Assembly and Analysis of the Complete Mitochondrial Genome of Eryngium foetidum L. (Apiaceae)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Extraction of Sample and Genome Sequencing
2.2. Assembly and Annotation of E. foetidum
2.3. Analysis of Codon Usage Bias and Selection Pressure
2.4. Repeat Sequence Identification and Prediction of RNA Editing Sites
2.5. Identification of Homologous Fragments and Collinearity Analysis
2.6. Construction of Maximum Likelihood Tree Based on the PCGs
3. Results
3.1. Mitochondrial Genome Sequencing and Assembly of E. foetidum
3.2. SSRs and Dispersed Repetitive Sequences
3.3. Characterization of Chloroplast Genome Transfer to Mitochondria in E. foetidum
3.4. Analysis of Relative Synonymous Codon Usage
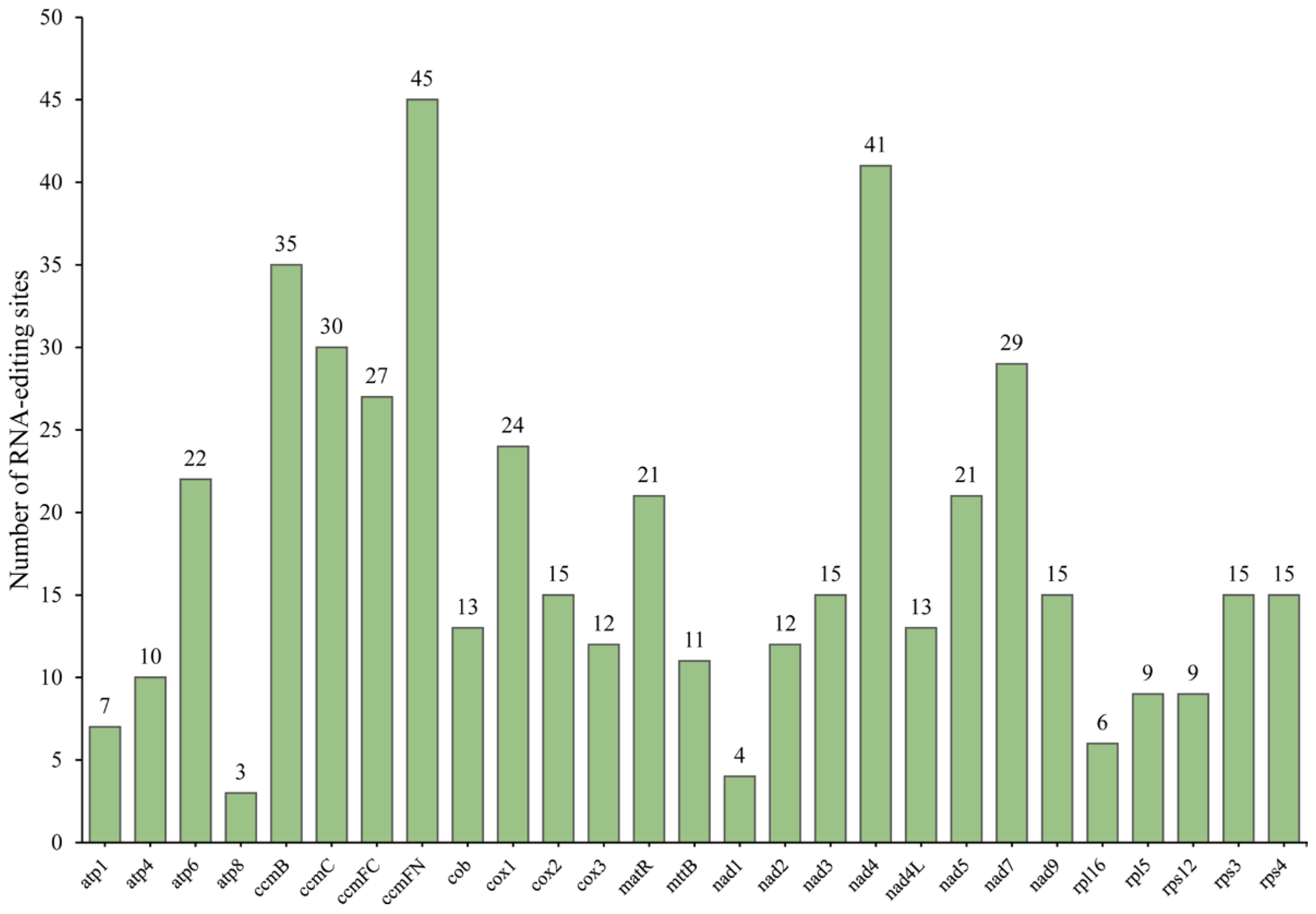
3.5. Predicted RNA Editing Sites
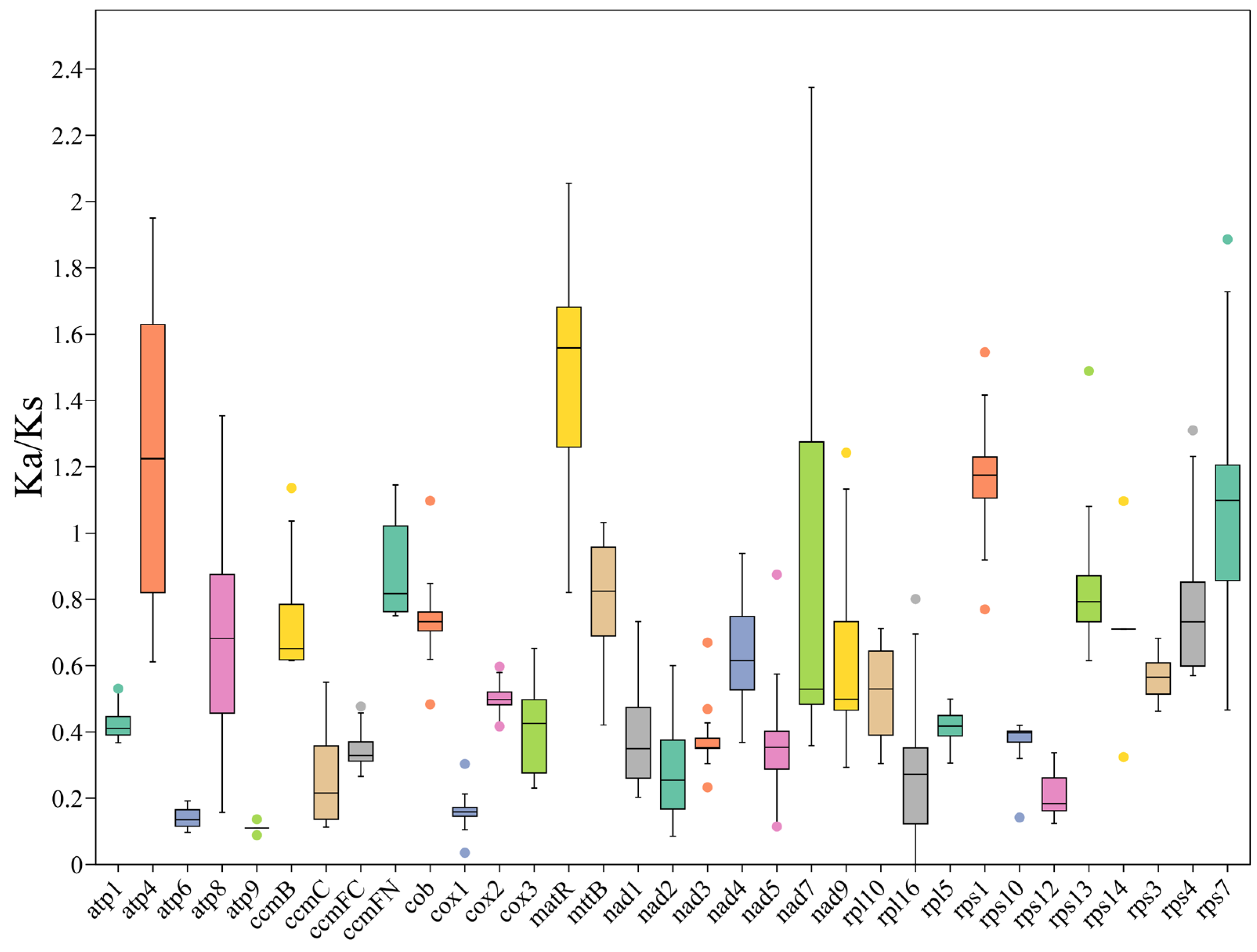
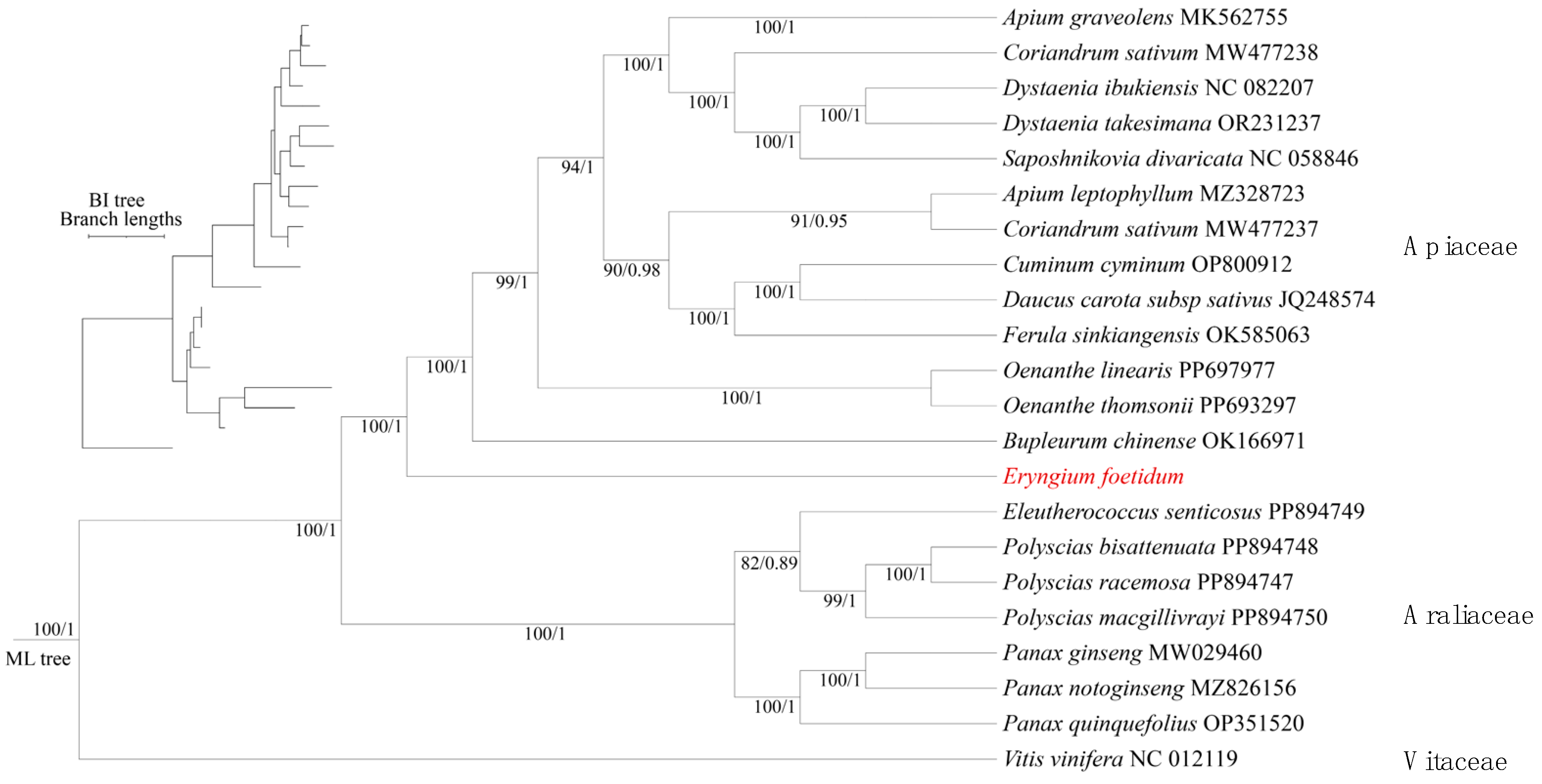
3.6. PCG Substitution Rates and Phylogenetic Tree Based on the PCGs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PCGs | Protein-coding gene |
| ATP | Adenosine triphosphate |
| CMS | Cytoplasmic male sterility |
| ORFs | Open reading frames |
| RSCU | Relative synonymous codon usage |
| ENC | Effective number of codons |
| SSRs | Simple sequence repeats |
| IGS | Intergenic spaces |
References
- Green, D.R.; Reed, J.C. Mitochondria and Apoptosis. Science 1998, 281, 1309–1312. [Google Scholar] [CrossRef]
- Xing, B.; Lin, L.; Wu, Q. Application of Mitochondrial Genomes to Species Identification and Evolution. Electron. J. Biotechnol. 2025, 76, 39–48. [Google Scholar] [CrossRef]
- Mower, J.P.; Sloan, D.B.; Alverson, A.J. Plant Mitochondrial Genome Diversity: The Genomics Revolution. In Plant Genome Diversity Volume 1: Plant Genomes, Their Residents, and Their Evolutionary Dynamics; Wendel, J.F., Greilhuber, J., Dolezel, J., Leitch, I.J., Eds.; Springer: Vienna, Austria, 2012; pp. 123–144. ISBN 978-3-7091-1130-7. [Google Scholar]
- Kubo, T.; Newton, K.J. Angiosperm Mitochondrial Genomes and Mutations. Mitochondrion 2008, 8, 5–14. [Google Scholar] [CrossRef]
- Gualberto, J.M.; Mileshina, D.; Wallet, C.; Niazi, A.K.; Weber-Lotfi, F.; Dietrich, A. The Plant Mitochondrial Genome: Dynamics and Maintenance. Biochimie 2014, 100, 107–120. [Google Scholar] [CrossRef]
- Day, D.A. Mitochondrial Structure and Function in Plants. In Plant Mitochondria: From Genome to Function; Springer: Dordrecht, The Netherlands, 2004; ISBN 978-1-4020-2400-9. [Google Scholar]
- Kazama, T.; Toriyama, K. Whole Mitochondrial Genome Sequencing and Re-Examination of a Cytoplasmic Male Sterility-Associated Gene in Boro-Taichung-Type Cytoplasmic Male Sterile Rice. PLoS ONE 2016, 11, e0159379. [Google Scholar] [CrossRef]
- Omelchenko, D.O.; Makarenko, M.S.; Kasianov, A.S.; Schelkunov, M.I.; Logacheva, M.D.; Penin, A.A. Assembly and Analysis of the Complete Mitochondrial Genome of Capsella Bursa-Pastoris. Plants 2020, 9, 469. [Google Scholar] [CrossRef]
- Gautam, R.; Shukla, P.; Kirti, P.B. Male Sterility in Plants: An Overview of Advancements from Natural CMS to Genetically Manipulated Systems for Hybrid Seed Production. Theor. Appl. Genet. 2023, 136, 195. [Google Scholar] [CrossRef]
- Kim, Y.-J.; Zhang, D. Molecular Control of Male Fertility for Crop Hybrid Breeding. Trends Plant Sci. 2018, 23, 53–65. [Google Scholar] [CrossRef]
- Wang, J.; Kan, S.; Liao, X.; Zhou, J.; Tembrock, L.R.; Daniell, H.; Jin, S.; Wu, Z. Plant Organellar Genomes: Much Done, Much More to Do. Trends Plant Sci. 2024, 29, 754–769. [Google Scholar] [CrossRef] [PubMed]
- Melo, S.M.F.D.; Marques, A.; Almeida, C. The Mitochondrial Genome Sequence of Syagrus coronata (Mart.) Becc. (Arecaceae) Is Characterized by Gene Insertion within Intergenic Spaces. Tree Genet. Genomes 2024, 20, 10. [Google Scholar] [CrossRef]
- Ekblom, R.; Wolf, J.B.W. A Field Guide to Whole-genome Sequencing, Assembly and Annotation. Evol. Appl. 2014, 7, 1026–1042. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De Novo Assembly of Organelle Genomes from Whole Genome Data. Nucleic Acids Res. 2016, 45, e18. [Google Scholar] [CrossRef]
- Li, X.; Lin, C.-Y.; Yang, J.-B.; Yu, W.-B. De Novo Assembling a Complete Mitochondrial Genome of Pedicularis Rex (Orobanchaceae) Using GetOrganelle Toolkit. Mitochondrial DNA Part B 2020, 5, 1056–1057. [Google Scholar] [CrossRef]
- Zou, Y.; Zhu, W.; Sloan, D.B.; Wu, Z. Long-read Sequencing Characterizes Mitochondrial and Plastid Genome Variants in Arabidopsis Msh1 Mutants. Plant J. 2022, 112, 738–755. [Google Scholar] [CrossRef]
- Hu, T.; Chitnis, N.; Monos, D.; Dinh, A. Next-Generation Sequencing Technologies: An Overview. Hum. Immunol. 2021, 82, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Kucuk, E.; Van Der Sanden, B.P.G.H.; O’Gorman, L.; Kwint, M.; Derks, R.; Wenger, A.M.; Lambert, C.; Chakraborty, S.; Baybayan, P.; Rowell, W.J.; et al. Comprehensive de Novo Mutation Discovery with HiFi Long-Read Sequencing. Genome Med. 2023, 15, 34. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Y.; Bollas, A.; Wang, Y.; Au, K.F. Nanopore Sequencing Technology, Bioinformatics and Applications. Nat. Biotechnol. 2021, 39, 1348–1365. [Google Scholar] [CrossRef] [PubMed]
- Treffer, R.; Deckert, V. Recent Advances in Single-Molecule Sequencing. Curr. Opin. Biotechnol. 2010, 21, 4–11. [Google Scholar] [CrossRef]
- Paul, J.H.A.; Seaforth, C.E.; Tikasingh, T. Eryngium foetidum L.: A Review. Fitoterapia 2011, 82, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.; Essien, E.; Ntuk, S.; Choudhary, M. Eryngium foetidum L. Essential Oils: Chemical Composition and Antioxidant Capacity. Medicines 2017, 4, 24. [Google Scholar] [CrossRef]
- Ignacimuthu, S.; Arockiasamy, S.; Antonysamy, M.; Ravichandran, P. Plant Regeneration through Somatic Embryogenesis from Mature Leaf Explants of Eryngium foetidum, a Condiment. Plant Cell Tissue Organ Cult. 1999, 56, 131–137. [Google Scholar] [CrossRef]
- Rodrigues, T.L.M.; Silva, M.E.P.; Gurgel, E.S.C.; Oliveira, M.S.; Lucas, F.C.A. Eryngium foetidum L. (Apiaceae): A Literature Review of Traditional Uses, Chemical Composition, and Pharmacological Activities. Evid.-Based Complement. Altern. Med. 2022, 2022, 2896895. [Google Scholar] [CrossRef]
- Singh, B.K.; Ramakrishna, Y.; Ngachan, S.V. Spiny Coriander (Eryngium foetidum L.): A Commonly Used, Neglected Spicing-Culinary Herb of Mizoram, India. Genet. Resour. Crop Evol. 2014, 61, 1085–1090. [Google Scholar] [CrossRef]
- Pedrosa, L.M.; Rosário, I.C.; de Castro, G.; Martins, C.C. Production of High-Quality Seeds in Eryngium foetidum: Optimizing Post-Harvest Resting Conditions for Sustainable Unconventional Food Systems. Agronomy 2025, 15, 185. [Google Scholar] [CrossRef]
- Acharya, G.C.; Mohanty, S.; Dasgupta, M.; Sahu, S.; Singh, S.; Koundinya, A.V.V.; Kumari, M.; Naresh, P.; Sahoo, M.R. Molecular Phylogeny, DNA Barcoding, and ITS2 Secondary Structure Predictions in the Medicinally Important Eryngium Genotypes of East Coast Region of India. Genes 2022, 13, 1678. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Guo, W.; Wei, C.; Wang, X.; Wang, Y.; Wang, J. Characterization and Phylogenetic Analysis of the Complete Mitochondrial Genome of Triplophysa microphthalma. Biology 2024, 13, 608. [Google Scholar] [CrossRef]
- Jinlu, L.; Shuo, W.; Jing, Y.; Ling, W.; Shiliang, Z. A Modified CTAB Protocol for Plant DNA Extraction. Chin. Bull. Bot. 2013, 48, 72–78. [Google Scholar] [CrossRef]
- Cai, Z.; Hu, J.; Yin, T.; Wang, D.; Shen, Q.; Ma, C.; Ou, D.; Xu, M.; Shi, X.; Li, Q.; et al. Long Amplicon HiFi Sequencing for Mitochondrial DNA Genomes. Mol. Ecol. Resour. 2023, 23, 1014–1022. [Google Scholar] [CrossRef]
- Bi, C.; Shen, F.; Han, F.; Qu, Y.; Hou, J.; Xu, K.; Xu, L.; He, W.; Wu, Z.; Yin, T. PMAT: An Efficient Plant Mitogenome Assembly Toolkit Using Low-Coverage HiFi Sequencing Data. Hortic. Res. 2024, 11, uhae023. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive Visualization of de Novo Genome Assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef]
- Chen, Y.; Ye, W.; Zhang, Y.; Xu, Y. High Speed BLASTN: An Accelerated MegaBLAST Search Tool. Nucleic Acids Res. 2015, 43, 7762–7768. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—Versatile and Accurate Annotation of Organelle Genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef]
- Li, J.; Ni, Y.; Lu, Q.; Chen, H.; Liu, C. PMGA: A Plant Mitochondrial Genome Annotator. Plant Commun. 2025, 6, 101191. [Google Scholar] [CrossRef]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved Detection and Functional Classification of Transfer RNA Genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Dunn, N.A.; Unni, D.R.; Diesh, C.; Munoz-Torres, M.; Harris, N.L.; Yao, E.; Rasche, H.; Holmes, I.H.; Elsik, C.G.; Lewis, S.E. Apollo: Democratizing Genome Annotation. PLoS Comput. Biol. 2019, 15, e1006790. [Google Scholar] [CrossRef]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) Version 1.3.1: Expanded Toolkit for the Graphical Visualization of Organellar Genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An Integrated and Scalable Desktop Platform for Streamlined Molecular Sequence Data Management and Evolutionary Phylogenetics Studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Ling, L.; Zhang, S.; Yang, T. Analysis of Codon Usage Bias in Chloroplast Genomes of Dryas octopetala Var. asiatica (Rosaceae). Genes 2024, 15, 899. [Google Scholar] [CrossRef]
- Parvathy, S.T.; Udayasuriyan, V.; Bhadana, V. Codon Usage Bias. Mol. Biol. Rep. 2022, 49, 539–565. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Luo, Y.; Lan, Z.; Qiu, D. Insights into Structure, Codon Usage, Repeats, and RNA Editing of the Complete Mitochondrial Genome of Perilla frutescens (Lamiaceae). Sci. Rep. 2024, 14, 13940. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-Web: A Web Server for Microsatellite Prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Benson, G. Tandem Repeats Finder: A Program to Analyze DNA Sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An Information Aesthetic for Comparative Genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Edera, A.A.; Small, I.; Milone, D.H.; Sanchez-Puerta, M.V. Deepred-Mt: Deep Representation Learning for Predicting C-to-U RNA Editing in Plant Mitochondria. Comput. Biol. Med. 2021, 136, 104682. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Chen, H.; Jiang, M.; Wang, L.; Wu, X.; Huang, L.; Liu, C. CPGAVAS2, an Integrated Plastome Sequence Annotator and Analyzer. Nucleic Acids Res. 2019, 47, W65–W73. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2017, 20, 1160–1166. [Google Scholar] [CrossRef]
- Yang, S.; De Angelis, D. Maximum Likelihood. In Computational Toxicology: Volume II; Reisfeld, B., Mayeno, A.N., Eds.; Humana Press: Totowa, NJ, USA, 2013; pp. 581–595. ISBN 978-1-62703-059-5. [Google Scholar]
- Zhu, J.; Wen, D.; Yu, Y.; Meudt, H.M.; Nakhleh, L. Bayesian Inference of Phylogenetic Networks from Bi-Allelic Genetic Markers. PLoS Comput. Biol. 2018, 14, e1005932. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian Phylogenetic Inference under Mixed Models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Kang, R.; Wang, Z.; Jiang, Y.; Zhou, H.; Abuduaini, A.; Suo, F.; Huang, L. The Complete Mitochondrial Genome of Cuminum cyminum (Apiales: Apiaceae) and Phylogenetic Analysis. Mitochondrial DNA B Resour. 2023, 8, 760–765. [Google Scholar] [CrossRef]
- Du, X.; Wang, K.; Tang, Y.; Wu, J.; Yang, X.; Zhang, H.; Liu, N.; Zhang, Z. Characterization and Phylogenetic Analysis of the Complete Mitochondrial Genome Sequence of Lagenaria siceraria, a Cucurbit Crop. Front. Plant Sci. 2025, 16, 1599596. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Y.; Liu, Y.Y.; Zeng, X.; Wu, P.; Li, Q.M.; Guo, S.X.; Hao, Z.G. Complete Mitochondrial Genome of Angelica dahurica and Its Implications on Evolutionary Analysis of Complex Mitochondrial Genome Architecture in Apiaceae. Front. Plant Sci. 2024, 15, 1367299. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Hwang, Y.; Kim, H.; Choi, K. Insights into the Nuclear-Organelle DNA Integration in Cicuta virosa (Apiaceae) Provided by Complete Plastid and Mitochondrial Genomes. BMC Genom. 2025, 26, 102. [Google Scholar] [CrossRef]
- Wynn, E.L.; Christensen, A.C. Repeats of Unusual Size in Plant Mitochondrial Genomes: Identification, Incidence and Evolution. G3 Genes|Genomes|Genet. 2019, 9, 549–559. [Google Scholar] [CrossRef]
- Ma, J.; Wang, S.; Zhu, X.; Sun, G.; Chang, G.; Li, L.; Hu, X.; Zhang, S.; Zhou, Y.; Song, C.-P.; et al. Major Episodes of Horizontal Gene Transfer Drove the Evolution of Land Plants. Mol. Plant 2022, 15, 857–871. [Google Scholar] [CrossRef]
- Anderson, S.; Bankier, A.; Barrell, B.; de Bruijn, M.; Coulson, A.; Drouin, J.; Eperon, I.; Nierlich, D.; Roe, B.; Sanger, F.; et al. Sequence and Organization of the Human Mitochondrial Genome. Nature 1981, 290, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Liu, C.; Zhao, A.; Tian, R.; Xie, D.; Xiao, Y.; Chen, H.; Zhou, S.; He, X.-J. Phylogeny and Diversification of Genus Sanicula L. (Apiaceae): Novel Insights from Plastid Phylogenomic Analyses. BMC Plant Biol. 2024, 24, 70. [Google Scholar] [CrossRef]
- Wang, L.; Liu, X.; Xu, Y.; Zhang, Z.; Wei, Y.; Hu, Y.; Zheng, C.; Qu, X. Assembly and Comparative Analysis of the First Complete Mitochondrial Genome of a Traditional Chinese Medicine Angelica biserrata (Shan et Yuan) Yuan et Shan. Int. J. Biol. Macromol. 2024, 257, 128571. [Google Scholar] [CrossRef]
- Li, X.; Li, M.; Li, W.; Zhou, J.; Han, Q.; Lu, W.; Luo, Q.; Zhu, S.; Xiong, A.; Tan, G.; et al. Comparative Analysis of the Complete Mitochondrial Genomes of Apium graveolens and Apium leptophyllum Provide Insights into Evolution and Phylogeny Relationships. Int. J. Mol. Sci. 2023, 24, 14615. [Google Scholar] [CrossRef]
- Fay, J.C.; Wu, C.I. Sequence Divergence, Functional Constraint, and Selection in Protein Evolution. Annu. Rev. Genom. Hum. Genet. 2003, 4, 213–235. [Google Scholar] [CrossRef]
- Pazos, F.; Valencia, A. Protein Co-Evolution, Co-Adaptation and Interactions. EMBO J. 2008, 27, 2648–2655. [Google Scholar] [CrossRef]
- Xie, D.F.; Huan-Xi, Y.U.; Price, M.; Xie, C.; He, X.J. Phylogeny of Chinese Allium Species in Section Daghestanica and Adaptive Evolution of Allium (Amaryllidaceae, Allioideae) Species Revealed by the Chloroplast Complete Genome. Front. Plant Sci. 2019, 10, 460. [Google Scholar] [CrossRef]
- Li, J.; Xu, Y.; Shan, Y.; Pei, X.; Yong, S.; Liu, C.; Yu, J. Assembly of the Complete Mitochondrial Genome of an Endemic Plant, Scutellaria tsinyunensis, Revealed the Existence of Two Conformations Generated by a Repeat-Mediated Recombination. Planta 2021, 254, 36. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Zhang, X.; Li, Z.; Song, Y.; Sun, Z. Assembly and Comparative Analysis of the Complete Mitochondrial Genome of Bupleurum chinense DC. BMC Genom. 2022, 23, 664. [Google Scholar] [CrossRef]
- Sun, Z.; Wu, Y.; Fan, P.; Guo, D.; Zhang, S.; Song, C. Assembly and Analysis of the Mitochondrial Genome of Prunella vulgaris. Front. Plant Sci. 2023, 14, 1237822. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Li, C.; Zhang, X.; Wang, C.; Roger, A.J.; Gao, F. Characterization and Comparative Analyses of Mitochondrial Genomes in Single-Celled Eukaryotes to Shed Light on the Diversity and Evolution of Linear Molecular Architecture. Int. J. Mol. Sci. 2021, 22, 2546. [Google Scholar] [CrossRef]
- Liu, S.-L.; Adams, K. Molecular Adaptation and Expression Evolution Following Duplication of Genes for Organellar Ribosomal Protein S13 in Rosids. BMC Evol. Biol. 2008, 8, 25. [Google Scholar] [CrossRef]
- Bijlsma, R.; Loeschcke, V. Genetic Erosion Impedes Adaptive Responses to Stressful Environments. Evol. Appl. 2012, 5, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Bergthorsson, U.; Adams, K.L.; Thomason, B.; Palmer, J.D. Widespread Horizontal Transfer of Mitochondrial Genes in Flowering Plants. Nature 2003, 424, 197–201. [Google Scholar] [CrossRef]
- Jiang, M.; Ni, Y.; Li, J.; Liu, C. Characterisation of the Complete Mitochondrial Genome of Taraxacum mongolicum Revealed Five Repeat-Mediated Recombinations. Plant Cell Rep. 2023, 42, 775–789. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Shan, Y.; Li, J.; Zhang, X.; Yu, J.; Wang, H. Assembly and Comparative Analysis of the Complete Mitochondrial Genome of Viburnum chinshanense. BMC Plant Biol. 2023, 23, 487. [Google Scholar] [CrossRef]
- Gallagher, L.; Betz, S.; Chase, C. Mitochondrial RNA Editing Truncates a Chimeric Open Reading Frame Associated with S Male-Sterility in Maize. Curr. Genet. 2002, 42, 179–184. [Google Scholar] [CrossRef]
- Kadowaki, K.; Ozawa, K.; Kazama, S.; Kubo, N.; Akihama, T. Creation of an Initiation Codon by RNA Editing in the Coxl Transcript from Tomato Mitochondria. Curr. Genet. 1995, 28, 415–422. [Google Scholar] [CrossRef]
- Quinones, V.; Zanlungo, S.; Holuigue, L.; Litvak, S.; Jordana, X. The Cox1 Initiation Codon Is Created by RNA Editing in Potato Mitochondria. Plant Physiol. 1995, 108, 1327–1328. [Google Scholar] [CrossRef]
- Liu, Y.J.; Xiu, Z.H.; Tan, M.B.C. Empty Pericarp5 Encodes a Pentatricopeptide Repeat Protein That Is Required for Mitochondrial RNA Editing and Seed Development in Maize. Plant Cell 2013, 25, 868–883. [Google Scholar] [CrossRef]
- Montaa-Lozano, P.; Balaguera-Reina, S.A.; Prada-Quiroga, C.F. Comparative Analysis of Codon Usage of Mitochondrial Genomes Provides Evolutionary Insights into Reptiles. Gene 2023, 851, 146999. [Google Scholar] [CrossRef]
- Wang, D.; Wu, Y.-W.; Shih, A.C.-C.; Wu, C.-S.; Wang, Y.-N.; Chaw, S.-M. Transfer of Chloroplast Genomic DNA to Mitochondrial Genome Occurred at Least 300 MYA. Mol. Biol. Evol. 2007, 24, 2040–2048. [Google Scholar] [CrossRef]
- Sloan, D.B.; Wu, Z. History of Plastid DNA Insertions Reveals Weak Deletion and AT Mutation Biases in Angiosperm Mitochondrial Genomes. Genome Biol. Evol. 2014, 12, 3210–3221. [Google Scholar] [CrossRef]
- Drouin, G.; Daoud, H.; Xia, J. Relative Rates of Synonymous Substitutions in the Mitochondrial, Chloroplast and Nuclear Genomes of Seed Plants. Mol. Phylogenet. Evol. 2008, 49, 827–831. [Google Scholar] [CrossRef] [PubMed]
- Zardoya, R. Recent Advances in Understanding Mitochondrial Genome Diversity. F1000Research 2020, 9, 270. [Google Scholar] [CrossRef] [PubMed]
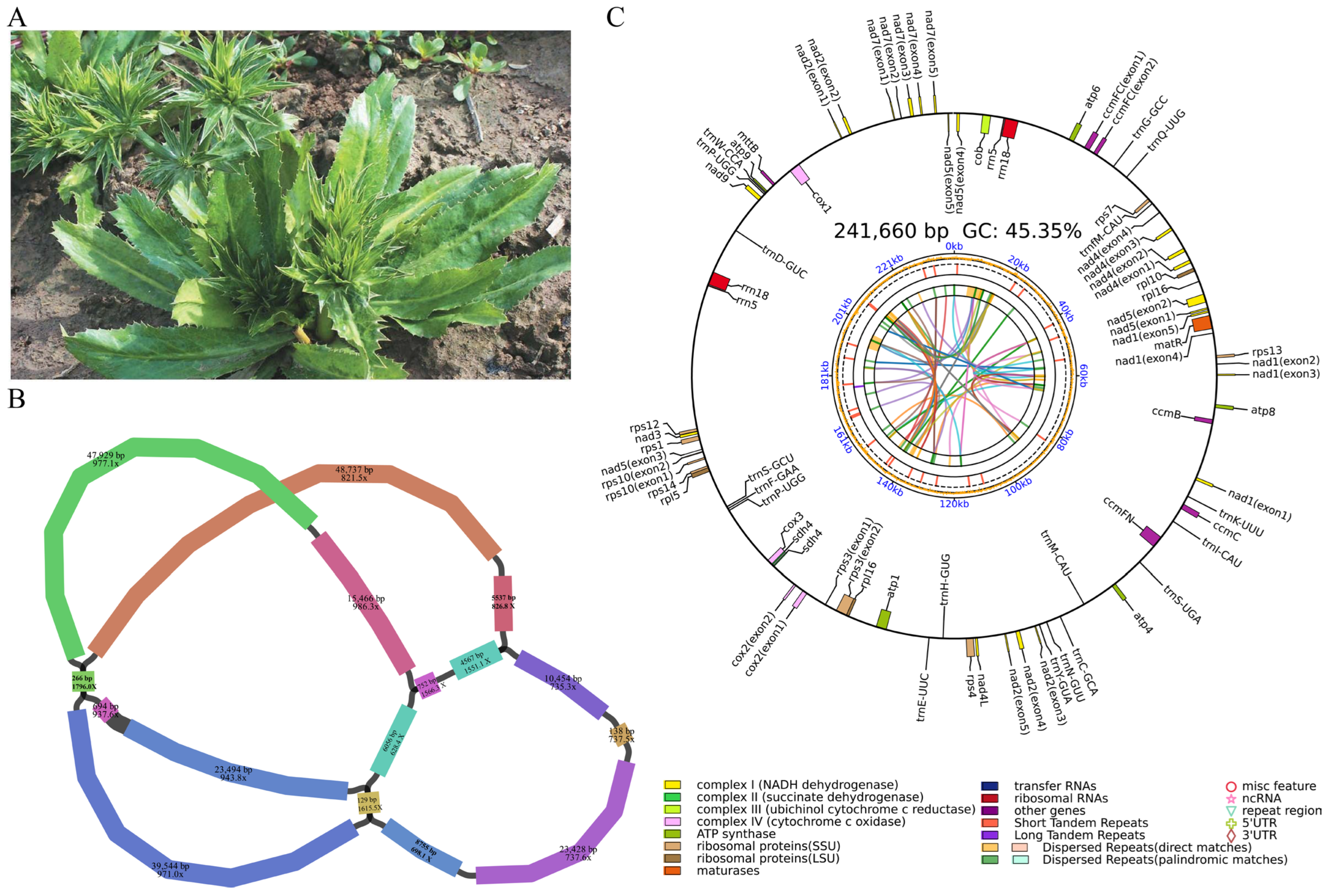
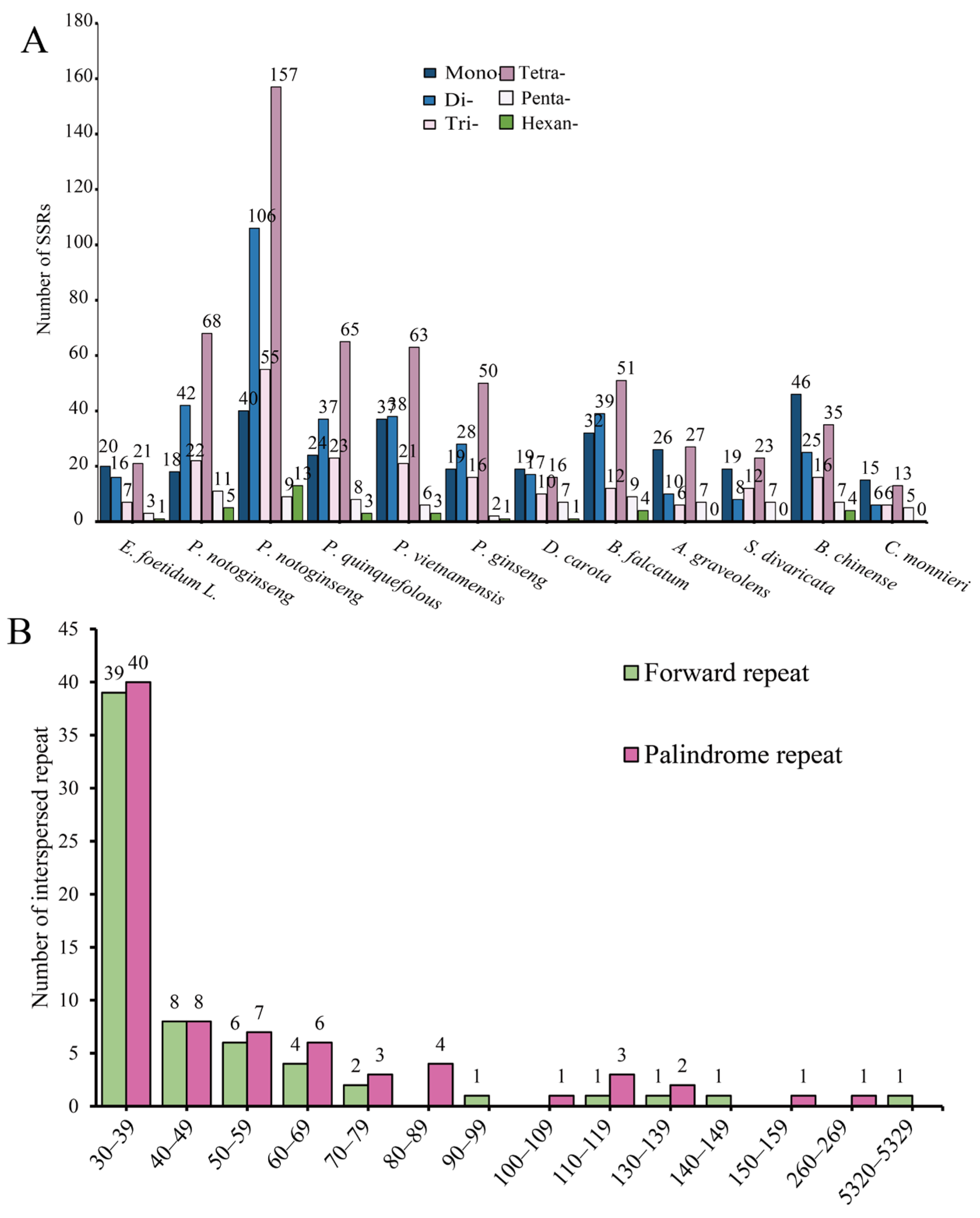
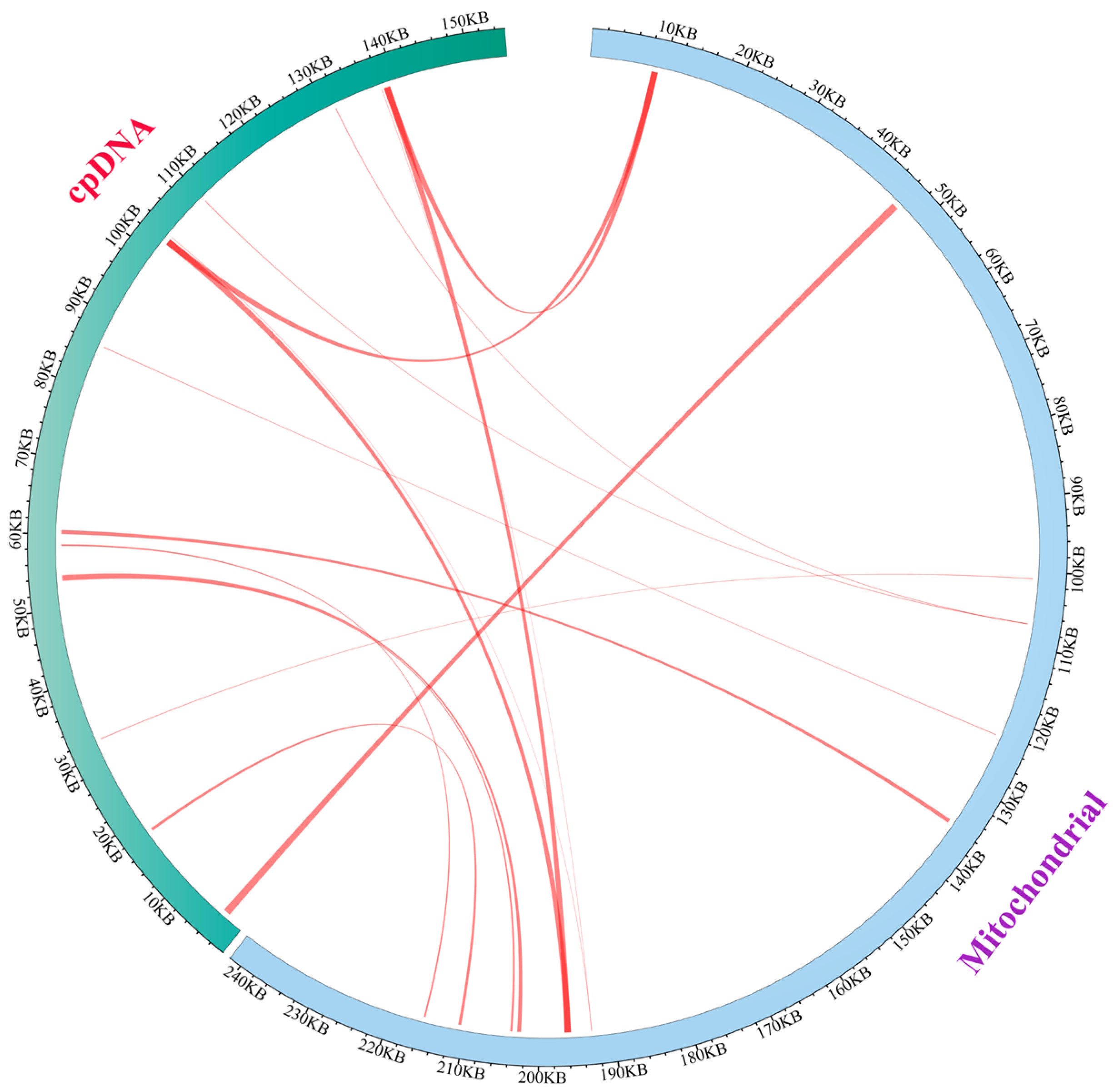
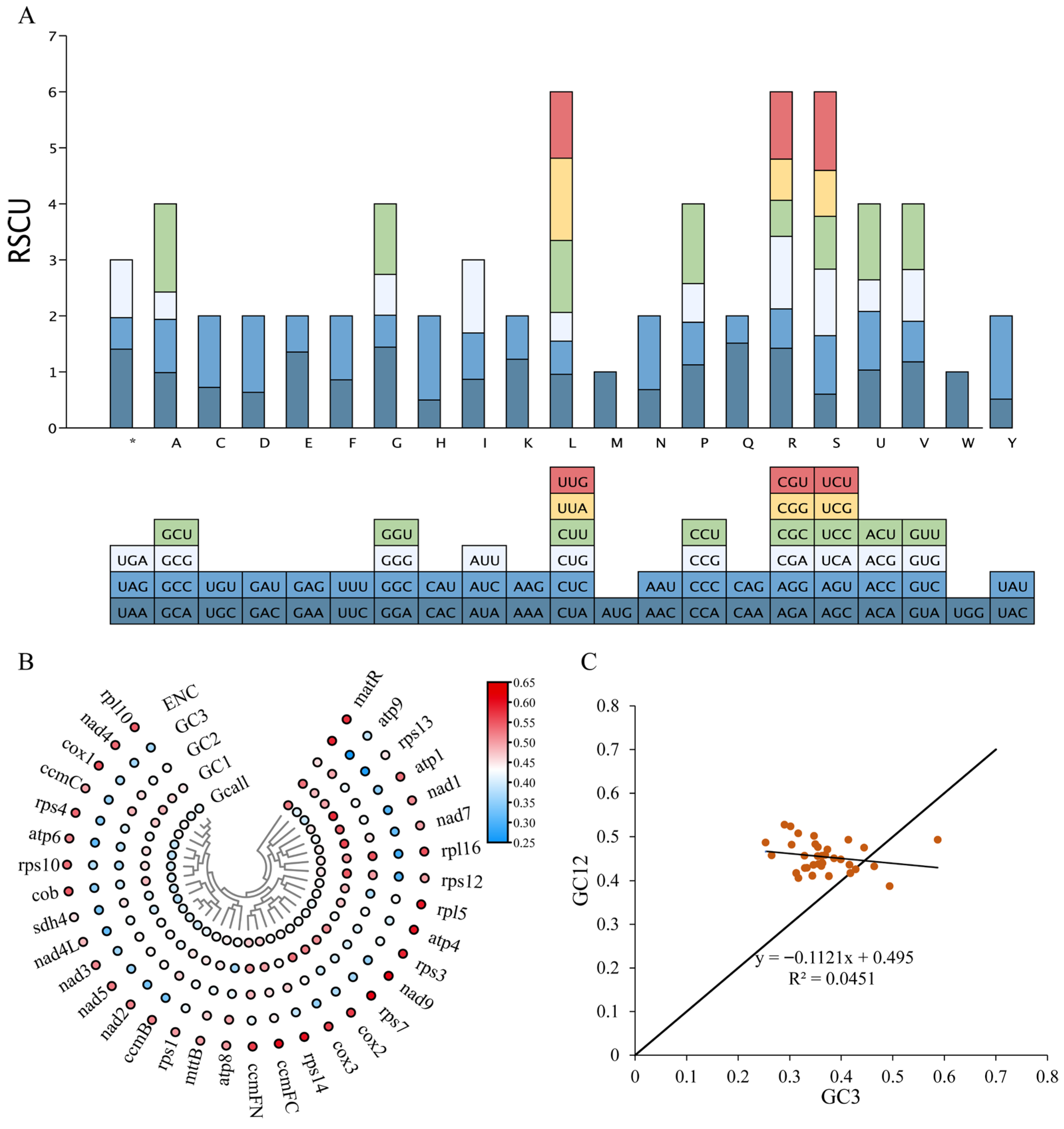
| Group of Genes | Gene Name |
|---|---|
| ATP synthase | atp1 atp4 atp6 atp8 atp9 |
| Cytochrome c biogenesis | ccmB ccmC ccmFC * ccmFN |
| Ubiquinol cytochrome c reductase | cob |
| Cytochrome c oxidase | cox1 cox2 * cox3 |
| Maturases | matR |
| Transport membrane protein | mttB |
| NADH dehydrogenase | nad1 **** nad2 **** nad3 nad4 **** nad4L nad5 **** nad7 **** nad9 |
| Ribosomal proteins (LSU) | rpl10 rpl16(2) rpl5 |
| Ribosomal proteins (SSU) | rps1 rps10 * rps12 rps13 rps14 rps3 * rps4 rps7 |
| Succinate dehydrogenase | sdh4(2) |
| Ribosomal RNAs | rrn18(2) rrn5(2) |
| Transfer RNAs | trnC-GCA trnD-GUC trnE-UUC trnF-GAA trnG-GCC trnH-GUG trnI-CAU trnK-UUU trnM-CAU trnN-GUU trnP-UGG(2) trnQ-UUG trnS-GCU trnS-UGA trnW-CCA trnY-GUA trnfM-CAU |
| No. | Length | Identity % | Mismatches | Gap Opens | cp Start | cp End | Gene (cp) | mt Start | mt End | Gene (mt) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 992 | 87.5 | 110 | 7 | 2465 | 3447 | rpl16 (26.65%)- rpl14 (55.23%) (IGS) | 45,655 | 46,641 | trnM-CAU (100%)- rpl2 (17.62%) (IGS) |
| 2 | 526 | 97.34 | 13 | 1 | 60,034 | 60,558 | rpoB (16.34%) | 135,392 | 135,917 | nad7 (8.48%) |
| 3 | 502 | 95.62 | 17 | 1 | 54,022 | 54,523 | trnD-GUC- psbM (IGS) | 202,447 | 202,943 | atp9-rps12 (IGS) |
| 4 | 276 | 97.10 | 5 | 3 | 53,748 | 54,023 | trnD-GUC (100%) | 203,615 | 203,887 | atp9-rps12 (IGS) |
| 5 | 215 | 93.95 | 10 | 3 | 58,651 | 58,438 | rpoB (6.66%) | 215,418 | 215,630 | cox3-trnS (GCU) |
| 6 | 887 | 74.07 | 179 | 37 | 101,982 | 102,845 | rrn16 (57.95) | 195,740 | 196,598 | atp4-nad6 (IGS) |
| 7 | 887 | 74.07 | 179 | 37 | 101,982 | 102,845 | rrn16 (57.95) | 8359 | 9217 | atp6-trnQ- UUG (IGS) |
| 8 | 887 | 74.07 | 179 | 37 | 139,163 | 138,300 | rrn16 (57.95) | 8359 | 9217 | atp6-trnQ- UUG (IGS) |
| 9 | 887 | 74.07 | 179 | 37 | 139163 | 138,300 | rrn16 (57.95) | 195,740 | 196,598 | atp4-nad6 (IGS) |
| 10 | 386 | 80.57 | 43 | 13 | 18,122 | 17,758 | trnP-UGG and trnW-CCA | 210,522 | 210,896 | rpl5 (65.45%) |
| 11 | 76 | 98.68 | 1 | 0 | 85,798 | 85,873 | trnH-GUG (100%) | 122,443 | 122,518 | Cox (9.52%) |
| 12 | 83 | 96.39 | 2 | 1 | 109,761 | 109,842 | trnN-GUU (100%) | 106,978 | 107,060 | nad2-rps12 (IGS) |
| 13 | 83 | 96.39 | 2 | 1 | 131,384 | 131,303 | trnN-GUU (100%) | 106,978 | 107,060 | nad2-rps12 (IGS) |
| 14 | 79 | 93.67 | 5 | 0 | 31,919 | 31,841 | trnM-CAU (100%) | 100,899 | 100,977 | nad2 (5.18%) |
| 15 | 38 | 97.37 | 1 | 0 | 103,140 | 103,177 | rrn16 (2.55%) | 192,948 | 192,985 | cob (3.17%) |
| 16 | 38 | 97. 367 | 1 | 0 | 138,005 | 137,968 | rrn16 (2.55%) | 192,948 | 192,985 | cob (3.17%) |
| Symbol | Codon | Count | RSCU | Symbol | Codon | Count | RSCU |
|---|---|---|---|---|---|---|---|
| Ter | UAA | 15 | 1.4062 | Met | AUG | 267 | 1 |
| Ter | UAG | 6 | 0.5625 | Asn | AAC | 110 | 0.6832 |
| Ter | UGA | 11 | 1.0312 | Asn | AAU | 212 | 1.3168 |
| Ala | GCA | 158 | 0.9875 | Pro | CCA | 163 | 1.1261 |
| Ala | GCC | 152 | 0.95 | Pro | CCC | 110 | 0.7599 |
| Ala | GCG | 78 | 0.4875 | Pro | CCG | 100 | 0.6908 |
| Ala | GCU | 252 | 1.575 | Pro | CCU | 206 | 1.4231 |
| Cys | UGC | 51 | 0.7234 | Gln | CAA | 218 | 1.5139 |
| Cys | UGU | 90 | 1.2766 | Gln | CAG | 70 | 0.4861 |
| Asp | GAC | 101 | 0.6352 | Arg | AGA | 168 | 1.4217 |
| Asp | GAU | 217 | 1.3648 | Arg | AGG | 83 | 0.7024 |
| Glu | GAA | 287 | 1.3538 | Arg | CGA | 153 | 1.2948 |
| Glu | GAG | 137 | 0.6462 | Arg | CGC | 76 | 0.6432 |
| Phe | UUC | 272 | 0.8594 | Arg | CGG | 87 | 0.7362 |
| Phe | UUU | 361 | 1.1406 | Arg | CGU | 142 | 1.2017 |
| Gly | GGA | 253 | 1.4416 | Ser | AGC | 94 | 0.6026 |
| Gly | GGC | 100 | 0.5698 | Ser | AGU | 163 | 1.0449 |
| Gly | GGG | 128 | 0.7293 | Ser | UCA | 185 | 1.1859 |
| Gly | GGU | 221 | 1.2593 | Ser | UCC | 147 | 0.9423 |
| His | CAC | 61 | 0.498 | Ser | UCG | 128 | 0.8205 |
| His | CAU | 184 | 1.502 | Ser | UCU | 219 | 1.4038 |
| Ile | AUA | 222 | 0.8672 | Thr | ACA | 130 | 1.0317 |
| Ile | AUC | 212 | 0.8281 | Thr | ACC | 132 | 1.0476 |
| Ile | AUU | 334 | 1.3047 | Thr | ACG | 71 | 0.5635 |
| Lys | AAA | 274 | 1.226 | Thr | ACU | 171 | 1.3571 |
| Lys | AAG | 173 | 0.774 | Val | GUA | 181 | 1.1792 |
| Leu | CUA | 163 | 0.9569 | Val | GUC | 111 | 0.7231 |
| Leu | CUC | 101 | 0.593 | Val | GUG | 142 | 0.9251 |
| Leu | CUG | 87 | 0.5108 | Val | GUU | 180 | 1.1726 |
| Leu | CUU | 219 | 1.2857 | Trp | UGG | 144 | 1 |
| Leu | UUA | 250 | 1.4677 | Tyr | UAC | 83 | 0.5139 |
| Leu | UUG | 202 | 1.1859 | Tyr | UAU | 240 | 1.4861 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Zhang, W.; Luo, Y.; Liu, J.; Li, Q.; Liu, Q. Assembly and Analysis of the Complete Mitochondrial Genome of Eryngium foetidum L. (Apiaceae). Biology 2025, 14, 1296. https://doi.org/10.3390/biology14091296
Zhang L, Zhang W, Luo Y, Liu J, Li Q, Liu Q. Assembly and Analysis of the Complete Mitochondrial Genome of Eryngium foetidum L. (Apiaceae). Biology. 2025; 14(9):1296. https://doi.org/10.3390/biology14091296
Chicago/Turabian StyleZhang, Lihong, Wenhu Zhang, Yongjian Luo, Jun Liu, Qing Li, and Qiongheng Liu. 2025. "Assembly and Analysis of the Complete Mitochondrial Genome of Eryngium foetidum L. (Apiaceae)" Biology 14, no. 9: 1296. https://doi.org/10.3390/biology14091296
APA StyleZhang, L., Zhang, W., Luo, Y., Liu, J., Li, Q., & Liu, Q. (2025). Assembly and Analysis of the Complete Mitochondrial Genome of Eryngium foetidum L. (Apiaceae). Biology, 14(9), 1296. https://doi.org/10.3390/biology14091296






