Lifetime Changes in Gut Microbiota and Metabolite Composition in High-Fat Diet-Induced Obesity in Apolipoprotein A-IV Gene Knockout Mice
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Development of ApoA-IV Knockout Mice and Maintenance of Animals
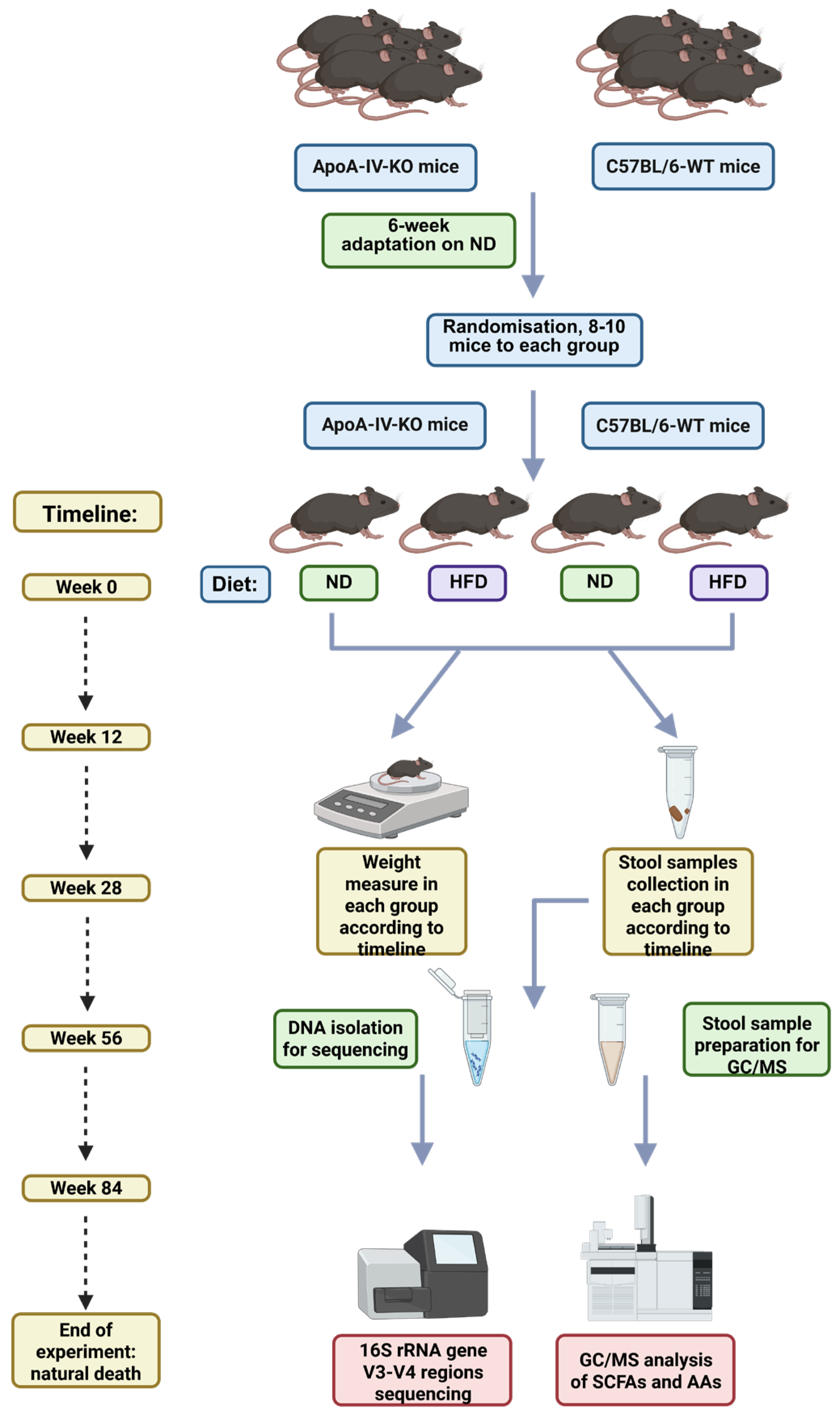
2.2. Experiment Design
2.3. DNA Extraction and 16S rRNA Gene Sequencing
2.4. Gas Chromatography–Mass Spectrometry Metabolite Analysis
2.5. Data Analysis
3. Results
3.1. Mice Characteristics
3.2. Metagenomic Sequencing
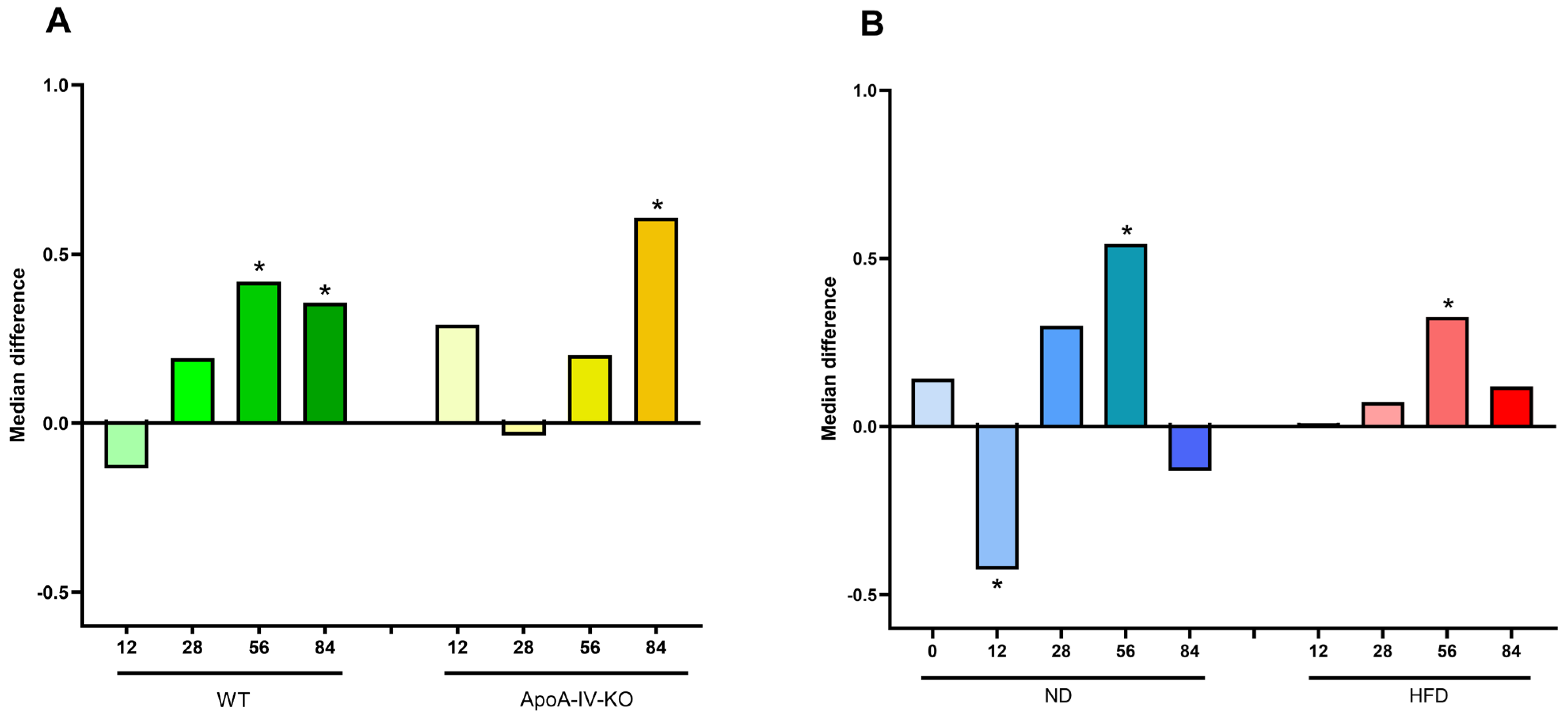
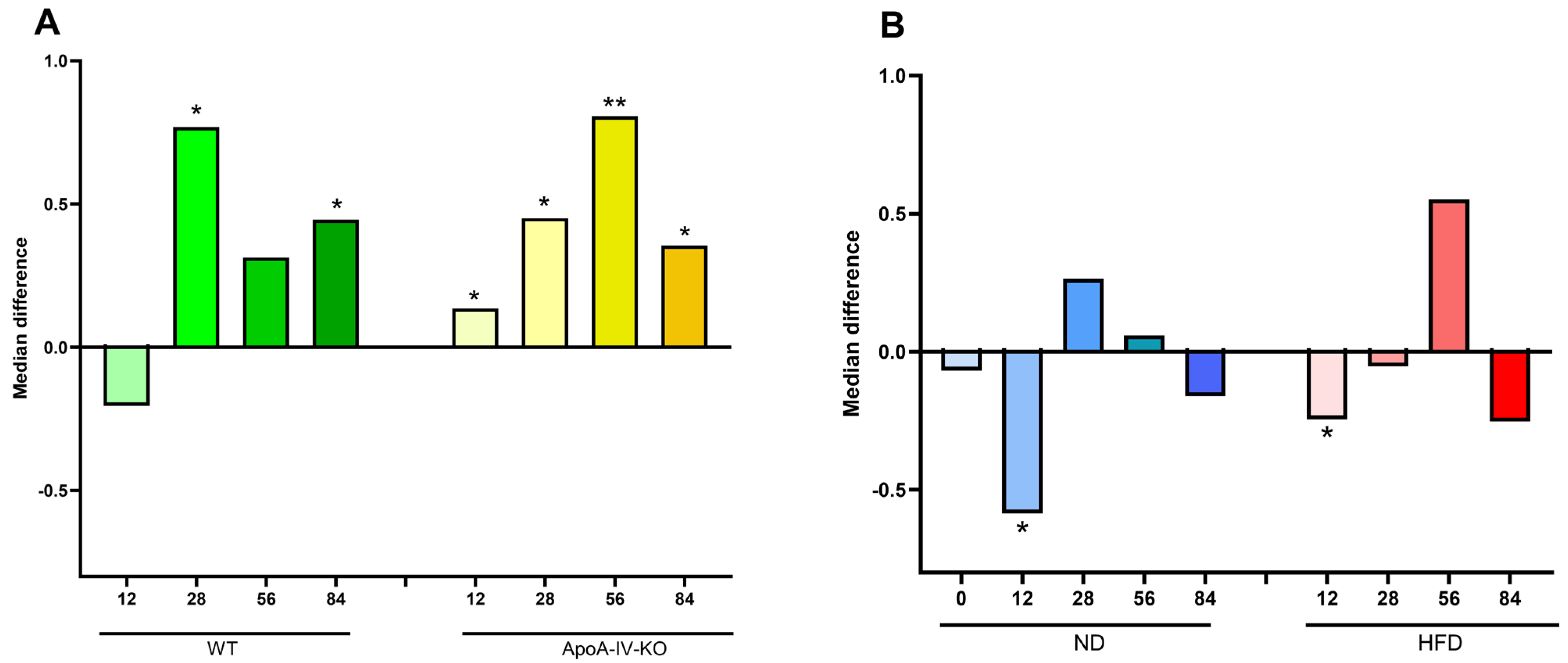
3.2.1. Microbiota Diversity and Ratio of Firmicutes to Bacteroidota
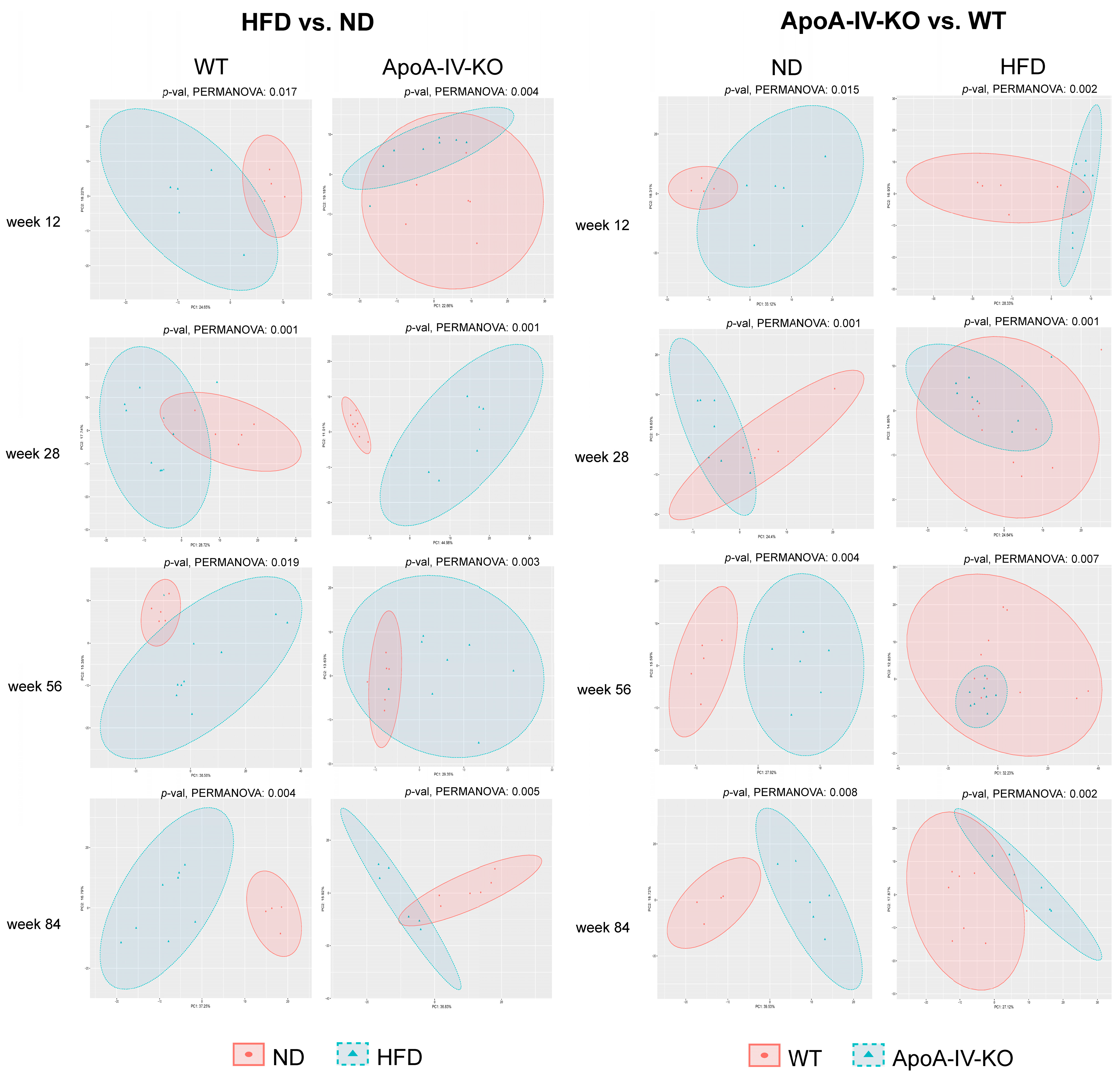
3.2.2. Pairwise Comparative Analyses of Gut Microbiome Composition
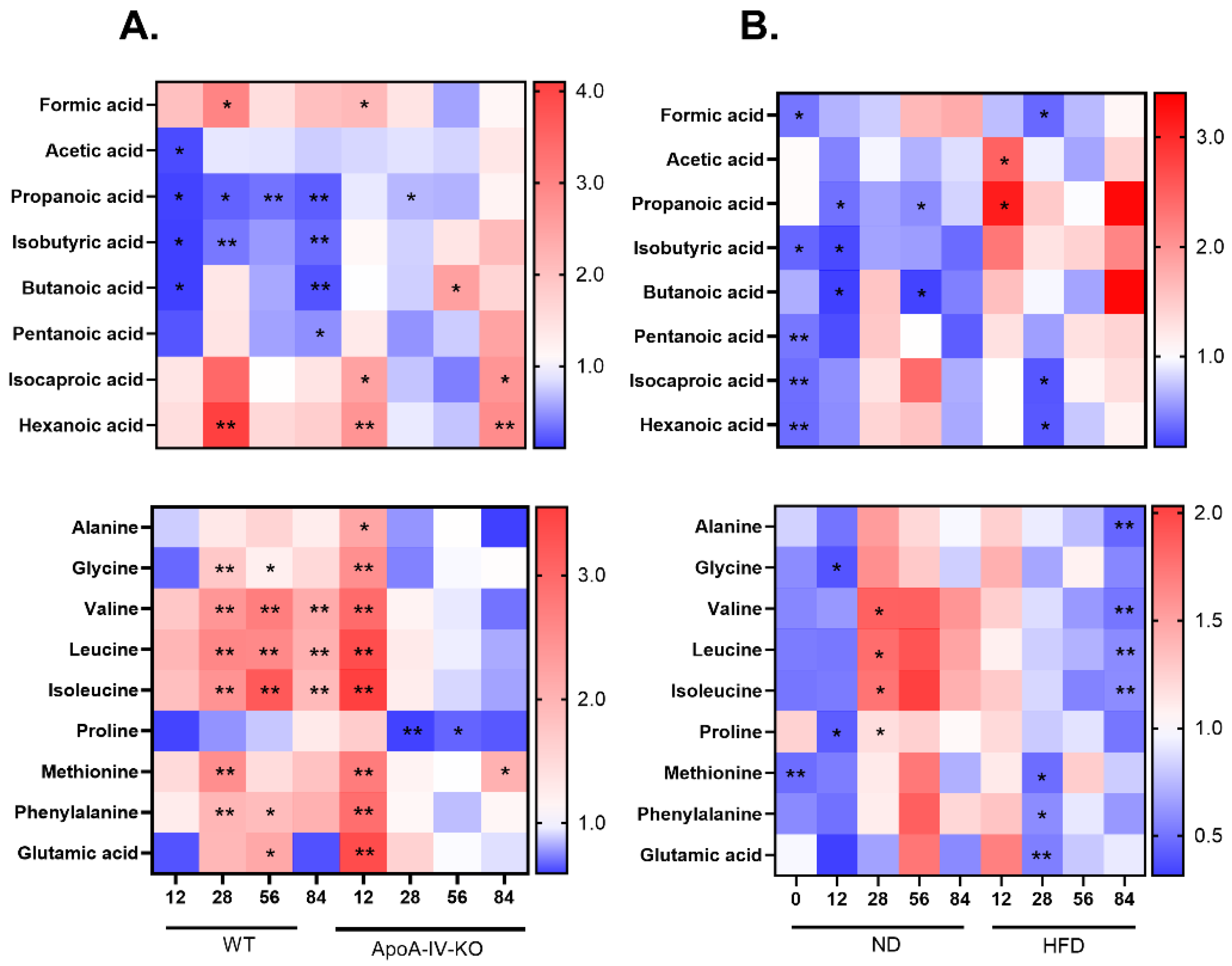
3.3. Metabolite Composition
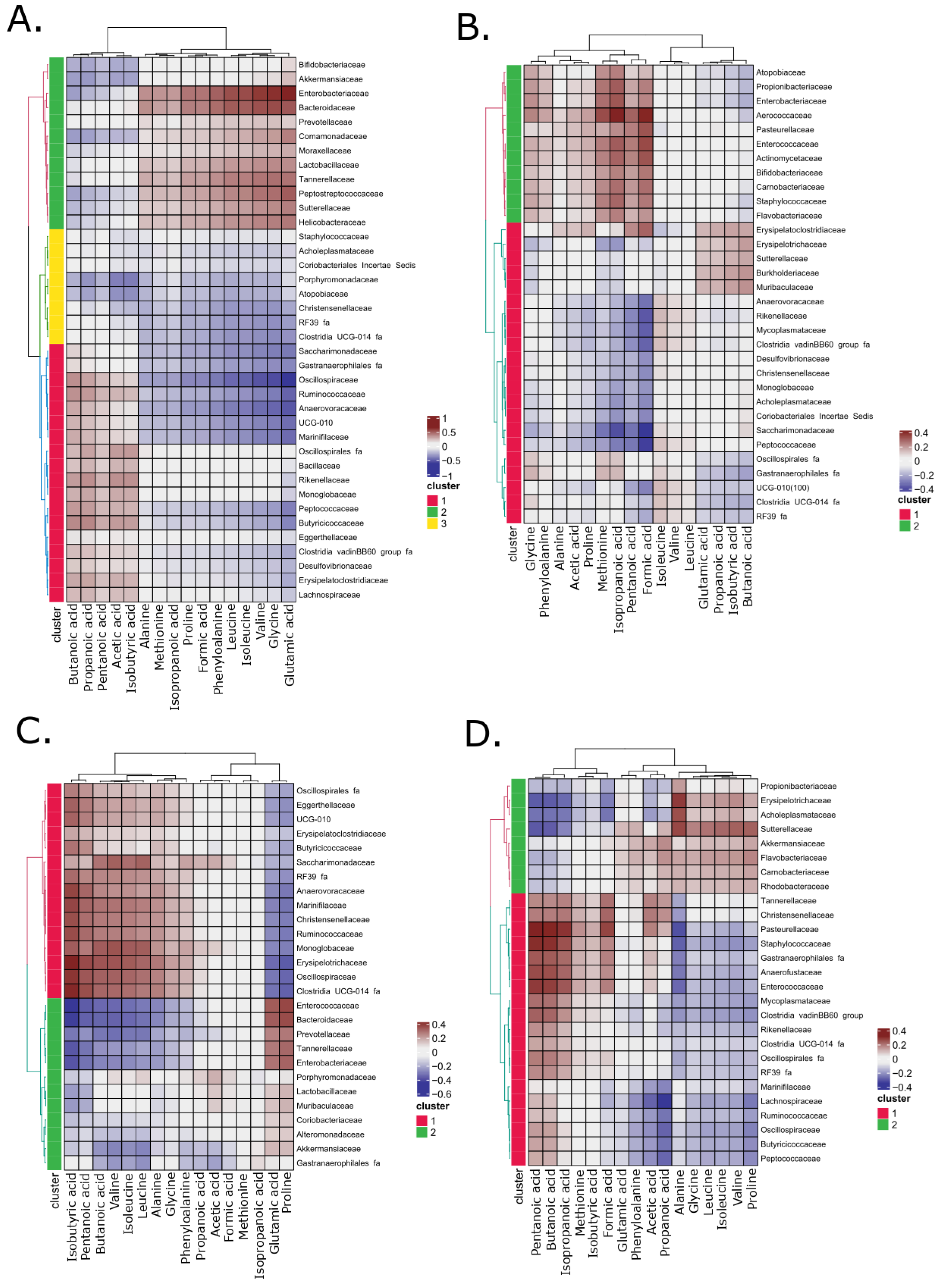
3.4. The Correlation Analysis on Identified Bacterial Taxa and Metabolite Concentrations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ApoA-IV | Apolipoprotein A-IV |
| HFDJ | High-fat diet |
| CCK | Cholecystokinin |
| KO | Knockout |
| ND | Normal diet |
| SCFA | Short-chain fatty acid |
| BCAA | Branched-chain amino acid |
| rCCA | Regularized canonical correlation analysis |
| WT | Wild type |
| GC/MS | Gas chromatography–mass spectrometry |
| F/B | Firmicutes to Bacteroidota |
| PCoA | Principal coordinate analysis |
| padj | Adjusted p-value |
References
- Liu, M.; Doi, T.; Tso, P. Regulation of Intestinal and Hypothalamic Apolipoprotein A-IV. Exp. Biol. Med. 2003, 228, 1181–1189. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, L.; Zhang, Z.; Feng, L.; Song, X.; Wu, J. Apolipoprotein A-IV Involves in Glucose and Lipid Metabolism of Rat. Nutr. Metab. 2019, 16, 41. [Google Scholar] [CrossRef]
- Tso, P.; Sun, W.; Liu, M. Gastrointestinal Satiety Signals IV. Apolipoprotein A-IV. Am. J. Physiol. Gastrointest. Liver Physiol. 2004, 286, G885–G890. [Google Scholar] [CrossRef]
- Pence, S.; Zhu, Q.; Binne, E.; Liu, M.; Shi, H.; Lo, C.C. Reduced Diet-Induced Thermogenesis in Apolipoprotein A-IV Deficient Mice. Int. J. Mol. Sci. 2019, 20, 3176. [Google Scholar] [CrossRef]
- Ostos, M.A.; Conconi, M.; Vergnes, L.; Baroukh, N.; Ribalta, J.; Girona, J.; Caillaud, J.M.; Ochoa, A.; Zakin, M.M. Antioxidative and Antiatherosclerotic Effects of Human Apolipoprotein A-IV in Apolipoprotein E-Deficient Mice. Arterioscler. Thromb. Vasc. Biol. 2001, 21, 1023–1028. [Google Scholar] [CrossRef]
- Xu, X.R.; Wang, Y.; Adili, R.; Ju, L.; Spring, C.M.; Jin, J.W.; Yang, H.; Neves, M.A.D.; Chen, P.; Yang, Y.; et al. Apolipoprotein A-IV Binds αIIbβ3 Integrin and Inhibits Thrombosis. Nat. Commun. 2018, 9, 3608. [Google Scholar] [CrossRef]
- Qu, J.; Ko, C.-W.; Tso, P.; Bhargava, A. Apolipoprotein A-IV: A Multifunctional Protein Involved in Protection against Atherosclerosis and Diabetes. Cells 2019, 8, 319. [Google Scholar] [CrossRef]
- Peng, J.; Li, X.-P. Apolipoprotein A-IV: A Potential Therapeutic Target for Atherosclerosis. Prostaglandins Other Lipid Mediat. 2018, 139, 87–92. [Google Scholar] [CrossRef]
- Tso, P.; Liu, M. Apolipoprotein A-IV, Food Intake, and Obesity. Physiol. Behav. 2004, 83, 631–643. [Google Scholar] [CrossRef]
- Wang, F.; Kohan, A.B.; Lo, C.-M.; Liu, M.; Howles, P.; Tso, P. Apolipoprotein A-IV: A Protein Intimately Involved in Metabolism. J. Lipid Res. 2015, 56, 1403–1418. [Google Scholar] [CrossRef]
- Kalogeris, T.J.; Painter, R.G. Adaptation of Intestinal Production of Apolipoprotein A-IV during Chronic Feeding of Lipid. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2001, 280, R1155–R1161. [Google Scholar] [CrossRef]
- Doi, T.; Liu, M.; Seeley, R.J.; Woods, S.C.; Tso, P. Effect of Leptin on Intestinal Apolipoprotein AIV in Response to Lipid Feeding. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2001, 281, R753–R759. [Google Scholar] [CrossRef][Green Version]
- Shen, L.; Tso, P.; Woods, S.C.; Sakai, R.R.; Davidson, W.S.; Liu, M. Hypothalamic Apolipoprotein A-IV Is Regulated by Leptin. Endocrinology 2007, 148, 2681–2689. [Google Scholar] [CrossRef]
- Wang, F.; Kohan, A.B.; Kindel, T.L.; Corbin, K.L.; Nunemaker, C.S.; Obici, S.; Woods, S.C.; Davidson, W.S.; Tso, P. Apolipoprotein A-IV Improves Glucose Homeostasis by Enhancing Insulin Secretion. Proc. Natl. Acad. Sci. USA 2012, 109, 9641–9646. [Google Scholar] [CrossRef]
- Li, X.; Xu, M.; Wang, F.; Kohan, A.B.; Haas, M.K.; Yang, Q.; Lou, D.; Obici, S.; Davidson, W.S.; Tso, P. Apolipoprotein A-IV Reduces Hepatic Gluconeogenesis through Nuclear Receptor NR1D1. J. Biol. Chem. 2014, 289, 2396–2404. [Google Scholar] [CrossRef]
- Lundsgaard, A.-M.; Del Giudice, R.; Kanta, J.M.; Larance, M.; Armour, S.L.; London, A.; Richter, M.M.; Andersen, N.R.; Nicolaisen, T.S.; Carl, C.S.; et al. Apolipoprotein A-IV Is Induced by High-Fat Diets and Mediates Positive Effects on Glucose and Lipid Metabolism. Mol. Metab. 2025, 95, 102119. [Google Scholar] [CrossRef]
- Zhan, J.; Weng, J.; Hunt, B.G.; Sean Davidson, W.; Liu, M.; Lo, C.C. Apolipoprotein A-IV Enhances Cholecystokinnin Secretion. Physiol. Behav. 2018, 188, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Covasa, M. Deficits in Gastrointestinal Responses Controlling Food Intake and Body Weight. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2010, 299, R1423–R1439. [Google Scholar] [CrossRef]
- Doumatey, A.P.; Zhou, J.; Zhou, M.; Prieto, D.; Rotimi, C.N.; Adeyemo, A. Proinflammatory and Lipid Biomarkers Mediate Metabolically Healthy Obesity: A Proteomics Study. Obesity 2016, 24, 1257–1265. [Google Scholar] [CrossRef]
- Cao, Z.; Lei, L.; Zhou, Z.; Xu, S.; Wang, L.; Gong, W.; Zhang, Q.; Pan, B.; Zhang, G.; Yuan, Q.; et al. Apolipoprotein A-IV and Its Derived Peptide, T55-121, Improve Glycemic Control and Increase Energy Expenditure. Life Metab. 2024, 3, loae010. [Google Scholar] [CrossRef]
- Heianza, Y.; Sun, D.; Li, X.; DiDonato, J.A.; Bray, G.A.; Sacks, F.M.; Qi, L. Gut Microbiota Metabolites, Amino Acid Metabolites and Improvements in Insulin Sensitivity and Glucose Metabolism: The POUNDS Lost Trial. Gut 2019, 68, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Vallianou, N.; Stratigou, T.; Christodoulatos, G.S.; Dalamaga, M. Understanding the Role of the Gut Microbiome and Microbial Metabolites in Obesity and Obesity-Associated Metabolic Disorders: Current Evidence and Perspectives. Curr. Obes. Rep. 2019, 8, 317–332. [Google Scholar] [CrossRef]
- Heiss, C.N.; Olofsson, L.E. Gut Microbiota-Dependent Modulation of Energy Metabolism. J. Innate Immun. 2018, 10, 163–171. [Google Scholar] [CrossRef] [PubMed]
- de la Cuesta-Zuluaga, J.; Mueller, N.T.; Álvarez-Quintero, R.; Velásquez-Mejía, E.P.; Sierra, J.A.; Corrales-Agudelo, V.; Carmona, J.A.; Abad, J.M.; Escobar, J.S. Higher Fecal Short-Chain Fatty Acid Levels Are Associated with Gut Microbiome Dysbiosis, Obesity, Hypertension and Cardiometabolic Disease Risk Factors. Nutrients 2018, 11, 51. [Google Scholar] [CrossRef]
- Yu, Y.; Ding, Y.; Wang, S.; Jiang, L. Gut Microbiota Dysbiosis and Its Impact on Type 2 Diabetes: From Pathogenesis to Therapeutic Strategies. Metabolites 2025, 15, 397. [Google Scholar] [CrossRef]
- Scheithauer, T.P.M.; Rampanelli, E.; Nieuwdorp, M.; Vallance, B.A.; Verchere, C.B.; Van Raalte, D.H.; Herrema, H. Gut Microbiota as a Trigger for Metabolic Inflammation in Obesity and Type 2 Diabetes. Front. Immunol. 2020, 11, 571731. [Google Scholar] [CrossRef]
- Shen, Y.; Fan, N.; Ma, S.; Cheng, X.; Yang, X.; Wang, G. Gut Microbiota Dysbiosis: Pathogenesis, Diseases, Prevention, and Therapy. MedComm 2025, 6, e70168. [Google Scholar] [CrossRef]
- Ahn, J.; Hayes, R.B. Environmental Influences on the Human Microbiome and Implications for Noncommunicable Disease. Annu. Rev. Public Health 2021, 42, 277–292. [Google Scholar] [CrossRef]
- Dethlefsen, L.; McFall-Ngai, M.; Relman, D.A. An Ecological and Evolutionary Perspective on Human-Microbe Mutualism and Disease. Nature 2007, 449, 811–818. [Google Scholar] [CrossRef] [PubMed]
- Sundekilde, U.K.; Yde, C.C.; Honore, A.H.; Caverly Rae, J.M.; Burns, F.R.; Mukerji, P.; Mawn, M.P.; Stenman, L.; Dragan, Y.; Glover, K.; et al. An Integrated Multi-Omics Analysis Defines Key Pathway Alterations in a Diet-Induced Obesity Mouse Model. Metabolites 2020, 10, 80. [Google Scholar] [CrossRef]
- Xu, C.; Liu, J.; Gao, J.; Wu, X.; Cui, C.; Wei, H.; Zheng, R.; Peng, J. Combined Soluble Fiber-Mediated Intestinal Microbiota Improve Insulin Sensitivity of Obese Mice. Nutrients 2020, 12, 351. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An Obesity-Associated Gut Microbiome with Increased Capacity for Energy Harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Fei, N.; Zhao, L. An Opportunistic Pathogen Isolated from the Gut of an Obese Human Causes Obesity in Germfree Mice. ISME J. 2013, 7, 880–884. [Google Scholar] [CrossRef]
- Kulecka, M.; Paziewska, A.; Zeber-Lubecka, N.; Ambrozkiewicz, F.; Kopczynski, M.; Kuklinska, U.; Pysniak, K.; Gajewska, M.; Mikula, M.; Ostrowski, J. Prolonged Transfer of Feces from the Lean Mice Modulates Gut Microbiota in Obese Mice. Nutr. Metab. 2016, 13, 57. [Google Scholar] [CrossRef]
- Li, Z.; Quan, G.; Jiang, X.; Yang, Y.; Ding, X.; Zhang, D.; Wang, X.; Hardwidge, P.R.; Ren, W.; Zhu, G. Effects of Metabolites Derived From Gut Microbiota and Hosts on Pathogens. Front. Cell. Infect. Microbiol. 2018, 8, 314. [Google Scholar] [CrossRef]
- den Besten, G.; van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.-J.; Bakker, B.M. The Role of Short-Chain Fatty Acids in the Interplay between Diet, Gut Microbiota, and Host Energy Metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef]
- Chambers, E.S.; Preston, T.; Frost, G.; Morrison, D.J. Role of Gut Microbiota-Generated Short-Chain Fatty Acids in Metabolic and Cardiovascular Health. Curr. Nutr. Rep. 2018, 7, 198–206. [Google Scholar] [CrossRef]
- Metges, C.C. Contribution of Microbial Amino Acids to Amino Acid Homeostasis of the Host. J. Nutr. 2000, 130, 1857S–1864S. [Google Scholar] [CrossRef]
- Neis, E.P.J.G.; Dejong, C.H.C.; Rensen, S.S. The Role of Microbial Amino Acid Metabolism in Host Metabolism. Nutrients 2015, 7, 2930–2946. [Google Scholar] [CrossRef]
- Dai, Z.-L.; Wu, G.; Zhu, W.-Y. Amino Acid Metabolism in Intestinal Bacteria: Links between Gut Ecology and Host Health. Front. Biosci. Landmark Ed. 2011, 16, 1768–1786. [Google Scholar] [CrossRef]
- Schwiertz, A.; Taras, D.; Schäfer, K.; Beijer, S.; Bos, N.A.; Donus, C.; Hardt, P.D. Microbiota and SCFA in Lean and Overweight Healthy Subjects. Obesity 2010, 18, 190–195. [Google Scholar] [CrossRef]
- Newgard, C.B.; An, J.; Bain, J.R.; Muehlbauer, M.J.; Stevens, R.D.; Lien, L.F.; Haqq, A.M.; Shah, S.H.; Arlotto, M.; Slentz, C.A.; et al. A Branched-Chain Amino Acid-Related Metabolic Signature That Differentiates Obese and Lean Humans and Contributes to Insulin Resistance. Cell Metab. 2009, 9, 311–326. [Google Scholar] [CrossRef]
- LI, X.; SHIMIZU, Y.; KIMURA, I. Gut Microbial Metabolite Short-Chain Fatty Acids and Obesity. Biosci. Microbiota Food Health 2017, 36, 135–140. [Google Scholar] [CrossRef]
- Rahat-Rozenbloom, S.; Fernandes, J.; Gloor, G.B.; Wolever, T.M.S. Evidence for Greater Production of Colonic Short-Chain Fatty Acids in Overweight than Lean Humans. Int. J. Obes. 2014, 38, 1525–1531. [Google Scholar] [CrossRef]
- Claesson, M.J.; Cusack, S.; O’Sullivan, O.; Greene-Diniz, R.; de Weerd, H.; Flannery, E.; Marchesi, J.R.; Falush, D.; Dinan, T.; Fitzgerald, G.; et al. Composition, Variability, and Temporal Stability of the Intestinal Microbiota of the Elderly. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4586–4591. [Google Scholar] [CrossRef]
- Fransen, F.; van Beek, A.A.; Borghuis, T.; Aidy, S.E.; Hugenholtz, F.; van der Gaast-de Jongh, C.; Savelkoul, H.F.J.; De Jonge, M.I.; Boekschoten, M.V.; Smidt, H.; et al. Aged Gut Microbiota Contributes to Systemical Inflammaging after Transfer to Germ-Free Mice. Front. Immunol. 2017, 8, 1385. [Google Scholar] [CrossRef]
- Weinstock, P.H.; Bisgaier, C.L.; Hayek, T.; Aalto-Setala, K.; Sehayek, E.; Wu, L.; Sheiffele, P.; Merkel, M.; Essenburg, A.D.; Breslow, J.L. Decreased HDL Cholesterol Levels but Normal Lipid Absorption, Growth, and Feeding Behavior in Apolipoprotein A-IV Knockout Mice. J. Lipid Res. 1997, 38, 1782–1794. [Google Scholar] [CrossRef] [PubMed]
- Unrug-Bielawska, K.; Sandowska-Markiewicz, Z.; Pyśniak, K.; Piątkowska, M.; Czarnowski, P.; Goryca, K.; Mróz, A.; Żeber-Lubecka, N.; Wójcik-Trechcińska, U.; Bałabas, A.; et al. Western Diet and Fecal Microbiota Transplantation Alter Phenotypic, Liver Fatty Acids, and Gut Metagenomics and Metabolomics in Mtarc2 Knockout Mice. Genes Nutr. 2025, 20, 13. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Krieger, A.M.; Yekutieli, D. Adaptive Linear Step-up Procedures That Control the False Discovery Rate. Biometrika 2006, 93, 491–507. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing Mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A Versatile Open Source Tool for Metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Palarea-Albaladejo, J.; Martín-Fernández, J.A. zCompositions—R Package for Multivariate Imputation of Left-Censored Data under a Compositional Approach. Chemom. Intell. Lab. Syst. 2015, 143, 85–96. [Google Scholar] [CrossRef]
- Zhou, H.; He, K.; Chen, J.; Zhang, X. LinDA: Linear Models for Differential Abundance Analysis of Microbiome Compositional Data. Genome Biol. 2022, 23, 95. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.-A. mixOmics: An R Package for ’omics Feature Selection and Multiple Data Integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef]
- Gu, Z. Complex Heatmap Visualization. iMeta 2022, 1, e43. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Zhang, B.; Horvath, S. Defining Clusters from a Hierarchical Cluster Tree: The Dynamic Tree Cut Package for R. Bioinforma. Oxf. Engl. 2008, 24, 719–720. [Google Scholar] [CrossRef]
- Breton, J.; Galmiche, M.; Déchelotte, P. Dysbiotic Gut Bacteria in Obesity: An Overview of the Metabolic Mechanisms and Therapeutic Perspectives of Next-Generation Probiotics. Microorganisms 2022, 10, 452. [Google Scholar] [CrossRef]
- Jian, Z.; Zeng, L.; Xu, T.; Sun, S.; Yan, S.; Zhao, S.; Su, Z.; Ge, C.; Zhang, Y.; Jia, J.; et al. The Intestinal Microbiome Associated with Lipid Metabolism and Obesity in Humans and Animals. J. Appl. Microbiol. 2022, 133, 2915–2930. [Google Scholar] [CrossRef]
- Kadyan, S.; Park, G.; Singh, T.P.; Patoine, C.; Singar, S.; Heise, T.; Domeier, C.; Ray, C.; Kumar, M.; Behare, P.V.; et al. Microbiome-Based Therapeutics towards Healthier Aging and Longevity. Genome Med. 2025, 17, 75. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Ko, C.-W.; Qu, J.; Wu, D.; Zhu, Q.; Liu, M.; Tso, P. Apolipoprotein A-IV-Deficient Mice in 129/SvJ Background Are Susceptible to Obesity and Glucose Intolerance. Nutrients 2023, 15, 4840. [Google Scholar] [CrossRef]
- Zhu, Y.; Chen, B.; Zhang, X.; Akbar, M.T.; Wu, T.; Zhang, Y.; Zhi, L.; Shen, Q. Exploration of the Muribaculaceae Family in the Gut Microbiota: Diversity, Metabolism, and Function. Nutrients 2024, 16, 2660. [Google Scholar] [CrossRef] [PubMed]
- Hodgkinson, K.; El Abbar, F.; Dobranowski, P.; Manoogian, J.; Butcher, J.; Figeys, D.; Mack, D.; Stintzi, A. Butyrate’s Role in Human Health and the Current Progress towards Its Clinical Application to Treat Gastrointestinal Disease. Clin. Nutr. Edinb. Scotl. 2023, 42, 61–75. [Google Scholar] [CrossRef]
- Mo, C.; Lou, X.; Xue, J.; Shi, Z.; Zhao, Y.; Wang, F.; Chen, G. The Influence of Akkermansia Muciniphila on Intestinal Barrier Function. Gut Pathog. 2024, 16, 41. [Google Scholar] [CrossRef]
- Gao, F.; Cheng, C.; Li, R.; Chen, Z.; Tang, K.; Du, G. The Role of Akkermansia Muciniphila in Maintaining Health: A Bibliometric Study. Front. Med. 2025, 12, 1484656. [Google Scholar] [CrossRef]
- Zeng, S.-Y.; Liu, Y.-F.; Liu, J.-H.; Zeng, Z.-L.; Xie, H.; Liu, J.-H. Potential Effects of Akkermansia Muciniphila in Aging and Aging-Related Diseases: Current Evidence and Perspectives. Aging Dis. 2023, 14, 2015–2027. [Google Scholar] [CrossRef]
- Ghotaslou, R.; Nabizadeh, E.; Memar, M.Y.; Law, W.M.H.; Ozma, M.A.; Abdi, M.; Yekani, M.; Kadkhoda, H.; Hosseinpour, R.; Bafadam, S.; et al. The Metabolic, Protective, and Immune Functions of Akkermansia Muciniphila. Microbiol. Res. 2023, 266, 127245. [Google Scholar] [CrossRef]
- Mruk-Mazurkiewicz, H.; Kulaszyńska, M.; Czarnecka, W.; Podkówka, A.; Ekstedt, N.; Zawodny, P.; Wierzbicka-Woś, A.; Marlicz, W.; Skupin, B.; Stachowska, E.; et al. Insights into the Mechanisms of Action of Akkermansia Muciniphila in the Treatment of Non-Communicable Diseases. Nutrients 2024, 16, 1695. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Qi, N.; Shen, L.; Lo, C.C.; Xu, M.; Duan, Q.; Ollberding, N.J.; Wu, Z.; Hui, D.Y.; Tso, P.; et al. Sexual Dimorphism in Lipid Metabolism and Gut Microbiota in Mice Fed a High-Fat Diet. Nutrients 2023, 15, 2175. [Google Scholar] [CrossRef]
- Kim, C.C.; Lunken, G.R.; Kelly, W.J.; Patchett, M.L.; Jordens, Z.; Tannock, G.W.; Sims, I.M.; Bell, T.J.; Hedderley, D.; Henrissat, B.; et al. Correction: Genomic Insights from Monoglobus Pectinilyticus: A Pectin-Degrading Specialist Bacterium in the Human Colon. ISME J. 2023, 17, 1520. [Google Scholar] [CrossRef]
- Wu, X.; Huang, X.; Ma, W.; Li, M.; Wen, J.; Chen, C.; Liu, L.; Nie, S. Bioactive Polysaccharides Promote Gut Immunity via Different Ways. Food Funct. 2023, 14, 1387–1400. [Google Scholar] [CrossRef]
- Bárcena, C.; Valdés-Mas, R.; Mayoral, P.; Garabaya, C.; Durand, S.; Rodríguez, F.; Fernández-García, M.T.; Salazar, N.; Nogacka, A.M.; Garatachea, N.; et al. Healthspan and Lifespan Extension by Fecal Microbiota Transplantation into Progeroid Mice. Nat. Med. 2019, 25, 1234–1242. [Google Scholar] [CrossRef]
- Enache, R.-M.; Profir, M.; Roşu, O.A.; Creţoiu, S.M.; Gaspar, B.S. The Role of Gut Microbiota in the Onset and Progression of Obesity and Associated Comorbidities. Int. J. Mol. Sci. 2024, 25, 12321. [Google Scholar] [CrossRef]
- Hu, Y.; Peng, J.; Li, F.; Wong, F.S.; Wen, L. Evaluation of Different Mucosal Microbiota Leads to Gut Microbiota-Based Prediction of Type 1 Diabetes in NOD Mice. Sci. Rep. 2018, 8, 15451. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Wu, D.; Ko, C.-W.; Zhu, Q.; Liu, M.; Tso, P. Deficiency of apoA-IV in Female 129X1/SvJ Mice Leads to Diet-Induced Obesity, Insulin Resistance, and Decreased Energy Expenditure. Nutrients 2023, 15, 4655. [Google Scholar] [CrossRef]
- Ghosh, T.S.; Shanahan, F.; O’Toole, P.W. The Gut Microbiome as a Modulator of Healthy Ageing. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 565–584. [Google Scholar] [CrossRef]
- Best, L.; Dost, T.; Esser, D.; Flor, S.; Gamarra, A.M.; Haase, M.; Kadibalban, A.S.; Marinos, G.; Walker, A.; Zimmermann, J.; et al. Metabolic Modelling Reveals the Aging-Associated Decline of Host-Microbiome Metabolic Interactions in Mice. Nat. Microbiol. 2025, 10, 973–991. [Google Scholar] [CrossRef] [PubMed]
- Kohan, A.B.; Wang, F.; Lo, C.-M.; Liu, M.; Tso, P. ApoA-IV: Current and Emerging Roles in Intestinal Lipid Metabolism, Glucose Homeostasis, and Satiety. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 308, G472–G481. [Google Scholar] [CrossRef]
- Hameed, A.; Mojsak, P.; Buczynska, A.; Suleria, H.A.R.; Kretowski, A.; Ciborowski, M. Altered Metabolome of Lipids and Amino Acids Species: A Source of Early Signature Biomarkers of T2DM. J. Clin. Med. 2020, 9, 2257. [Google Scholar] [CrossRef]
- Mansoori, S.; Ho, M.Y.-M.; Ng, K.K.-W.; Cheng, K.K.-Y. Branched-Chain Amino Acid Metabolism: Pathophysiological Mechanism and Therapeutic Intervention in Metabolic Diseases. Obes. Rev. Off. J. Int. Assoc. Study Obes. 2025, 26, e13856. [Google Scholar] [CrossRef] [PubMed]
- Khaledi, M.; Poureslamfar, B.; Alsaab, H.O.; Tafaghodi, S.; Hjazi, A.; Singh, R.; Alawadi, A.H.; Alsaalamy, A.; Qasim, Q.A.; Sameni, F. The Role of Gut Microbiota in Human Metabolism and Inflammatory Diseases: A Focus on Elderly Individuals. Ann. Microbiol. 2024, 74, 1. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeber-Lubecka, N.; Kulecka, M.; Balabas, A.; Czarnowski, P.; Pyśniak, K.; Dąbrowska, M.; Ostrowski, J.; Hennig, E.E. Lifetime Changes in Gut Microbiota and Metabolite Composition in High-Fat Diet-Induced Obesity in Apolipoprotein A-IV Gene Knockout Mice. Biology 2025, 14, 1278. https://doi.org/10.3390/biology14091278
Zeber-Lubecka N, Kulecka M, Balabas A, Czarnowski P, Pyśniak K, Dąbrowska M, Ostrowski J, Hennig EE. Lifetime Changes in Gut Microbiota and Metabolite Composition in High-Fat Diet-Induced Obesity in Apolipoprotein A-IV Gene Knockout Mice. Biology. 2025; 14(9):1278. https://doi.org/10.3390/biology14091278
Chicago/Turabian StyleZeber-Lubecka, Natalia, Maria Kulecka, Aneta Balabas, Pawel Czarnowski, Kazimiera Pyśniak, Michalina Dąbrowska, Jerzy Ostrowski, and Ewa E. Hennig. 2025. "Lifetime Changes in Gut Microbiota and Metabolite Composition in High-Fat Diet-Induced Obesity in Apolipoprotein A-IV Gene Knockout Mice" Biology 14, no. 9: 1278. https://doi.org/10.3390/biology14091278
APA StyleZeber-Lubecka, N., Kulecka, M., Balabas, A., Czarnowski, P., Pyśniak, K., Dąbrowska, M., Ostrowski, J., & Hennig, E. E. (2025). Lifetime Changes in Gut Microbiota and Metabolite Composition in High-Fat Diet-Induced Obesity in Apolipoprotein A-IV Gene Knockout Mice. Biology, 14(9), 1278. https://doi.org/10.3390/biology14091278






