Comprehensive Analysis of TaNCED Gene Family in Wheat Vernalization Process
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Physicochemical Properties Analysis of TaNCED Members
2.2. Construction of Phylogenetic Trees
2.3. Motif Composition, Conserved Domains and Gene Structure Analysis of TaNCED Members
2.4. Chromosomal Location, Gene Duplication and Synteny Analysis of TaNCED Members
2.5. Cis-Acting Elements Analysis of TaNCED Members
2.6. Spatiotemporal Expression Profiling of TaNCED Members During Vernalization Response
2.7. Dual-Luciferase Reporter Assays
3. Results
3.1. Identification and Molecular Features of TaNCED Members
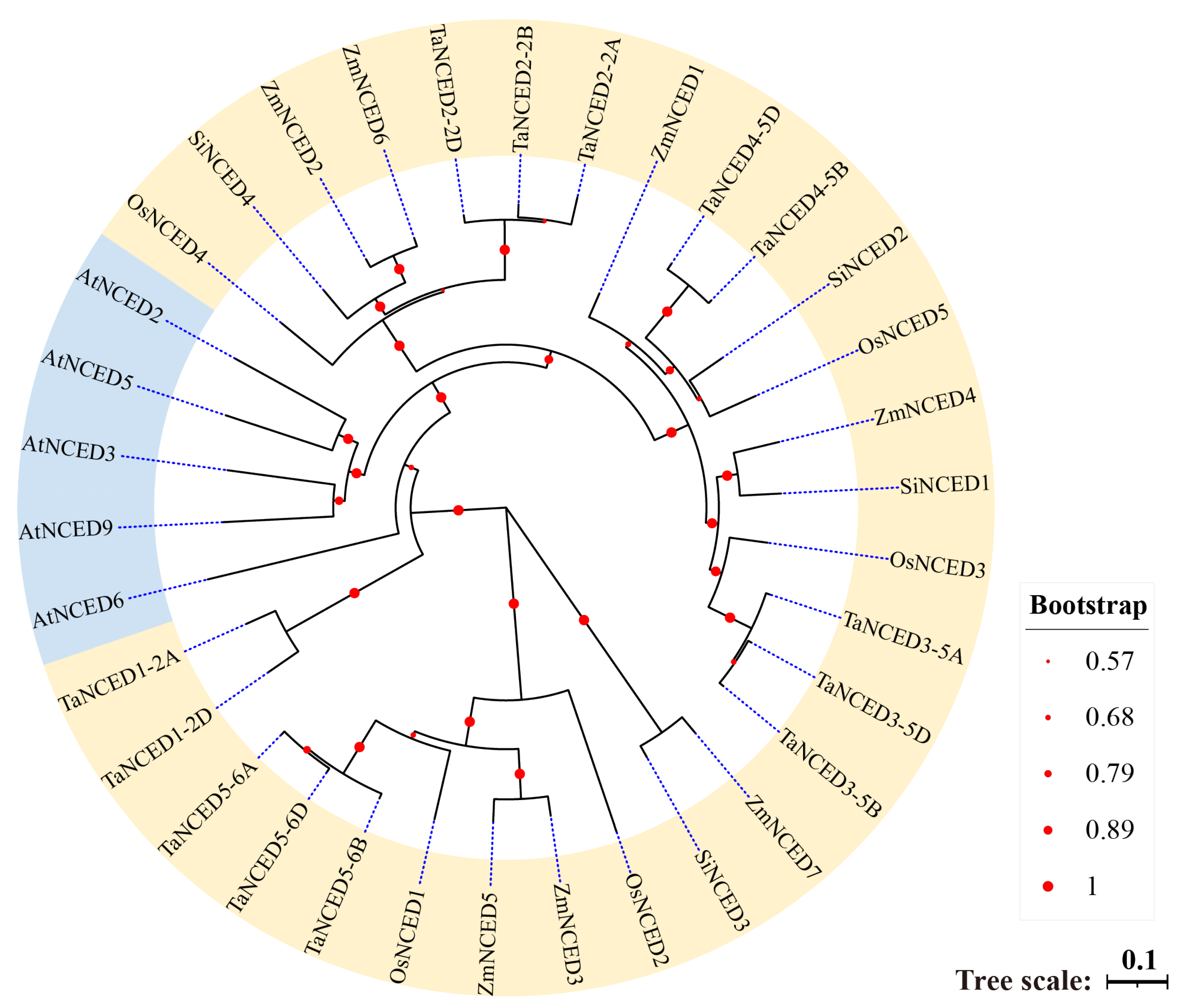
3.2. Phylogenetic Analysis of NCED Genes Across Different Plant Species
3.3. Conserved Motifs, Domains, and Gene Structure Analysis of TaNCED Members
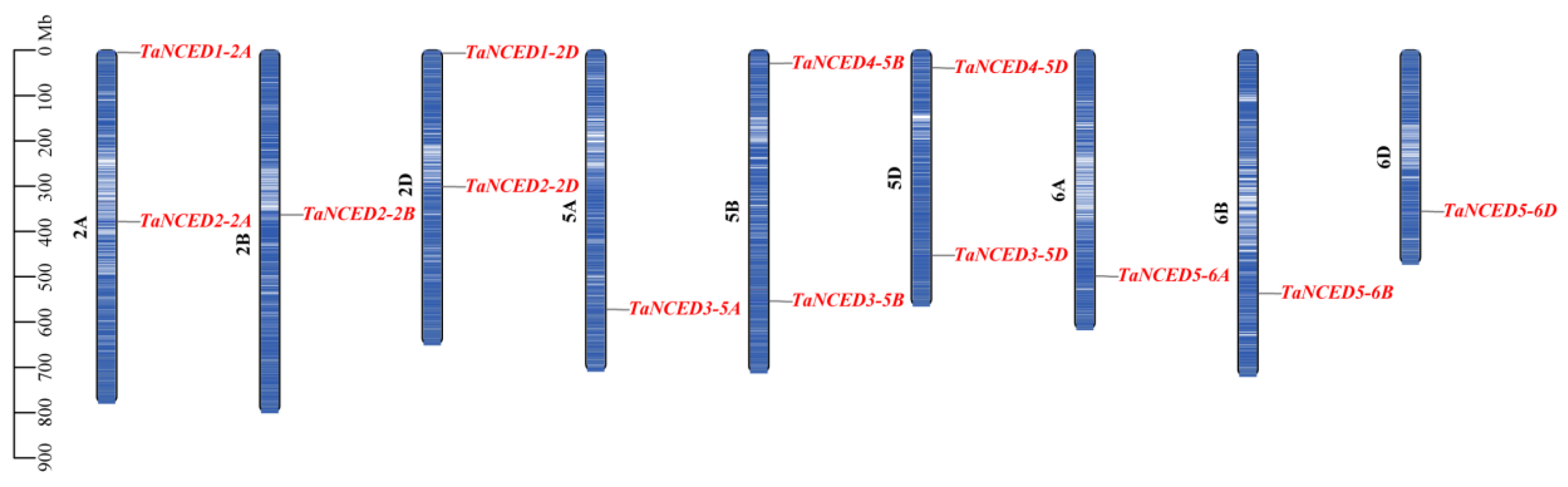
3.4. Chromosomal Distribution and Collinearity Analysis of TaNCED Members
3.5. Genomic Landscape of Cis-Acting Elements in TaNCED Members
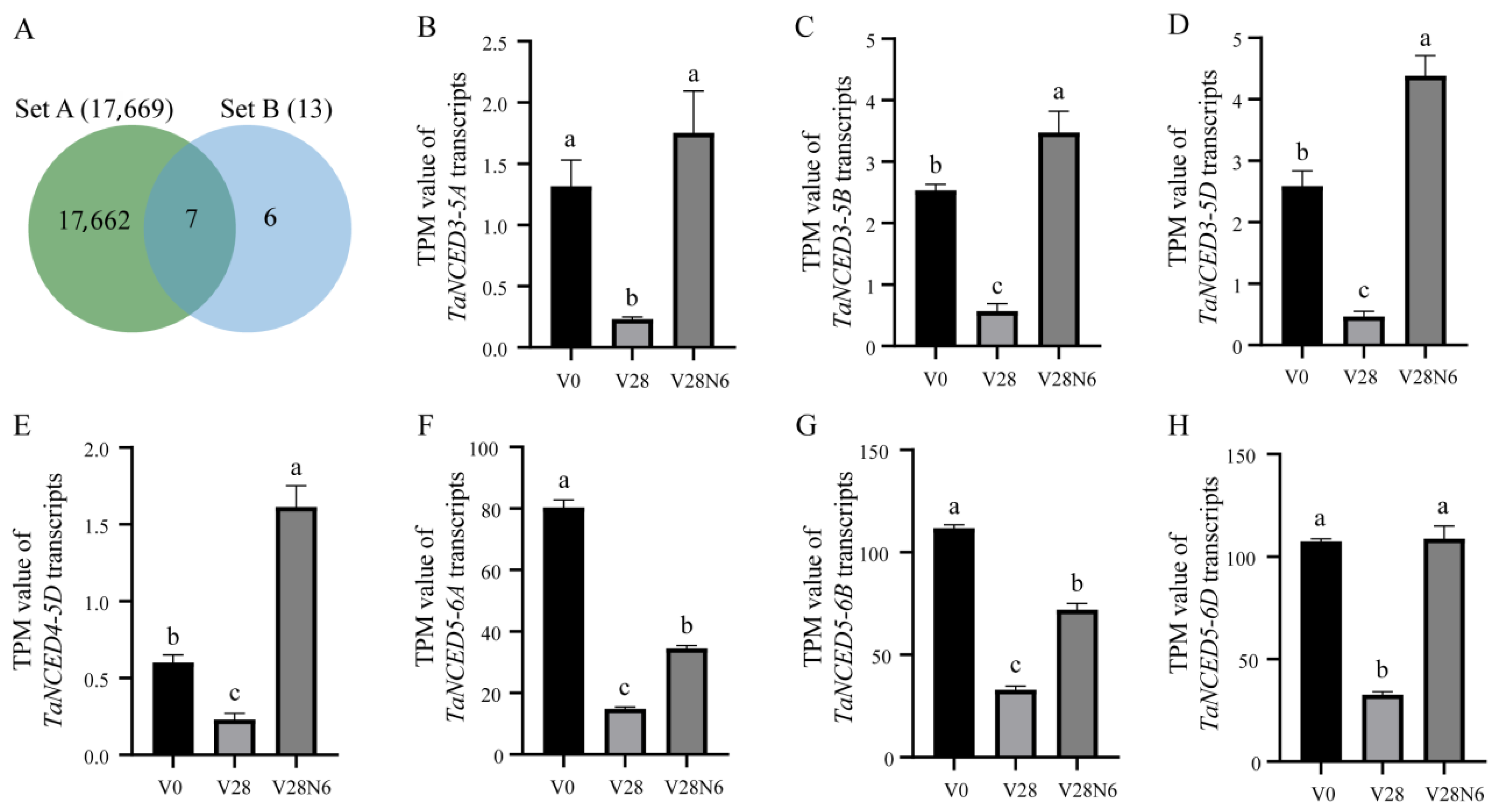
3.6. TaNCED Members Exhibit Differential Spatiotemporal Expression During Vernalization
4. Discussion
4.1. Structural Diversification and Functional Specialization of TaNCED Members
4.2. Regulatory Architecture Governing Spatiotemporal Expression
4.3. Functional Implications for Vernalization Response
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kavi Kishor, P.B.; Tiozon, R.N., Jr.; Fernie, A.R.; Sreenivasulu, N. Abscisic acid and its role in the modulation of plant growth, development, and yield stability. Trends Plant Sci. 2022, 27, 1283–1295. [Google Scholar] [CrossRef]
- Sajeev, N.; Koornneef, M.; Bentsink, L. A commitment for life: Decades of unraveling the molecular mechanisms behind seed dormancy and germination. Plant Cell 2024, 36, 1358–1376. [Google Scholar] [CrossRef]
- Ali, F.; Qanmber, G.; Li, F.G.; Wang, Z. Updated role of ABA in seed maturation, dormancy, and germination. J. Adv. Res. 2021, 35, 199–214. [Google Scholar] [CrossRef]
- Yang, D.Y.; Zhao, F.L.; Zhu, D.L.; Chen, X.; Kong, X.X.; Wu, Y.F.; Chen, M.; Du, J.M.; Qu, L.J.; Wu, Z. Progressive chromatin silencing of ABA biosynthesis genes permits seed germination in Arabidopsis. Plant Cell 2022, 34, 2871–2891. [Google Scholar] [CrossRef]
- Nonogaki, H. Seed germination and dormancy: The classic story, new puzzles, and evolution. J. Integr. Plant Biol. 2019, 61, 541–563. [Google Scholar] [CrossRef]
- Yao, C.; Finlayson, S.A. Abscisic acid is a general negative regulator of Arabidopsis axillary bud growth. Plant Physiol. 2015, 169, 611–626. [Google Scholar] [CrossRef] [PubMed]
- González-Grandío, E.; Pajoro, A.; Franco-Zorrilla, J.M.; Tarancón, C.; Immink, R.G.; Cubas, P. Abscisic acid signaling is controlled by a BRANCHED1/HD-ZIP I cascade in Arabidopsis axillary buds. Proc. Natl. Acad. Sci. USA 2017, 114, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Maurya, J.P.; Azeez, A.; Miskolczi, P.; Tylewicz, S.; Stojkovič, K.; Delhomme, N.; Busov, V.; Bhalerao, R.P. A genetic network mediating the control of bud break in hybrid aspen. Nat. Commun. 2018, 9, 4173. [Google Scholar] [CrossRef]
- Takahashi, F.; Suzuki, T.; Osakabe, Y.; Betsuyaku, S.; Kondo, Y.; Dohmae, N.; Fukuda, H.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature 2018, 556, 235–238. [Google Scholar] [CrossRef]
- Rowe, J.; Grangé-Guermente, M.; Exposito-Rodriguez, M.; Wimalasekera, R.; Lenz, M.O.; Shetty, K.N.; Cutler, S.R.; Jones, A.M. Next-generation ABACUS biosensors reveal cellular ABA dynamics driving root growth at low aerial humidity. Nat. Plants 2023, 9, 1103–1115. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Zareen, S.; Park, J.; Khan, H.A.; Lim, C.J.; Bader, Z.E.; Hussain, S.; Chung, W.S.; Gechev, T.; Pardo, J.M.; et al. ABA INSENSITIVE 2 promotes flowering by inhibiting OST1/ABI5-dependent FLOWERING LOCUS C transcription in Arabidopsis. J. Exp. Bot. 2024, 75, 2481–2493. [Google Scholar] [CrossRef]
- Waadt, R.; Seller, C.A.; Hsu, P.K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.M.; Zhu, J.H.; Gong, Z.Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef]
- Seo, M.; Koshiba, T. Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 2002, 7, 41–48. [Google Scholar] [CrossRef]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef]
- Guo, Q.Q.; Li, X.; Niu, L.; Jameson, P.E.; Zhou, W.B. Transcription-associated metabolomic adjustments in maize occur during combined drought and cold stress. Plant Physiol. 2021, 186, 677–695. [Google Scholar] [CrossRef]
- Van Zelm, E.; Zhang, Y.X.; Testerink, C. Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.B.; Guo, Y.F.; Kang, L.P.; Yin, C.B.; Bi, A.Y.; Xu, D.X.; Zhang, Z.L.; Zhang, J.J.; Yang, X.H.; Xu, J.; et al. Population genomics unravels the Holocene history of bread wheat and its relatives. Nat. Plants 2023, 9, 403–419. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Miao, F.; Jia, H.Y.; Li, G.Q.; Powers, C.; Nagarajan, R.; Alderman, P.D.; Carver, B.F.; Ma, Z.Q.; Yan, L.L. O-linked N-acetylglucosamine transferase is involved in fine regulation of flowering time in winter wheat. Nat. Commun. 2021, 12, 2303. [Google Scholar] [CrossRef] [PubMed]
- Cha, J.K.; O’Connor, K.; Alahmad, S.; Lee, J.H.; Dinglasan, E.; Park, H.; Lee, S.M.; Hirsz, D.; Kwon, S.W.; Kwon, Y.; et al. Speed vernalization to accelerate generation advance in winter cereal crops. Mol. Plant 2022, 15, 1300–1309. [Google Scholar] [CrossRef]
- Xu, S.J.; Chong, K. Remembering winter through vernalisation. Nat. Plants 2018, 4, 997–1009. [Google Scholar] [CrossRef]
- Luo, X.; He, Y.H. Experiencing winter for spring flowering: A molecular epigenetic perspective on vernalization. J. Integr. Plant Biol. 2020, 62, 104–117. [Google Scholar] [CrossRef]
- Niu, D.; Gao, Z.; Cui, B.W.; Zhang, Y.X.; He, Y.H. A molecular mechanism for embryonic resetting of winter memory and restoration of winter annual growth habit in wheat. Nat. Plants 2024, 10, 37–52. [Google Scholar] [CrossRef]
- Yan, L.; Loukoianov, A.; Tranquilli, G.; Helguera, M.; Fahima, T.; Dubcovsky, J. Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. USA 2003, 100, 6263–6268. [Google Scholar] [CrossRef]
- Yan, L.; Fu, D.; Li, C.; Blechl, A.; Tranquilli, G.; Bonafede, M.; Sanchez, A.; Valarik, M.; Yasuda, S.; Dubcovsky, J. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. USA 2006, 103, 19581–19586. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bo, Y.; Han, L.Y.; He, J.E.; Lanczycki, C.J.; Lu, S.N.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, 200–203. [Google Scholar] [CrossRef]
- Lu, S.N.; Wang, J.Y.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, 265–268. [Google Scholar] [CrossRef]
- Wang, J.Y.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.N.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, 384–388. [Google Scholar] [CrossRef]
- Chen, Y.M.; Song, W.J.; Xie, X.M.; Wang, Z.H.; Guan, P.F.; Peng, H.R.; Jiao, Y.N.; Ni, Z.F.; Sun, Q.X.; Guo, W.L. A collinearity-incorporating homology inference strategy for connecting emerging assemblies in the Triticeae tribe as a pilot practice in the plant Pangenomic era. Mol. Plant 2020, 13, 1694–1708. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C.; Shen, H. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12: Molecular evolutionary genetic analysis version 12 for adaptive and green computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, 78–82. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, 39–49. [Google Scholar] [CrossRef]
- Wang, D.P.; Zhang, Y.B.; Zhang, Z.; Zhu, J.; Yu, J. KaKs Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Xu, X.T.; He, C.; Jin, L.J.; Zhou, Z.R.; Gao, J.; Guo, M.R.; Wang, X.; Chen, C.Y.; Ayaad, M.; et al. Chromatin loops gather targets of upstream regulators together for efficient gene transcription regulation during vernalization in wheat. Genome Biol. 2024, 25, 306. [Google Scholar] [CrossRef]
- Liu, Y.H.; Liu, P.; Gao, L.F.; Li, Y.S.; Ren, X.N.; Jia, J.Z.; Wang, L.; Zheng, X.; Tong, Y.P.; Pei, H.C.; et al. Epigenomic identification of vernalization cis-regulatory elements in winter wheat. Genome Biol. 2024, 25, 200. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Kidokoro, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulatory networks in plant responses to drought and cold stress. Plant Physiol. 2024, 195, 170–189. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.P.; Duan, X.B.; Luo, L.; Dai, S.J.; Ding, Z.J.; Xia, G.M. How plant hormones mediate salt stress responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef]
- Liang, X.Y.; Li, J.F.; Yang, Y.Q.; Jiang, C.F.; Guo, Y. Designing salt stress-resilient crops: Current progress and future challenges. J. Integr. Plant Biol. 2024, 66, 303–329. [Google Scholar] [CrossRef]
- Ma, Y.L.; Cao, J.; He, J.H.; Chen, Q.Q.; Li, X.F.; Yang, Y. Molecular mechanism for the regulation of ABA homeostasis during plant development and stress responses. Int. J. Mol. Sci. 2018, 19, 3643. [Google Scholar] [CrossRef]
- Brookbank, B.P.; Patel, J.; Gazzarrini, S.; Nambara, E. Role of basal ABA in plant growth and development. Genes 2021, 12, 1936. [Google Scholar] [CrossRef]
- Sharma, A.; Gupta, A.; Ramakrishnan, M.; Ha, C.V.; Zheng, B.S.; Bhardwaj, M.; Tran, L.P. Roles of abscisic acid and auxin in plants during drought: A molecular point of view. Plant Physiol. Biochem. 2023, 204, 108129. [Google Scholar] [CrossRef]
- Zhao, H.Y.; Zhang, Y.M.; Zheng, Y. Integration of ABA, GA, and light signaling in seed germination through the regulation of ABI5. Front. Plant Sci. 2022, 13, 1000803. [Google Scholar] [CrossRef]
- Liao, Z.G.; Zhang, Y.C.; Yu, Q.; Fang, W.C.; Chen, M.Y.; Li, T.F.; Liu, Y.; Liu, Z.C.; Chen, L.; Yu, S.W.; et al. Coordination of growth and drought responses by GA-ABA signaling in rice. New Phytol. 2023, 240, 1149–1161. [Google Scholar] [CrossRef]
- Xiang, Z.P.; Zhang, L.; Long, Y.X.; Zhang, M.Z.; Yao, Y.X.; Deng, H.L.; Quan, C.B.; Lu, M.F.; Cui, B.L.; Wang, D. An ABA biosynthesis enzyme gene OsNCED4 regulates NaCl and cold stress tolerance in rice. Sci. Rep. 2024, 14, 26711. [Google Scholar] [CrossRef] [PubMed]
- Bao, G.G.; Zhuo, C.L.; Qian, C.M.; Xiao, T.; Guo, Z.F.; Lu, S.Y. Co-expression of NCED and ALO improves vitamin C level and tolerance to drought and chilling in transgenic tobacco and stylo plants. Plant Biotechnol. J. 2016, 14, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Azeez, A.; Zhao, Y.C.; Singh, R.K.; Yordanov, Y.S.; Dash, M.; Miskolczi, P.; Stojkovič, K.; Strauss, S.H.; Bhalerao, R.P.; Busov, V.B. EARLY BUD-BREAK 1 and EARLY BUD-BREAK 3 control resumption of poplar growth after winter dormancy. Nat. Commun. 2021, 12, 1123. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Miskolczi, P.; Maurya, J.P.; Bhalerao, R.P. A tree ortholog of SHORT VEGETATIVE PHASE floral repressor mediates photoperiodic control of bud dormancy. Curr. Biol. 2019, 29, 128–133. [Google Scholar] [CrossRef] [PubMed]




| Gene Name | AA (aa) | MW (Da) | pI | Instability Index | Aliphatic Index | GRAVY | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| TaNCED1-2A | 567 | 60,602 | 6.41 | 44.46 | 85.59 | −0.039 | Chloroplast |
| TaNCED1-2D | 603 | 65,075 | 5.88 | 44.95 | 86.9 | −0.049 | Chloroplast |
| TaNCED2-2A | 593 | 64,382 | 6.59 | 44.76 | 79.12 | −0.307 | Chloroplast and Cytoplasm |
| TaNCED2-2B | 596 | 64,739 | 6.64 | 41.27 | 78.88 | −0.288 | Chloroplast and Cytoplasm |
| TaNCED2-2D | 582 | 63,022 | 5.9 | 41.21 | 81.63 | −0.244 | Chloroplast and Cytoplasm |
| TaNCED3-5A | 591 | 64,488 | 5.41 | 40.43 | 80.73 | −0.193 | Chloroplast and Cytoplasm |
| TaNCED3-5B | 595 | 64,878 | 5.51 | 40.91 | 80.35 | −0.195 | Chloroplast, Cytoplasm and Mitochondrial |
| TaNCED3-5D | 595 | 64,828 | 5.41 | 40.99 | 81.01 | −0.176 | Chloroplast and Cytoplasm |
| TaNCED4-5B | 614 | 66,620 | 5.52 | 38.74 | 76.32 | −0.2 | Chloroplast and Cytoplasm |
| TaNCED4-5D | 615 | 66,683 | 5.55 | 36.22 | 75.87 | −0.2 | Chloroplast and Cytoplasm |
| TaNCED5-6A | 639 | 68,768 | 5.93 | 46.55 | 74.18 | −0.207 | Chloroplast and Cytoplasm |
| TaNCED5-6B | 643 | 69,262 | 5.92 | 46.06 | 75.09 | −0.194 | Chloroplast and Cytoplasm |
| TaNCED5-6D | 639 | 68,792 | 5.86 | 46.09 | 74.63 | −0.201 | Chloroplast and Cytoplasm |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, G.; Cheng, H. Comprehensive Analysis of TaNCED Gene Family in Wheat Vernalization Process. Biology 2025, 14, 1293. https://doi.org/10.3390/biology14091293
Cui G, Cheng H. Comprehensive Analysis of TaNCED Gene Family in Wheat Vernalization Process. Biology. 2025; 14(9):1293. https://doi.org/10.3390/biology14091293
Chicago/Turabian StyleCui, Guoqing, and Hao Cheng. 2025. "Comprehensive Analysis of TaNCED Gene Family in Wheat Vernalization Process" Biology 14, no. 9: 1293. https://doi.org/10.3390/biology14091293
APA StyleCui, G., & Cheng, H. (2025). Comprehensive Analysis of TaNCED Gene Family in Wheat Vernalization Process. Biology, 14(9), 1293. https://doi.org/10.3390/biology14091293






