP/C (p Value) | Log2 FC HLP/HCP (p Value) | |
|---|---|---|---|---|---|---|---|---|
| Glycolysis enzymes | PGM | phosphoglucomutase | U/D | U/D | D | - | - | −5.789 (0.001) |
| GPI | glucose-6-phosphate isomerase | U | U/D | D | 0.949 (0.005) | - | −10.46 (0.000) | |
| galM | aldose-1-epimerase | U/D | U | U | - | 16.90 (0.000) | 4.231 (0.000) | |
| ald | fructose biphosphate aldolase | U/D | U/D | D | - | - | −9.600 (0.014) | |
| gapA | glyceraldhyde-3-phosphate dehydrogenase | U/D | U/D | D | - | - | −10.00 (0.008) | |
| gapN | glyceraldhyde-3-phosphate dehydrogenase NADP+ | U | N | D | 12.24 (0.000) | - | −11.80 (0.000) | |
| PGK | phosphoglycerate kinase | U/D | U/D | D | - | - | −4.200 (0.000) | |
| gpmI | 2,3 biphosphoglycerate-independent phosphoglycerate mutase | U/D | U/D | D | - | - | −10.10 (0.005) | |
| gpmA | 2,3biphosphoglycerate-dependent phosphoglycerate mutase | U | U | N | 7.700 (0.001) | 8.800 (0.020) | - | |
| ENO | enolase | U/D | U/D | D | - | - | −8.000 (0.000) | |
| PEPCKA | phosphoenol pyruvate carboxykinase ATP | U/D | U/D | U | - | - | 10.90 (0.000) | |
| PEPCKG | phosphoenol pyruvate carboxykinase GTP | U/D | D | D | - | −10.20 (0.005) | −9.900 (0.000) | |
| PK | pyruvate kinase | U/D | U/D | U | - | - | 10.70 (0.004) | |
| PDC | pyruvate decarboxylase | U/D | U/D | U | - | - | 3.000 (0.000) | |
| E1.2.1.5 | aldehyde dehydrogenase (NAD(P)+) | D | D | N | −16.30 (0.000) | −16.30 (0.000) | - | |
| exaA | alcohol dehydrogenase (cytochrome c) | U/D | U | U | - | 9.900 (0.005) | 10.70 (0.000) | |
| ME-NAD | NAD-dependent malic enzyme, mitochondrial | U/D | U/D | U | - | - | 2.200 (0.020) | |
| ME-NADP | malic enzyme-NADP dependent | U | U | D | 10.56 (0.001) | 11.95 (0.000) | −9.078 (0.014) | |
| MDH1 | malate dehydrogenase mitochondrial | D | D | U | −9.969 (0.000) | −16.98 (0.000) | 2.039 (0.035) | |
| MDH2 | malate dehydrogenase, NAD-dependent (cytoplasmic) | U/D | U/D | U | - | - | 2.541 (0.002) | |
| ACL | beta subunit citrate lyase | U | U | N | 6.705 (0.000) | 7.944 (0.000) | - | |
| TCA cycle | CS | citrate synthase | U/D | U/D | D | - | - | −11.02 (0.000) |
| IDH | Isocitrate dehydrogenase | D | D | N | −12.62 (0.000) | −12.39 (0.000) | - | |
| sucD | Succinyl-CoA synthetase alpha subunit | U | N | D | 10.71 (0.014) | - | −1.898 (0.004) | |
| frdA | fumarate reductase flavoprotein subunit | N | D | D | - | −9.681 (0.014) | −9.590 (0.000) | |
| PPP | zwf/G6PD | glucose-6-phosphate1-dehydrogenase | U/D | U/D | D | - | - | −10.87 (0.005) |
| GND | 6-phosphogluconate dehydrogenase | U/D | U/D | D | - | - | −10.06 (0.000) | |
| rpiA | ribose5-phosphate isomerase | U | N | D | 11.39 (0.000) | - | −10.18 (0.005) | |
| rbsK | ribokinase | U/D | U/D | U | - | - | 2.364 (0.000) | |
| RPE | ribulose-phosphate-3-epimerase | N | N | D | - | - | −9.230 (0.008) | |
| tktA | transketolase | N | N | D | - | - | −10.68 (0.004) |
| Pathway | Symbol | Definition | HCP/C | HLP/C | HLP/HCP | Log2FC HCP/C (p Value) | Log2FC HLP/C (p Value) | Log2FC HLP/HCP (p Value) |
|---|---|---|---|---|---|---|---|---|
| Fatty acid synthesis | ACC | Acetyl CoA carboxylase | U | U/D | U | 10.06 (0.000) | - | 5.033 (0.000) |
| FAS1 | Fatty acid synthase subunit beta, fungi type | U/D | U/D | U | - | - | 4.853 (0.000) | |
| FAS2 | Fatty acid synthase subunit alpha, fungi type | U | U | U | 11.16 (0.000) | 18.18 (0.000) | 2.990 (0.000) | |
| Fatty acid elongation | ACAC2 | acetyl-CoA acyltransferase 2 | U | U | N | 7.019 (0.000) | 8.874 (0.000) | - |
| HADH | 3-hydroxyacyl-CoA dehydrogenase | N | U | N | 7.872 (0.000) | 8.803 (0.000) | - | |
| MECR | mitochondrial enoyl-[acyl-carrier protein] reductase/trans-2-enoyl-CoA reductase | U | U | N | 14.20 (0.000) | 15.24 (0.000) | - | |
| PPT | palmitoyl-protein thioesterase | N | N | U | - | - | 2.782 (0.001) | |
| ACOT1_2_4 | acyl-coenzyme A thioesterase 1/2/4 | D | D | N | −13.35 (0.000) | −13.35 (0.000) | - | |
| KCS | 3-ketoacyl-CoA synthase | N | D | D | - | −12.24 (0.004) | −11.64 (0.000) | |
| PHS1 | very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase | D | D | N | −12.91 (0.000) | −12.91 (0.000) | - | |
| ACSL, fadD | long-chain acyl-CoA synthetase | N | D | D | - | −10.19 (0.000) | −9.288 (0.002) | |
| Fatty acid degradation | ACOX1 | acyl-CoA oxidase | U/D | U/D | U | - | - | 2.043 (0.038) |
| acd | acyl-CoA dehydrogenase | N | D | D | - | −12.11 (0.008) | −10.55 (0.024) | |
| echA | enoyl-CoA hydratase | N | D | D | - | −11.53 (0.008) | −10.51 (0.004) | |
| HADH | 3-hydroxyacyl-CoA dehydrogenase | U | U | N | 7.872 (0.000) | 8.803 (0.000) | - | |
| fadA | acetyl-CoA acyltransferase | N | N | D | - | - | −9.703 (0.043) | |
| atoB | acetyl-CoA C-acetyltransferase | N | N | D | - | - | −10.62 (0.000) | |
| Glycerophospholipid pathway | GPD1 | glycerol-3-phosphate dehydrogenase (NAD+) | N | N | D | - | - | −8.774 (0.008) |
| glpA | glycerol-3-phosphate dehydrogenase | N | N | D | - | - | −9.185 (0.004) | |
| pgsA | CDP-diacylglycerol---glycerol-3-phosphate 3-phosphatidyltransferase | D | D | N | −15.34 (0.000) | −15.34 (0.000) | - | |
| GEP4 | phosphatidylglycerophosphatase GEP4 | N | N | D | - | - | −2.357 (0.007) | |
| plc | phospholipase C | D | D | U | −12.39 (0.000) | −12.39 (0.000) | 2.414 (0.006) | |
| GDE1 | glycerophosphodiester phosphodiesterase | N | N | U | - | - | 2.357 (0.001) | |
| CKT1 | choline kinase | N | N | U | - | - | 3.400 (0.000) | |
| clsA_B | cardiolipin synthase A/B | N | N | D | - | - | −10.02 (0.001) | |
| PCYT1 | choline-phosphate cytidylyltransferase | D | D | N | −11.94 (0.000) | −11.94 (0.000) | - |
| Pathway | Symbol | Definition | HCP/C | HLP/C | HLP/HCP | Log2FC HCP/C (p Value) | Log2FC HLP/C (p Value) | Log2FC HLP/HCP (p Value) |
|---|---|---|---|---|---|---|---|---|
| Mevalonate | atoB | acetyl-CoA C-acetyltransferase | N | N | D | - | - | −10.62 (0.000) |
| HMGCR | hydroxymethylglutaryl-CoA reductase (NADPH) | U/D | U/D | U | - | - | 2.284 (0.008) | |
| MVK, mvaK1 | mevalonate kinase | D | D | N | −15.04 (0.000) | −15.04 (0.000) | - | |
| mvaK2 | phosphomevalonate kinase | U | U | U | 14.89 (0.000) | 14.92 (0.000) | 0.026 (0.000) | |
| MVD, mvaD | diphosphomevalonate decarboxylase | D | D | N | −12.63 (0.000) | −12.63 (0.000) | - | |
| Isoprenoid | IDI | isopentenyl-diphosphate delta-isomerase | U/D | D | D | - | −8.907 (0.025) | −9.527 (0.038) |
| FDPS | farnesyl diphosphate synthase | U/D | U/D | D | - | - | −0.87 | |
| hexPS, COQ1 | prenyl cysteine oxidase/farnesylcysteine lyase | U/D | U/D | U | - | - | 2.219 (0.018) | |
| Carotenoids | crtB | 15-cis-phytoene synthase | D | D | N | −12.40 (0.000) | −12.40 (0.000) | - |
| AL1 | phytoene desaturase (3,4-didehydrolycopene-forming) | D | D | N | −14.16 (0.000) | −14.16 (0.000) | - | |
| AL2 | 15-cis-phytoene synthase/lycopene beta-cyclase | U/D | U/D | U | - | - | 0.125 (0.047) | |
| crtZ | beta-carotene 3-hydroxylase | N | D | D | - | −10.80 (0.043) | −10.65 (0.001) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Elfeky, N.; Yang, Y.; Zhu, G.; Bao, Y. Prospective Mapping of Transcriptional Changes Associated with Lipid and Carotenoid Production in Rhodotorula glutinis Using Different Feeding Approaches. Biology 2026, 15, 60. https://doi.org/10.3390/biology15010060
Elfeky N, Yang Y, Zhu G, Bao Y. Prospective Mapping of Transcriptional Changes Associated with Lipid and Carotenoid Production in Rhodotorula glutinis Using Different Feeding Approaches. Biology. 2026; 15(1):60. https://doi.org/10.3390/biology15010060
Chicago/Turabian StyleElfeky, Nora, Yongheng Yang, Guoping Zhu, and Yongming Bao. 2026. "Prospective Mapping of Transcriptional Changes Associated with Lipid and Carotenoid Production in Rhodotorula glutinis Using Different Feeding Approaches" Biology 15, no. 1: 60. https://doi.org/10.3390/biology15010060
APA StyleElfeky, N., Yang, Y., Zhu, G., & Bao, Y. (2026). Prospective Mapping of Transcriptional Changes Associated with Lipid and Carotenoid Production in Rhodotorula glutinis Using Different Feeding Approaches. Biology, 15(1), 60. https://doi.org/10.3390/biology15010060






