Hepatocyte-Specific Transcriptional Responses to Liver-Targeted Delivery of a Soluble Epoxide Hydrolase Inhibitor in a Mouse Model of Alcohol-Associated Liver Disease
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of t-TUCB Fusogenic Lipid Vesicles (t-TUCB-FLVs)
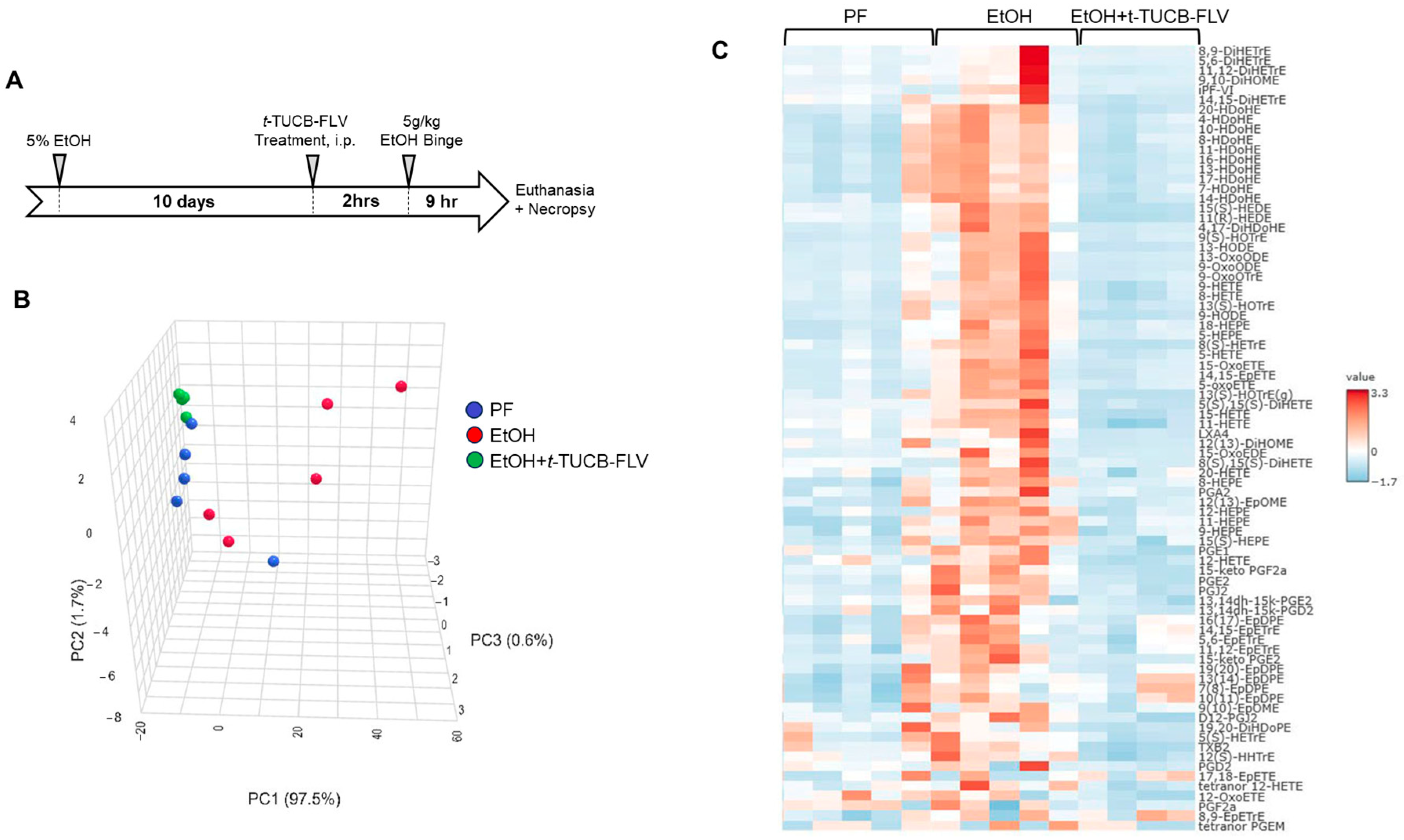
2.2. Animal Studies
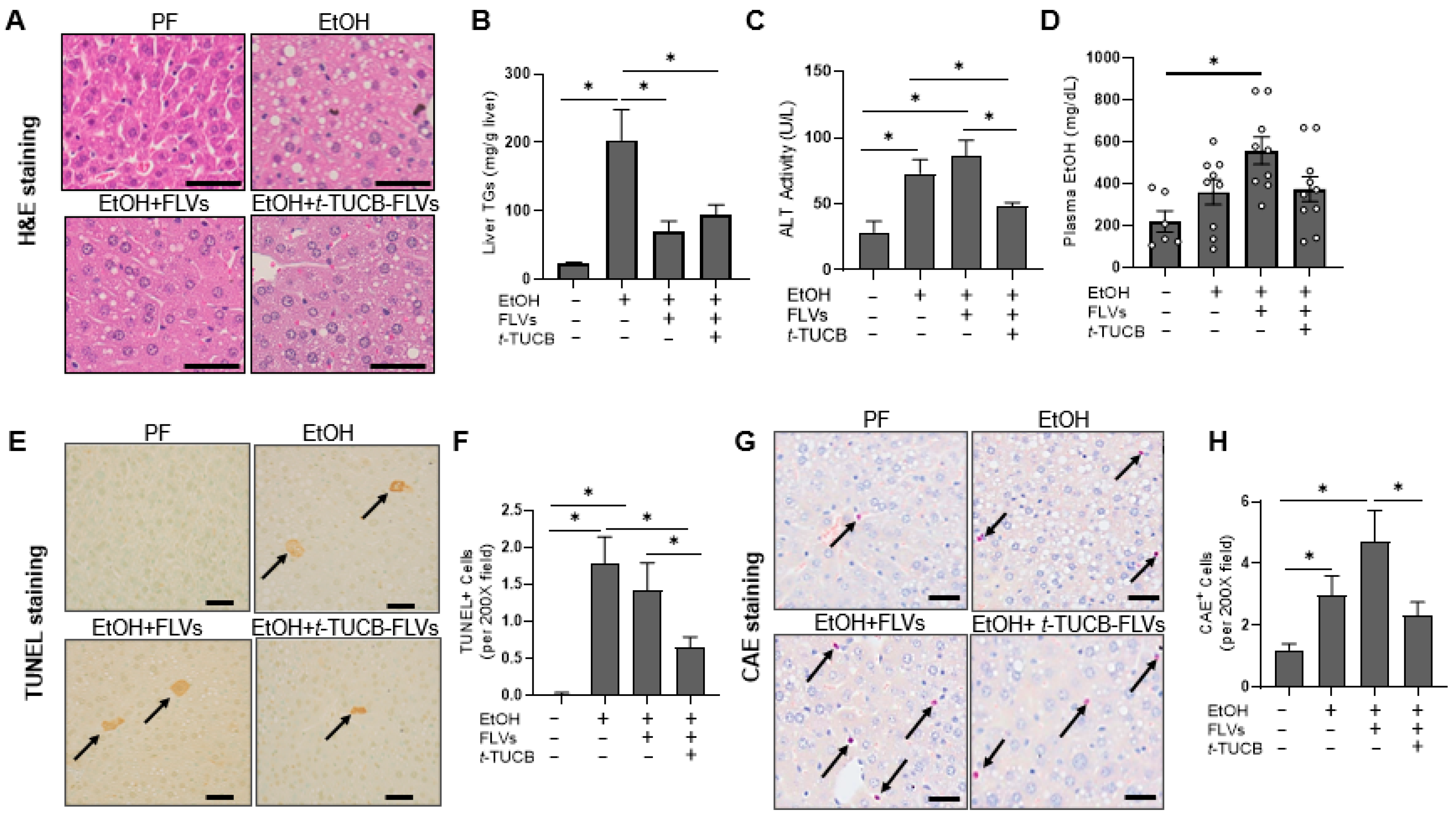
2.3. Assessment of Liver Damage
2.4. Measurement of Plasma EtOH Concentration
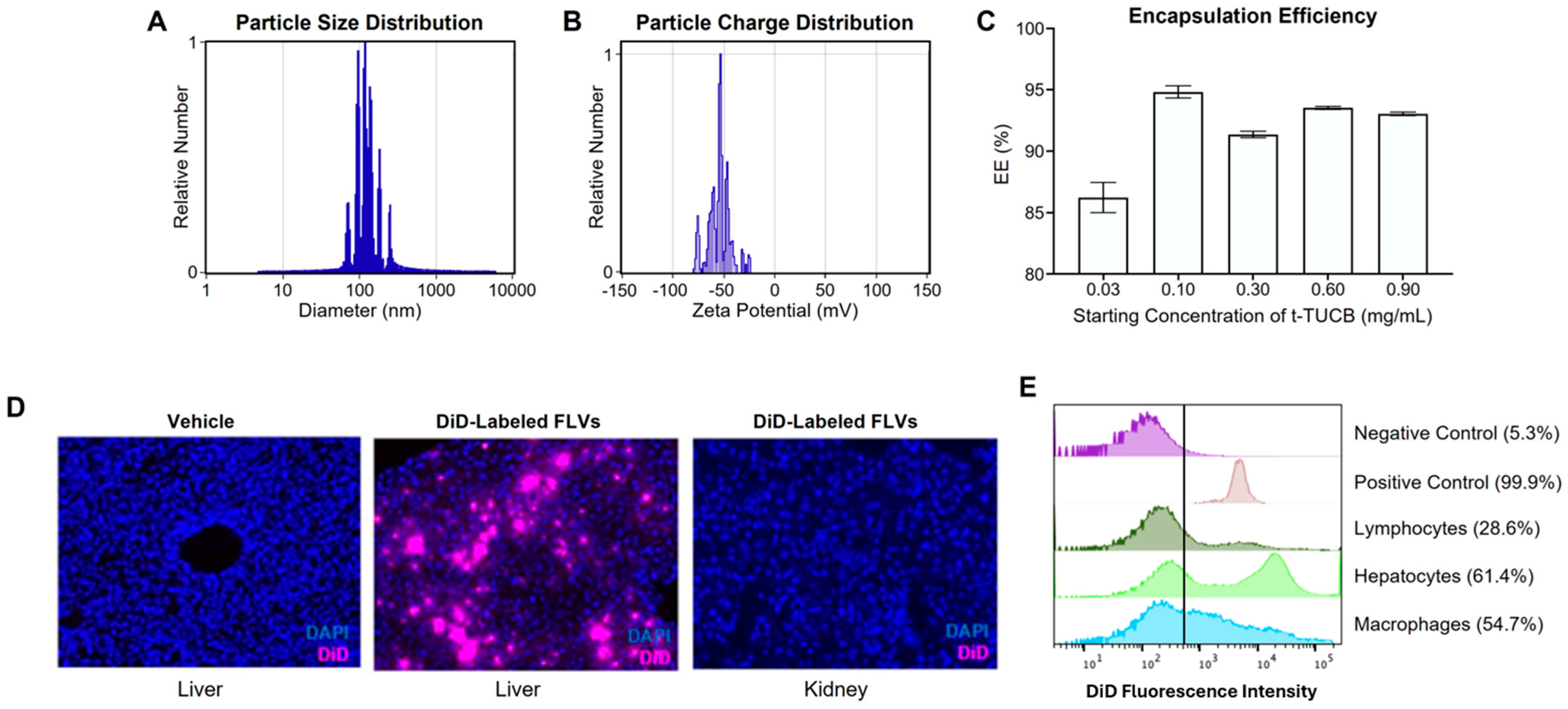
2.5. t-TUCB-FLV Tissue and Cell Distribution Visualization
2.6. Liver Lipidomics and Triglyceride (TG) Analyses
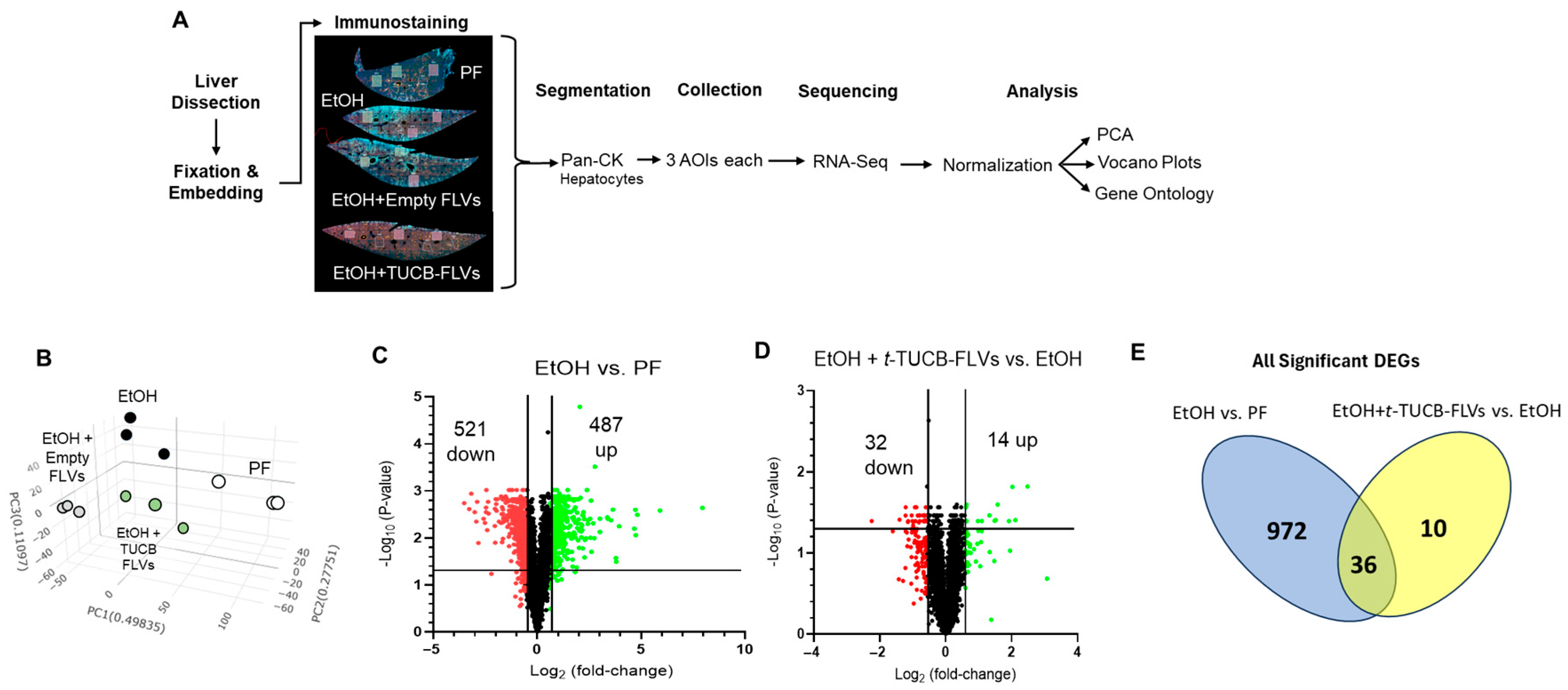
2.7. NanoString Liver Spatial Transcriptomics Analysis
2.8. Statistical Analysis
3. Results
3.1. Characterization of a Liver-Targeted s-EH Inhibitor Delivery Platform
3.2. Effects of t-TUCB-FLVs on an Animal Model of ALD
3.2.1. t-TUCB-FLVs Induce Significant Changes in the Hepatic Lipidomic Profile
3.2.2. t-TUCB-FLVs Attenuated EtOH-Induced Liver Damage
3.3. Hepatocyte-Specific Transcriptional Responses to EtOH and t-TUCB-FLVs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALD | Alcohol-associated liver disease |
| s-EH | Soluble epoxide hydrolase |
| EtOH | Ethanol |
| t-TUCB | trans-4-[4-(3-adamantan-1-yl-ureido)-cyclohexyloxy]-benzoic acid |
| Ep-FAs | Epoxy fatty acids |
| Dh-FAs | Dihydroxy fatty acids |
| PUFAs | Polyunsaturated fatty acids |
| DOPC | 1,2-dioleoyl-sn-glycero-3-phosphocholine |
| POPA | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate |
| FLVs | Fusogenic lipid vesicles |
| DiD | 1,1′-Dioctadecyl-3,3,3′,3′-Tetramethylindodicarbocyanine, 4-Chlorobenzenesulfonate |
| PF | Pair fed |
| ALT | Alanine aminotransferase |
| TUNEL | Terminal deoxynucleotidyl transferase dUTP nick end labeling |
| CAE | Chloroacetate esterase |
| TGs | Triglycerides |
| DSP | Digital Spatial Profiling |
| AOI | Areas of Interest |
| WTA | Whole transcriptome analysis |
| GSEA | Gene Set Enrichment Analysis |
| GO | Gene Ontology |
| SEM | Standard error of the mean |
| ANOVA | Analysis of variance |
| DEG | Differentially expressed gene |
References
- Tan, H.K.; Yates, E.; Lilly, K.; Dhanda, A.D. Oxidative stress in alcohol-related liver disease. World J. Hepatol. 2020, 12, 332–349. [Google Scholar] [CrossRef]
- Seitz, H.K.; Bataller, R.; Cortez-Pinto, H.; Gao, B.; Gual, A.; Lackner, C.; Mathurin, P.; Mueller, S.; Szabo, G.; Tsukamoto, H. Alcoholic liver disease. Nat. Rev. Dis. Primers 2018, 4, 16. [Google Scholar] [CrossRef] [PubMed]
- Warner, D.R.; Warner, J.B.; Abdelfadil, Y.; Hardesty, J.E.; Treves, R.; Lei, C.; Hanford, H.E.; McClain, C.J.; Kirpich, I.A. Effects of soluble epoxide hydrolase inhibition on liver injury and gut microbiota in mice chronically fed ethanol. Alcohol Clin. Exp. Res. 2025, 49, 1730–1743. [Google Scholar] [CrossRef] [PubMed]
- Warner, J.B.; Hardesty, J.E.; Song, Y.L.; Floyd, A.T.; Deng, Z.; Jebet, A.; He, L.; Zhang, X.; McClain, C.J.; Hammock, B.D.; et al. Hepatic Transcriptome and Its Regulation Following Soluble Epoxide Hydrolase Inhibition in Alcohol-Associated Liver Disease. Am. J. Pathol. 2024, 194, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Mello, A.; Hsu, M.F.; Koike, S.; Chu, B.; Cheng, J.; Yang, J.; Morisseau, C.; Torok, N.J.; Hammock, B.D.; Haj, F.G. Soluble Epoxide Hydrolase Hepatic Deficiency Ameliorates Alcohol-Associated Liver Disease. Cell. Mol. Gastroenterol. Hepatol. 2021, 11, 815–830. [Google Scholar] [CrossRef] [PubMed]
- Node, K.; Huo, Y.; Ruan, X.; Yang, B.; Spiecker, M.; Ley, K.; Zeldin, D.C.; Liao, J.K. Anti-inflammatory properties of cytochrome P450 epoxygenase-derived eicosanoids. Science 1999, 285, 1276–1279. [Google Scholar] [CrossRef]
- Lopez-Vicario, C.; Alcaraz-Quiles, J.; Garcia-Alonso, V.; Rius, B.; Hwang, S.H.; Titos, E.; Lopategi, A.; Hammock, B.D.; Arroyo, V.; Claria, J. Inhibition of soluble epoxide hydrolase modulates inflammation and autophagy in obese adipose tissue and liver: Role for omega-3 epoxides. Proc. Natl. Acad. Sci. USA 2015, 112, 536–541. [Google Scholar] [CrossRef]
- Gilroy, D.W.; Edin, M.L.; De Maeyer, R.P.; Bystrom, J.; Newson, J.; Lih, F.B.; Stables, M.; Zeldin, D.C.; Bishop-Bailey, D. CYP450-derived oxylipins mediate inflammatory resolution. Proc. Natl. Acad. Sci. USA 2016, 113, E3240–E3249. [Google Scholar] [CrossRef]
- Panigrahy, D.; Kalish, B.T.; Huang, S.; Bielenberg, D.R.; Le, H.D.; Yang, J.; Edin, M.L.; Lee, C.R.; Benny, O.; Mudge, D.K.; et al. Epoxyeicosanoids promote organ and tissue regeneration. Proc. Natl. Acad. Sci. USA 2013, 110, 13528–13533. [Google Scholar] [CrossRef]
- Wang, W.; Yang, J.; Zhang, J.; Wang, Y.; Hwang, S.H.; Qi, W.; Wan, D.; Kim, D.; Sun, J.; Sanidad, K.Z.; et al. Lipidomic profiling reveals soluble epoxide hydrolase as a therapeutic target of obesity-induced colonic inflammation. Proc. Natl. Acad. Sci. USA 2018, 115, 5283–5288. [Google Scholar] [CrossRef]
- Zhang, G.; Kodani, S.; Hammock, B.D. Stabilized epoxygenated fatty acids regulate inflammation, pain, angiogenesis and cancer. Prog. Lipid Res. 2014, 53, 108–123. [Google Scholar] [CrossRef]
- Samokhvalov, V.; Jamieson, K.L.; Darwesh, A.M.; Keshavarz-Bahaghighat, H.; Lee, T.Y.T.; Edin, M.; Lih, F.; Zeldin, D.C.; Seubert, J.M. Deficiency of Soluble Epoxide Hydrolase Protects Cardiac Function Impaired by LPS-Induced Acute Inflammation. Front. Pharmacol. 2018, 9, 1572. [Google Scholar] [CrossRef]
- Bettaieb, A.; Koike, S.; Chahed, S.; Zhao, Y.; Bachaalany, S.; Hashoush, N.; Graham, J.; Fatima, H.; Havel, P.J.; Gruzdev, A.; et al. Podocyte-specific soluble epoxide hydrolase deficiency in mice attenuates acute kidney injury. FEBS J. 2017, 284, 1970–1986. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Cai, H.; Song, J.; Chang, Q. The effects of sEH inhibitor on depression-like behavior and neurogenesis in male mice. J. Neurosci. Res. 2017, 95, 2483–2492. [Google Scholar] [CrossRef]
- Chen, Z.; Tang, Y.; Yu, J.; Dong, R.; Yang, Y.; Fu, M.; Luo, J.; Hu, S.; Wang, D.W.; Tu, L.; et al. sEH Inhibitor TPPU Ameliorates Cecal Ligation and Puncture-Induced Sepsis by Regulating Macrophage Functions. Shock 2019, 53, 761–771. [Google Scholar] [CrossRef]
- Guo, Y.; Luo, F.; Zhang, X.; Chen, J.; Shen, L.; Zhu, Y.; Xu, D. TPPU enhanced exercise-induced epoxyeicosatrienoic acid concentrations to exert cardioprotection in mice after myocardial infarction. J. Cell. Mol. Med. 2018, 22, 1489–1500. [Google Scholar] [CrossRef]
- Bettaieb, A.; Nagata, N.; AbouBechara, D.; Chahed, S.; Morisseau, C.; Hammock, B.D.; Haj, F.G. Soluble epoxide hydrolase deficiency or inhibition attenuates diet-induced endoplasmic reticulum stress in liver and adipose tissue. J. Biol. Chem. 2013, 288, 14189–14199. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Hu, T.; Xiong, W.; Jiang, X.; Cao, Y.; Li, Z.; Jiang, H.; Wang, X. Soluble epoxide hydrolase deficiency promotes liver regeneration and ameliorates liver injury in mice by regulating angiocrine factors and angiogenesis. Biochim. Biophys. Acta Gen. Subj. 2023, 1867, 130394. [Google Scholar] [CrossRef]
- Zhang, C.Y.; Tan, X.H.; Yang, H.H.; Jin, L.; Hong, J.R.; Zhou, Y.; Huang, X.T. COX-2/sEH Dual Inhibitor Alleviates Hepatocyte Senescence in NAFLD Mice by Restoring Autophagy through Sirt1/PI3K/AKT/mTOR. Int. J. Mol. Sci. 2022, 23, 8267. [Google Scholar] [CrossRef] [PubMed]
- Dang, J.; Cai, T.; Tuo, Y.; Peng, S.; Wang, J.; Gu, A.; Li, J.; Ding, L.; Du, S.; Wang, L. Corn Peptides Alleviate Nonalcoholic Fatty Liver Fibrosis in Mice by Inhibiting NLRP3 Inflammasome Activation and Regulating Gut Microbiota. J. Agric. Food Chem. 2024, 72, 19378–19394. [Google Scholar] [CrossRef] [PubMed]
- Helmstadter, M.; Schmidt, J.; Kaiser, A.; Weizel, L.; Proschak, E.; Merk, D. Differential Therapeutic Effects of FXR Activation, sEH Inhibition, and Dual FXR/sEH Modulation in NASH in Diet-Induced Obese Mice. ACS Pharmacol. Transl. Sci. 2021, 4, 966–979. [Google Scholar] [CrossRef]
- Warner, J.; Hardesty, J.; Zirnheld, K.; McClain, C.; Warner, D.; Kirpich, I. Soluble Epoxide Hydrolase Inhibition in Liver Diseases: A Review of Current Research and Knowledge Gaps. Biology 2020, 9, 124. [Google Scholar] [CrossRef] [PubMed]
- Kodani, S.D.; Bhakta, S.; Hwang, S.H.; Pakhomova, S.; Newcomer, M.E.; Morisseau, C.; Hammock, B.D. Identification and optimization of soluble epoxide hydrolase inhibitors with dual potency towards fatty acid amide hydrolase. Bioorg. Med. Chem. Lett. 2018, 28, 762–768. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Zhu, Y.; Lin, J.; Zheng, L.; Zhang, C.; Luo, M. Inhibition of soluble epoxide hydrolase lowers portal hypertension in cirrhotic rats by ameliorating endothelial dysfunction and liver fibrosis. Prostaglandins Other Lipid Mediat. 2017, 131, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.H.; Zheng, L.; Gui, L.; Lin, J.Y.; Zhu, Y.M.; Deng, W.S.; Luo, M. Soluble epoxide hydrolase inhibition with t-TUCB alleviates liver fibrosis and portal pressure in carbon tetrachloride-induced cirrhosis in rats. Clin. Res. Hepatol. Gastroenterol. 2018, 42, 118–125. [Google Scholar] [CrossRef]
- Kirpich, I.A.; Gobejishvili, L.N.; Bon Homme, M.; Waigel, S.; Cave, M.; Arteel, G.; Barve, S.S.; McClain, C.J.; Deaciuc, I.V. Integrated hepatic transcriptome and proteome analysis of mice with high-fat diet-induced nonalcoholic fatty liver disease. J. Nutr. Biochem. 2011, 22, 38–45. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstrale, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef]
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Adler, P.; Vilo, J.; Peterson, H. g:Profiler-interoperable web service for functional enrichment analysis and gene identifier mapping (2023 update). Nucleic Acids Res. 2023, 51, W207–W212. [Google Scholar] [CrossRef]
- Rodriguez, W.E.; Wahlang, B.; Wang, Y.; Zhang, J.; Vadhanam, M.V.; Joshi-Barve, S.; Bauer, P.; Cannon, R.; Ahmadi, A.R.; Sun, Z.; et al. Phosphodiesterase 4 Inhibition as a Therapeutic Target for Alcoholic Liver Disease: From Bedside to Bench. Hepatology 2019, 70, 1958–1971. [Google Scholar] [CrossRef]
- Miyata, T.; Nagy, L.E. Programmed cell death in alcohol-associated liver disease. Clin. Mol. Hepatol. 2020, 26, 618–625. [Google Scholar] [CrossRef]
- Yu, J.H.; Choi, M.G.; Lee, N.Y.; Kwon, A.; Lee, E.; Koo, J.H. Hepatocyte GPCR signaling regulates IRF3 to control hepatic stellate cell transdifferentiation. Cell Commun. Signal. 2024, 22, 48. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, N.; Yan, K.; Gandrass, M.; Muller, M.; Krisp, C.; Hasler, R.; Carambia, A.; Nofer, J.R.; Bernardes, J.P.; Khouja, M.; et al. Cell-autonomous hepatocyte-specific GP130 signaling is sufficient to trigger a robust innate immune response in mice. J. Hepatol. 2021, 74, 407–418. [Google Scholar] [CrossRef]
- Hwang, S.H.; Wagner, K.M.; Yang, J.; Morrisseau, C.; Wan, D.; Fettinger, J.C.; Olmstead, M.M.; Hammock, B.D. Coamorphous Solid Dispersion of a Soluble Epoxide Hydrolase Inhibitor t-TUCB with Amino Acid L-Arginine. Org. Process Res. Dev. 2025, 29, 1523–1530. [Google Scholar] [CrossRef]
- Warner, J.B.; Guenthner, S.C.; Hardesty, J.E.; McClain, C.J.; Warner, D.R.; Kirpich, I.A. Liver-specific drug delivery platforms: Applications for the treatment of alcohol-associated liver disease. World J. Gastroenterol. 2022, 28, 5280–5299. [Google Scholar] [CrossRef]
- Satishchandran, A.; Ambade, A.; Rao, S.; Hsueh, Y.C.; Iracheta-Vellve, A.; Tornai, D.; Lowe, P.; Gyongyosi, B.; Li, J.; Catalano, D.; et al. MicroRNA 122, Regulated by GRLH2, Protects Livers of Mice and Patients From Ethanol-Induced Liver Disease. Gastroenterology 2018, 154, 238–252.e237. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, G.; Perriotte-Olson, C.; Casey, C.A.; Donohue, T.M., Jr.; Talmon, G.A.; Harris, E.N.; Kabanov, A.V.; Saraswathi, V. Effect of nanoformulated copper/zinc superoxide dismutase on chronic ethanol-induced alterations in liver and adipose tissue. Alcohol 2019, 79, 71–79. [Google Scholar] [CrossRef]
- Hendrikx, T.; Duan, Y.; Wang, Y.; Oh, J.H.; Alexander, L.M.; Huang, W.; Starkel, P.; Ho, S.B.; Gao, B.; Fiehn, O.; et al. Bacteria engineered to produce IL-22 in intestine induce expression of REG3G to reduce ethanol-induced liver disease in mice. Gut 2019, 68, 1504–1515. [Google Scholar] [CrossRef]
- Luria, A.; Morisseau, C.; Tsai, H.J.; Yang, J.; Inceoglu, B.; De Taeye, B.; Watkins, S.M.; Wiest, M.M.; German, J.B.; Hammock, B.D. Alteration in plasma testosterone levels in male mice lacking soluble epoxide hydrolase. Am. J. Physiol. Endocrinol. Metab. 2009, 297, E375–E383. [Google Scholar] [CrossRef]
- Zuloaga, K.L.; Zhang, W.; Roese, N.E.; Alkayed, N.J. Soluble epoxide hydrolase gene deletion improves blood flow and reduces infarct size after cerebral ischemia in reproductively senescent female mice. Front. Pharmacol. 2014, 5, 290. [Google Scholar] [CrossRef]
- Lazaar, A.L.; Yang, L.; Boardley, R.L.; Goyal, N.S.; Robertson, J.; Baldwin, S.J.; Newby, D.E.; Wilkinson, I.B.; Tal-Singer, R.; Mayer, R.J.; et al. Pharmacokinetics, pharmacodynamics and adverse event profile of GSK2256294, a novel soluble epoxide hydrolase inhibitor. Br. J. Clin. Pharmacol. 2016, 81, 971–979. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Deng, Z.B.; Mu, J.; Zhang, L.; Yan, J.; Miller, D.; Feng, W.; McClain, C.J.; Zhang, H.G. Ginger-derived nanoparticles protect against alcohol-induced liver damage. J. Extracell. Vesicles 2015, 4, 28713. [Google Scholar] [CrossRef]
- Landgraf, D.; Wang, L.L.; Diemer, T.; Welsh, D.K. NPAS2 Compensates for Loss of CLOCK in Peripheral Circadian Oscillators. PLoS Genet. 2016, 12, e1005882. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.T., IV; Voigt, R.M.; Shaikh, M.; Forsyth, C.B.; Keshavarzian, A. Circadian Mechanisms in Alcohol Use Disorder and Tissue Injury. Alcohol. Clin. Exp. Res. 2018, 42, 668–677. [Google Scholar] [CrossRef]
- Yang, T.; Yuan, P.; Yang, Y.; Liang, N.; Wang, Q.; Li, J.; Lu, R.; Zhang, H.; Mu, J.; Yan, Z.; et al. NPAS2 Contributes to Liver Fibrosis by Direct Transcriptional Activation of Hes1 in Hepatic Stellate Cells. Mol. Ther. Nucleic Acids 2019, 18, 1009–1022. [Google Scholar] [CrossRef]
- Gratton, M.O.; Torban, E.; Jasmin, S.B.; Theriault, F.M.; German, M.S.; Stifani, S. Hes6 promotes cortical neurogenesis and inhibits Hes1 transcription repression activity by multiple mechanisms. Mol. Cell. Biol. 2003, 23, 6922–6935. [Google Scholar] [CrossRef]
- Park, J.E.; Lee, M.; Kim, S.C.; Zhang, Y.; Hardwick, J.P.; Lee, Y.K. Hairy and enhancer of split 6 prevents hepatic lipid accumulation through inhibition of Pparg2 expression. Hepatol. Commun. 2017, 1, 1085–1098. [Google Scholar] [CrossRef][Green Version]
- Wang, Y.; Wang, P.; Yu, Y.; Huang, E.; Yao, Y.; Guo, D.; Peng, H.; Tian, B.; Zheng, Q.; Jia, M.; et al. Hepatocyte Ninjurin2 promotes hepatic stellate cell activation and liver fibrosis through the IGF1R/EGR1/PDGF-BB signaling pathway. Metabolism 2023, 140, 155380. [Google Scholar] [CrossRef]
- Thakur, V.; McMullen, M.R.; Pritchard, M.T.; Nagy, L.E. Regulation of macrophage activation in alcoholic liver disease. J. Gastroenterol. Hepatol. 2007, 22 (Suppl. 1), S53–S56. [Google Scholar] [CrossRef]
- McMullen, M.R.; Pritchard, M.T.; Wang, Q.; Millward, C.A.; Croniger, C.M.; Nagy, L.E. Early growth response-1 transcription factor is essential for ethanol-induced fatty liver injury in mice. Gastroenterology 2005, 128, 2066–2076. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y. Erdr1 Drives Macrophage Programming via Dynamic Interplay with YAP1 and Mid1. Immunohorizons 2024, 8, 198–213. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, H.; Tsubota, T.; Kanki, K.; Shiota, G. All-trans retinoic acid ameliorates hepatic stellate cell activation via suppression of thioredoxin interacting protein expression. J. Cell. Physiol. 2018, 233, 607–616. [Google Scholar] [CrossRef]
- Choi, E.H.; Park, S.J. TXNIP: A key protein in the cellular stress response pathway and a potential therapeutic target. Exp. Mol. Med. 2023, 55, 1348–1356. [Google Scholar] [CrossRef]
- Batista, T.M.; Dagdeviren, S.; Carroll, S.H.; Cai, W.; Melnik, V.Y.; Noh, H.L.; Saengnipanthkul, S.; Kim, J.K.; Kahn, C.R.; Lee, R.T. Arrestin domain-containing 3 (Arrdc3) modulates insulin action and glucose metabolism in liver. Proc. Natl. Acad. Sci. USA 2020, 117, 6733–6740. [Google Scholar] [CrossRef]
- Tang, Y.; Zhou, H.; Pan, X.; Zhong, Z.; Liu, H.; Guo, Y. Arrestin domain containing 3 promotes alcohol-induced liver steatosis by reducing stearoyl-CoA desaturase-1 ubiquitinated degradation. Metabolism 2025, 167, 156175. [Google Scholar] [CrossRef]
- Herrera-Lopez, E.E.; Guerrero-Escalera, D.; Aguirre-Maldonado, I.; Lopez-Hernandez, A.; Montero, H.; Gutierrez-Nava, M.A.; Del Pozo-Yauner, L.; Arellanes-Robledo, J.; Camacho, J.; Perez-Carreon, J.I. Annexins A2 and A5 are potential early biomarkers of hepatocarcinogenesis. Sci. Rep. 2023, 13, 6948. [Google Scholar] [CrossRef] [PubMed]
- Fulham, M.A.; Mandrekar, P. Sexual Dimorphism in Alcohol Induced Adipose Inflammation Relates to Liver Injury. PLoS ONE 2016, 11, e0164225. [Google Scholar] [CrossRef] [PubMed]

| Lipid | EtOH vs. PF | EtOH+t-TUCB-FLV vs. EtOH | |||
|---|---|---|---|---|---|
| Fold-Change | p-Value | Fold-Change | p-Value | ||
| PGE1 | 1.75 | 0.095 | −4.19 | 0.007 | |
| PGE2 | 2.53 | 0.004 | −4.13 | 0.001 | |
| 15-keto PGE2 | 8.85 | 0.012 | −25.32 | 0.011 | |
| 13,14dh-15k-PGE2 | 2.39 | <0.001 | −4.06 | <0.001 | |
| PGA2 | 1.67 | 0.603 | −3.39 | 0.195 | |
| tetranor PGEM | 1.25 | 0.686 | −1.33 | 0.596 | |
| PGD2 | 1.81 | 0.178 | −3.05 | 0.049 | |
| PGJ2 | 2.32 | 0.066 | −4.10 | 0.022 | |
| D12-PGJ2 | 2.29 | 0.025 | −3.20 | 0.037 | |
| 13,14dh-15k-PGD2 | 1.81 | 0.107 | −2.29 | 0.053 | |
| PGF2a | 1.21 | 0.684 | −1.93 | 0.107 | |
| 15-keto PGF2a | 2.11 | 0.015 | −4.22 | 0.002 | |
| iPF-VI | 5.11 | 0.216 | −11.50 | 0.412 | |
| TXB2 | 1.35 | 0.394 | −5.63 | 0.005 | |
| 5(S),15(S)-DiHETE | 2.80 | 0.145 | −5.60 | 0.312 | |
| 8(S),15(S)-DiHETE | 4.33 | 0.079 | −10.11 | 0.084 | |
| 9-HODE | 1.89 | 0.043 | −3.54 | 0.005 | |
| 13-HODE | 2.39 | 0.033 | −4.70 | 0.008 | |
| 9(S)-HOTrE | 3.64 | 0.048 | −5.73 | 0.034 | |
| 13(S)-HOTrE | 2.34 | 0.081 | −4.89 | 0.023 | |
| 11(R)-HEDE | 2.15 | 0.005 | −5.67 | <0.001 | |
| 15(S)-HEDE | 2.54 | 0.007 | −8.87 | 0.001 | |
| 8(S)-HETrE | 2.54 | 0.010 | −7.07 | 0.001 | |
| 5(S)-HETrE | 1.19 | 0.865 | −3.24 | 0.138 | |
| 5-HETE | 2.92 | 0.034 | −4.73 | 0.018 | |
| 8-HETE | 1.83 | 0.023 | −2.86 | 0.004 | |
| 9-HETE | 2.12 | 0.008 | −3.28 | 0.002 | |
| 11-HETE | 1.38 | 0.007 | −2.08 | <0.001 | |
| 12-HETE | 1.86 | 0.085 | −4.92 | 0.007 | |
| 15-HETE | 1.87 | 0.023 | −3.69 | 0.002 | |
| 20-HETE | 1.75 | 0.107 | −2.01 | 0.072 | |
| tetranor 12-HETE | 2.04 | 0.178 | −3.60 | 0.090 | |
| 12(S)-HHTrE | 1.36 | 0.115 | −2.30 | 0.003 | |
| 5-HEPE | 4.64 | 0.008 | −4.37 | 0.013 | |
| 8-HEPE | 2.50 | 0.028 | −2.56 | 0.026 | |
| 9-HEPE | 2.14 | 0.022 | −3.06 | 0.006 | |
| 11-HEPE | 2.14 | 0.010 | −1.83 | 0.035 | |
| 12-HEPE | 3.16 | <0.001 | −3.50 | <0.001 | |
| 15(S)-HEPE | 2.00 | 0.116 | −2.50 | 0.101 | |
| 18-HEPE | 3.54 | 0.006 | −3.25 | 0.011 | |
| 4-HDoHE | 3.53 | 0.004 | −4.04 | 0.004 | |
| 7-HDoHE | 2.37 | 0.034 | −3.24 | 0.018 | |
| 8-HDoHE | 2.49 | 0.012 | −2.95 | 0.009 | |
| 10-HDoHE | 2.47 | 0.009 | −3.33 | 0.004 | |
| 11-HDoHE | 2.31 | 0.015 | −3.45 | 0.005 | |
| 13-HDoHE | 2.00 | 0.031 | −2.89 | 0.010 | |
| 14-HDoHE | 2.37 | 0.009 | −4.09 | 0.002 | |
| 16-HDoHE | 2.38 | 0.008 | −3.39 | 0.003 | |
| 17-HDoHE | 2.14 | 0.033 | −3.53 | 0.009 | |
| 20-HDoHE | 2.51 | 0.005 | −4.46 | 0.001 | |
| s-EH Substrates and Products | 9(10)-EpOME | 1.38 | 0.731 | −2.26 | 0.349 |
| 12(13)-EpOME | 2.13 | 0.175 | −3.84 | 0.065 | |
| 5(6)-EpETrE | 1.63 | 0.286 | −1.84 | 0.222 | |
| 8(9)-EpETrE | 1.79 | 0.320 | −1.04 | 0.992 | |
| 11(12)-EpETrE | 2.24 | 0.073 | −2.23 | 0.075 | |
| 14(15)-EpETrE | 1.91 | 0.159 | −1.60 | 0.335 | |
| 14(15)-EpETE | 5.00 | 0.029 | −4.69 | 0.102 | |
| 17(18)-EpETE | 1.17 | 0.958 | −1.05 | 0.995 | |
| 7(8)-EpDPE | 1.60 | 0.506 | −0.88 | 0.923 | |
| 10(11)-EpDPE | 1.86 | 0.329 | −1.13 | 0.930 | |
| 13(14)-EpDPE | 1.57 | 0.564 | −1.03 | 0.997 | |
| 16(17)-EpDPE | 2.58 | 0.113 | −2.03 | 0.238 | |
| 19(20)-EpDPE | 1.55 | 0.583 | −2.00 | 0.401 | |
| 9(10)-DiHOME | 3.78 | 0.345 | −13.34 | 0.237 | |
| 12(13)-DiHOME | 1.23 | 0.928 | −7.76 | 0.280 | |
| 5(6)-DiHETrE | 7.29 | 0.240 | −18.83 | 0.220 | |
| 8(9)-DiHETrE | 4.15 | 0.310 | −9.97 | 0.240 | |
| 11(12)-DiHETrE | 2.51 | 0.303 | −5.97 | 0.150 | |
| 14(15)-DiHETrE | 1.40 | 0.667 | −3.82 | 0.129 | |
| 19(20)-DiHDoPE | −1.22 | 0.860 | −4.18 | 0.243 | |
| 9-OxoODE | 4.39 | 0.046 | −14.54 | 0.024 | |
| 13-OxoODE | 6.03 | 0.038 | −16.41 | 0.028 | |
| 9-OxoOTrE | 3.72 | 0.102 | −8.87 | 0.059 | |
| 15-OxoEDE | 5.95 | 0.072 | −18.75 | 0.077 | |
| 5-oxoETE | 7.58 | 0.009 | −6.97 | 0.014 | |
| 12-OxoETE | 1.03 | 0.994 | −3.69 | 0.221 | |
| 15-OxoETE | 4.41 | 0.011 | −5.41 | 0.011 | |
| LXA4 | 3.37 | 0.207 | −13.40 | 0.373 | |
| 4,17-DiHDoHE | 8.20 | 0.017 | −17.57 | 0.026 | |
| 9(10)EpOME:DiHOME | −1.45 | 0.649 | 3.24 | 0.003 | |
| 12(13)EpOME:DiHOME | 2.65 | 0.028 | 1.33 | 0.329 | |
| 5(6)-EpETrE:DiHETrE | −2.13 | 0.418 | 4.50 | 0.006 | |
| 8(9)-EpETrE:DiHETrE | −1.17 | 0.924 | 3.93 | <0.001 | |
| 11(12)-EpETrE:DiHETrE | 1.48 | 0.551 | 1.71 | 0.093 | |
| 14(15)-EpETrE:DiHETrE | 1.90 | 0.149 | 1.73 | 0.031 | |
| 19(20)-EpDPE:DiHDoPE | 2.70 | 0.053 | 1.73 | 0.034 | |
| All Ep-FAs:Diols | 1.03 | 0.995 | 1.83 | 0.003 | |
| EtOH vs. PF | EtOH+t-TUCB FLV vs. EtOH | |||
|---|---|---|---|---|
| Gene | log2FC | p Adj | log2FC | p Adj |
| Up-regulated by EtOH and Down-regulated by t-TUCB-FLVs | ||||
| Aass | 2.086 | 0.001 | −0.661 | 0.034 |
| Arl6ip5 | 0.981 | 0.004 | −0.675 | 0.040 |
| Crcp | 1.074 | 0.002 | −0.678 | 0.040 |
| Dop1b | 1.170 | 0.006 | −0.990 | 0.046 |
| Egr1 | 3.553 | 0.004 | −2.248 | 0.040 |
| Errfi1 | 0.610 | 0.009 | −0.689 | 0.034 |
| Gm3776 | 1.039 | 0.005 | −1.218 | 0.035 |
| Gstt3 | 1.443 | 0.004 | −1.013 | 0.040 |
| Il1r1 | 1.897 | 0.001 | −0.800 | 0.034 |
| Klf10 | 1.169 | 0.004 | −1.033 | 0.034 |
| Mt2 | 7.955 | 0.002 | −1.142 | 0.040 |
| Npas2 | 0.701 | 0.011 | −0.620 | 0.039 |
| Pctp | 1.247 | 0.001 | −0.632 | 0.041 |
| Ppp1r3b | 0.900 | 0.002 | −0.935 | 0.027 |
| Sgk2 | 1.093 | 0.003 | −0.791 | 0.040 |
| Slc10a2 | 0.683 | 0.003 | −0.787 | 0.034 |
| Slc39a14 | 1.641 | 0.001 | −0.893 | 0.027 |
| Slc7a2 | 0.789 | 0.017 | −0.916 | 0.040 |
| Tm4sf4 | 1.100 | 0.003 | −0.594 | 0.040 |
| Tns1 | 0.722 | 0.002 | −0.606 | 0.027 |
| Zbtb16 | 0.919 | 0.004 | −0.977 | 0.049 |
| Down-regulated by EtOH and Up-regulated by t-TUCB-FLVs | ||||
| C6 | −1.685 | 0.002 | 1.095 | 0.040 |
| Cxcl12 | −1.483 | 0.001 | 0.630 | 0.040 |
| Cyp7b1 | −1.111 | 0.008 | 1.918 | 0.040 |
| Hes6 | −1.465 | 0.004 | 0.630 | 0.040 |
| Mup13 | −2.027 | 0.002 | 1.517 | 0.039 |
| Nudt7 | −2.581 | 0.004 | 1.481 | 0.040 |
| Slco1a1 | −2.460 | 0.002 | 1.328 | 0.034 |
| Regulated in same direction by EtOH and t-TUCB-FLVs | ||||
| Akr1c19 | −1.024 | 0.010 | −0.815 | 0.042 |
| Apcs | 2.781 | <0.001 | 0.630 | 0.025 |
| Ces1d | −0.746 | 0.004 | −0.984 | 0.041 |
| Fads2 | −1.651 | 0.005 | −0.655 | 0.034 |
| Orm1 | 1.658 | 0.010 | 0.978 | 0.040 |
| Orm2 | 1.759 | 0.002 | 2.112 | 0.039 |
| Saa1 | 4.679 | 0.006 | 2.025 | 0.015 |
| Saa2 | 4.743 | 0.009 | 2.481 | 0.015 |
| Uniquely regulated by t-TUCB-FLVs | ||||
| Anxa5 | −0.632 | 0.034 | ||
| Arrdc3 | −1.218 | 0.027 | ||
| Erdr1 | 1.577 | 0.025 | ||
| Gck | −0.662 | 0.035 | ||
| Gmppa | 0.618 | 0.027 | ||
| Rgs16 | −1.402 | 0.039 | ||
| Rhobtb1 | −0.639 | 0.040 | ||
| Sult1d1 | −0.850 | 0.035 | ||
| Txnip | −1.151 | 0.041 | ||
| Ugp2 | −0.586 | 0.040 | ||
| GO:BP Term | GO Term ID | Adj. p-Value | Genes |
|---|---|---|---|
| Bile acid biosynthetic process | GO:0006699 | 0.011 | Errfi1, Cyp7b1, Ces1d |
| Small molecule biosynthetic process | GO:0044283 | 0.012 | Aass, Egr1, Errfi1, Slc39a14, Cyp7b1, Ces1d, Fads2 |
| Circadian rhythm | GO:0007623 | 0.016 | Egr1, Klf10, Npas2, Cyp7b1, Ces1D, Hes6 |
| Organic anion transport | GO:0015711 | 0.029 | Arl6ip5, Slc10a2, Slc39a14, Slc7a2, SlcO1a1, Ces1d |
| Bile acid metabolic process | GO:0008206 | 0.042 | Errfi1, Cyp7b1, Ces1d |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Warner, D.R.; Warner, J.B.; Hardesty, J.E.; Abdelfadil, Y.; Soni, C.; Bauer, P.; Maldonado, C.; McClain, C.J.; Kirpich, I.A. Hepatocyte-Specific Transcriptional Responses to Liver-Targeted Delivery of a Soluble Epoxide Hydrolase Inhibitor in a Mouse Model of Alcohol-Associated Liver Disease. Biology 2025, 14, 1267. https://doi.org/10.3390/biology14091267
Warner DR, Warner JB, Hardesty JE, Abdelfadil Y, Soni C, Bauer P, Maldonado C, McClain CJ, Kirpich IA. Hepatocyte-Specific Transcriptional Responses to Liver-Targeted Delivery of a Soluble Epoxide Hydrolase Inhibitor in a Mouse Model of Alcohol-Associated Liver Disease. Biology. 2025; 14(9):1267. https://doi.org/10.3390/biology14091267
Chicago/Turabian StyleWarner, Dennis R., Jeffrey B. Warner, Josiah E. Hardesty, Yasmeen Abdelfadil, Chirag Soni, Philip Bauer, Claudio Maldonado, Craig J. McClain, and Irina A. Kirpich. 2025. "Hepatocyte-Specific Transcriptional Responses to Liver-Targeted Delivery of a Soluble Epoxide Hydrolase Inhibitor in a Mouse Model of Alcohol-Associated Liver Disease" Biology 14, no. 9: 1267. https://doi.org/10.3390/biology14091267
APA StyleWarner, D. R., Warner, J. B., Hardesty, J. E., Abdelfadil, Y., Soni, C., Bauer, P., Maldonado, C., McClain, C. J., & Kirpich, I. A. (2025). Hepatocyte-Specific Transcriptional Responses to Liver-Targeted Delivery of a Soluble Epoxide Hydrolase Inhibitor in a Mouse Model of Alcohol-Associated Liver Disease. Biology, 14(9), 1267. https://doi.org/10.3390/biology14091267






