Droplet Digital PCR Assay for Detection and Quantification of ‘Candidatus Phytoplasma solani’ in Grapevine Samples
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Sources
2.2. DNA Extraction
2.3. Quantitative PCR Assay
2.4. Droplet Digital PCR Assay
2.5. Assays Linearity, Limit of Detection (LOD), and Limit of Quantitation (LOQ)
3. Results
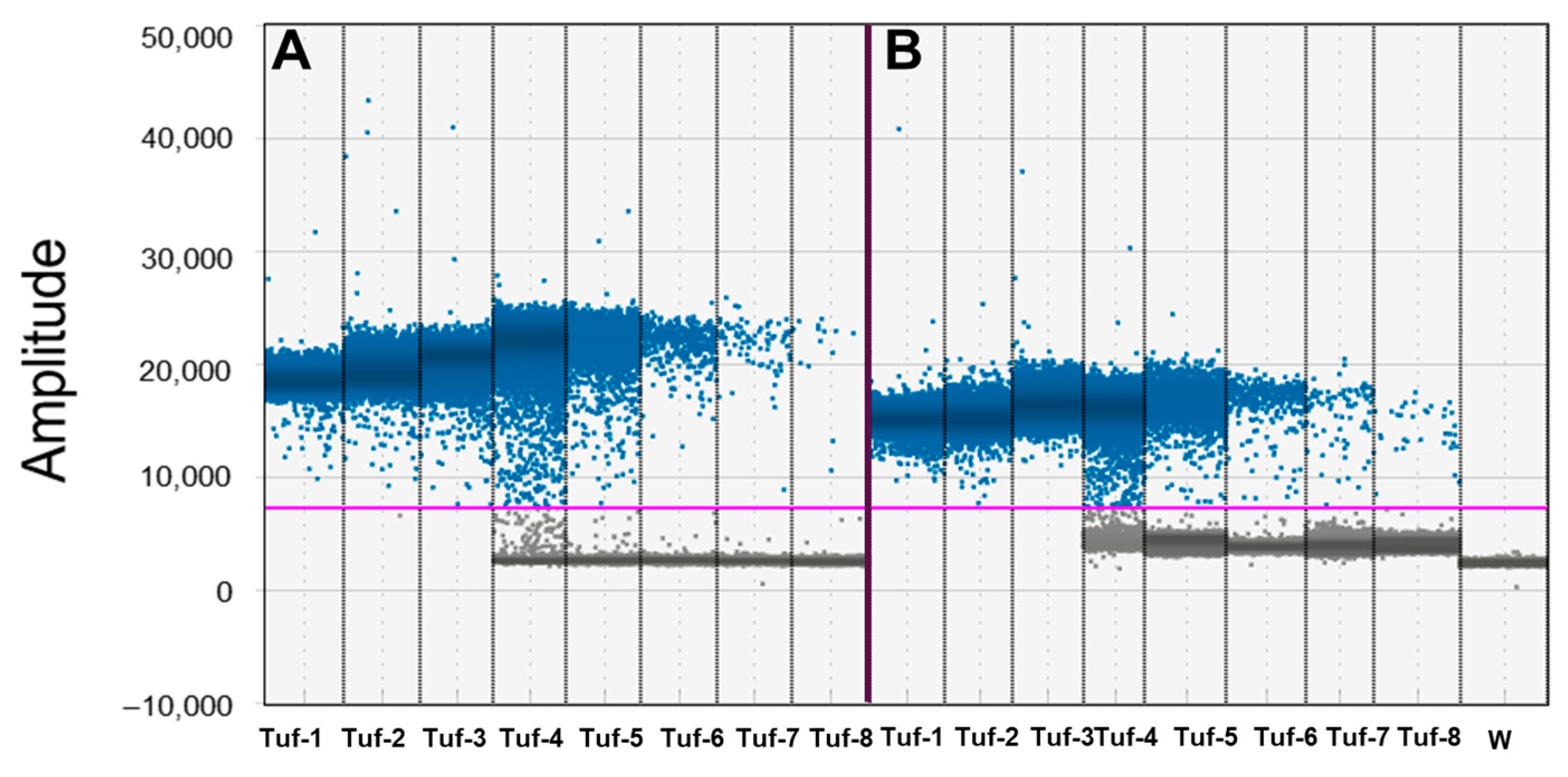
3.1. Assessment of Linear Dynamic Range and Precision of ddPCR and qPCR Techniques
| Purified tuf qPCR Amplicon from Periwinkle Inoculated with P7 Isolate (P7) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| ID Sample | DNA (ng) | qPCR (Cq) Mean ± SD | qPCR CV (%) | ddPCR (Copies Per 20 μL/Reaction) Mean ± SD | ddPCR CV(%) | ||||
| P7 | P7+ 250 mg of DNA from Roots | P7 | P7+ 250 mg of DNA from Roots | P7 | P7+ 250 mg of DNA from Roots | P7 | P7+ 250 mg of DNA from Roots | ||
| Tuf-1 | 1.44 × 10−2 | 13.6 ± 0.37 | 15.4 ± 0.24 | 3.58 | 1.94 | saturated | saturated | nd | nd |
| Tuf-2 | 1.44 × 10−3 | 16.7 ± 0.30 | 18.9 ± 0.32 | 2.2 | 1.82 | saturated | saturated | nd | nd |
| Tuf-3 | 1.44 × 10−4 | 19.9 ± 0.43 | 21.6 ± 0.20 | 2.4 | 1.02 | saturated | saturated | nd | nd |
| Tuf-4 | 1.44 × 10−5 | 23.3 ± 0.2 | 25.6 ± 0.16 | 0.94 | 0.68 | 52,340 ± 1126 | 54,067 ± 2338 | 2.15 | 4.32 |
| Tuf-5 | 1.44 × 10−6 | 26.5 ± 0.49 | 28.4 ± 0.24 | 1.99 | 0.86 | 6871 ± 126.7 | 6755 ± 141.1 | 1.84 | 2.06 |
| Tuf-6 | 1.44 × 10−7 | 31.3 ± 0.55 | 29.5 * ± 0.25 | 1.97 | 0.84 | 687 ± 37.5 | 584 ± 28.9 | 6.058 | 4.84 |
| Tuf-7 | 1.44 × 10−8 | 33.9 ± 0.86 | 30.5 * ± 036 | 2.78 | 1.20 | 65± 8.0 | 64 ± 10.4 | 11.51 | 16.13 |
| Tuf-8 | 1.44 × 10−9 | − | − | − | − | 5.7 ± 0.6 | 7.1 ± 0.8 | 11.21 | 11.33 |
| Statistics of qPCR: Standard Curve Performance | Statistics of ddPCR: Linear Regression | ||||||||
| Slope | −3.372 | −2.605 | y = 4 × 109x ± 414.33 | y = 4 × 109x ± 347.09 | |||||
| Value of fit (R2) | 0.999 | 0.925 | Value of fit (R2) | 0.999 | R2 0.9993 | ||||
| Efficiency | 97.9% | 142.5% | |||||||
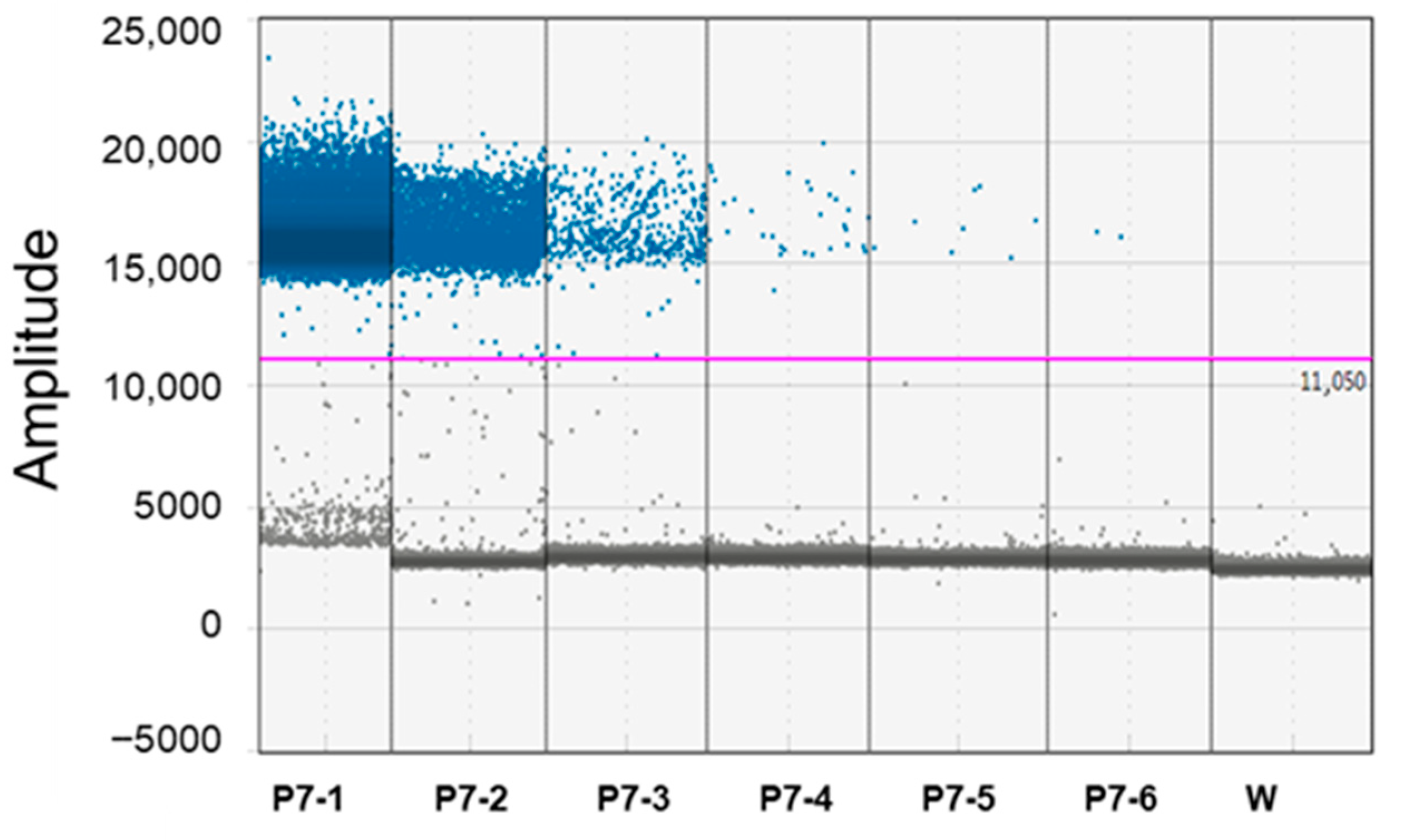
| Periwinkle Infected by ‘Candidatus Phytoplasma solani’ for P7 Isolate | |||||
|---|---|---|---|---|---|
| ID Sample | Total DNA (ng) | Cq (qPCR) Mean ± SD | CV (%) (qPCR) | Copies Per 20 μL Well (ddPCR) Mean ± SD | CV (%) (ddPCR) |
| P7-1 | 1 | 22.0 ± 0.1 | 0.44 | 80,773 ± 5216 | 6.46 |
| P7-2 | 10−1 | 24.8 ± 0.15 | 0.60 | 9195.5 ± 256 | 2.82 |
| P7-3 | 10−2 | 28.3 ± 0.32 | 1.09 | 948.3 ± 51 | 5.40 |
| P7-4 | 10−3 | 31.7 ± 0.20 | 0.64 | 104.6 ± 9.7 | 8.66 |
| P7-5 | 10−4 | 34.1 * ± 0.32 | 0.58 | 19. ± 1.9 | 10.52 |
| P7-6 | 10−5 | − | − | 3.4 ± 0.31 | 9.36 |
| Statistics of qPCR: standard curve performance Slope: −3.226 Efficiency: 104.2 Value of fit (R2): 0.987 | Statistics of ddPCR: linear regression y = 72,554x + 233.97 Value of fit R2 = 0.9998 | ||||
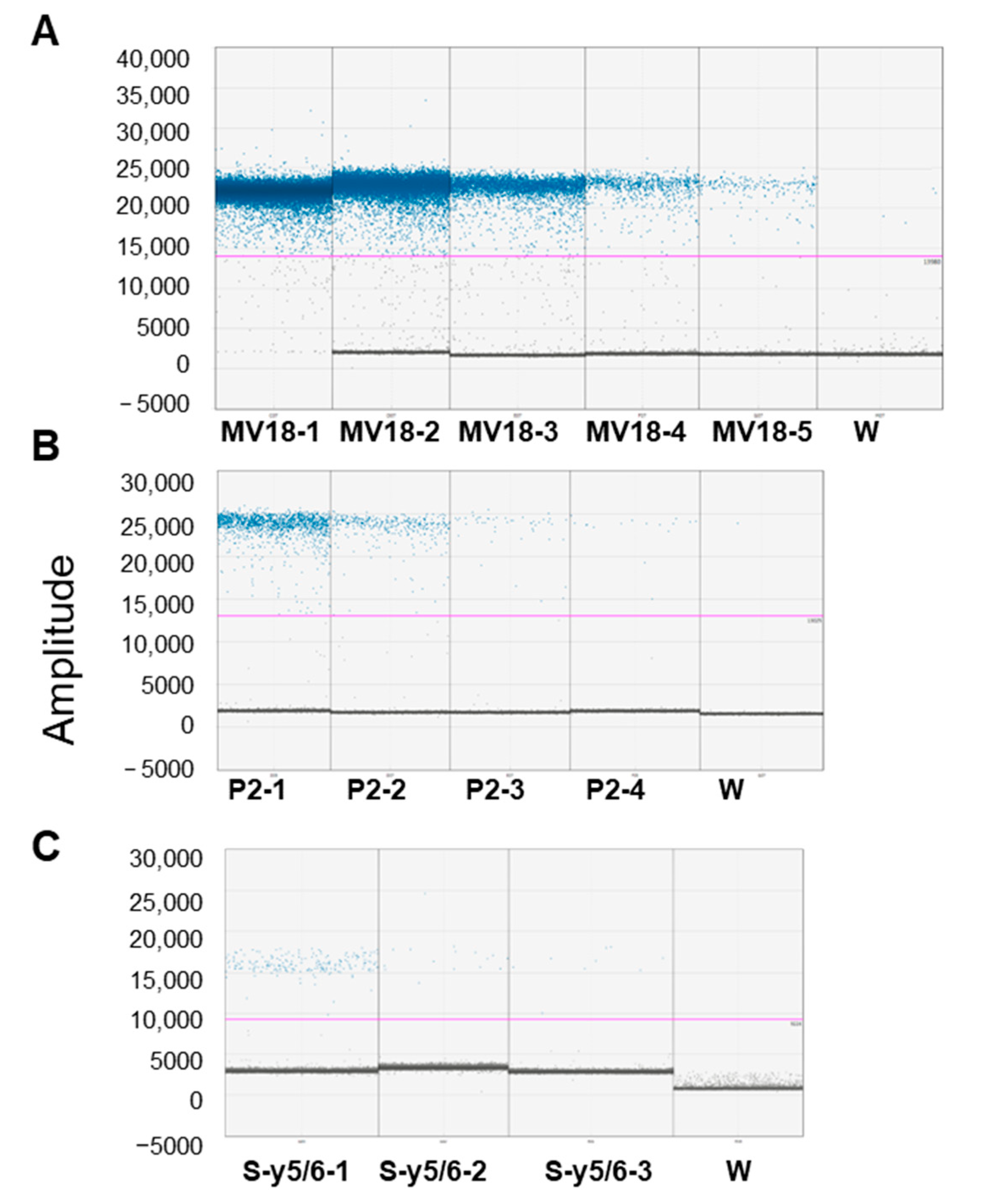
| Symptomatic Leaf Tissue Samples (MV18, P2) and Roots from Symptomatic Plants (S-y5/6) | |||||
|---|---|---|---|---|---|
| ID Sample | Total DNA (ng) | Cq (qPCR) Mean ± SD | CV (%) (qPCR) | Copies Per 20-μL Well (ddPCR) Mean ± SD | CV (%) (ddPCR) |
| MV18-1 | 50 | 22.2 ± 0.3 | 1.33 | saturated | − |
| MV18-2 | 10 | 23.8 ± 0.34 | 1.48 | 37,572 ± 643.2 | 1.71 |
| MV18-3 | 2 | 24.2 ± 0.41 | 0.94 | 5540 ± 370.4 | 6.68 |
| MV18-4 | 0.4 | 27.0 ± 0.28 | 0.65 | 1230 ± 101.5 | 8.25 |
| MV18-5 | 0.08 | 30.2 ± 0.34 | 0.81 | 279 ± 31 | 11.09 |
| P2-1 | 200 | 27.9 ± 0.21 | 0.86 | 1487 ± 170.6 | 11.47 |
| P2-2 | 40 | 29.3 ± 0.31 | 1.19 | 287 ± 25.98 | 9.070 |
| P2-3 | 8 | 31.1 ± 0.75 | 2.67 | 73 ± 5.68 | 7.82 |
| P2-4 | 1.6 | 32.8 ± 0.51 | 1.73 | 24 ± 3.62 | 15.02 |
| S-y5/6-1 | 50 | 30.1 ± 0.45 | 1.51 | 190 ± 20.04 | 10.52 |
| S-y5/6-2 | 5 | 31.9 ± 0.51 | 1.62 | 34 ± 3.6 | 10.6 |
| S-y5/6-3 | 2 | − | 8 ± 1.5 | 19.9 | |
3.2. ‘Ca. P. solani’ Detection on Grapevine Samples: ddPCR vs. qPCR
| qPCR (Cq) Mean ±SD | ddPCR (Copies Per 20-μL Well) Mean ±SD | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N° | Plant Code | S/R/A * | S *-Leaf | S *-Root | A *-Leaf | A *-Root | S *-Leaf | S *-Root | A *-Leaf | A *-Root |
| 1 | P1 | S | 26.8 ± 0.52 | ne | − | na | 3836.8 ± 232 | ne | 0 | na |
| 2 | P2 | S | 30.4 ± 0.48 | ne | − | na | 1492 ± 211 | ne | 9 ± 1.2 | na |
| 3 | P3 | S | 28.4 ± 0.32 | ne | − | na | 622.4 ± 65 | ne | 17.8 ± 3 | na |
| 4 | P4 | S | 27.7 ± 0.27 | ne | 30.2 ± 0.21 | na | 1296 ± 253 | ne | 188 ± 16 | na |
| 5 | MG14 | S | 25.9 ± 0.12 | ne | ne | na | 11,276 ± 434 | ne | ne | na |
| 6 | MG15 | S | 26.6 ± 0.65 | ne | ne | na | 4532 ± 121 | ne | ne | na |
| 7 | MG16 | S | 33.1 ± 0.53 | ne | ne | na | 36.2 ± 2.5 | ne | ne | na |
| 8 | MV16 | S | 30.1 ± 0.58 | ne | ne | na | 690 ± 32 | ne | ne | na |
| 9 | MV18 | S | 23.8 ± 0.42 | ne | ne | na | 37,572 ± 453 | ne | ne | na |
| 10 | MV4 | S | 30.8 ± 0.61 | ne | ne | na | 287 ± 23 | ne | ne | na |
| 11 | P99 | S | 30.1 ± 0.38 | ne | ne | na | 324 ± 44 | ne | ne | na |
| 12 | P105 | S | 30.8 ± 0.44 | ne | ne | na | 218 ± 12 | ne | ne | na |
| 13 | NT4 | S | 26.6 ± 0.64 | ne | ne | na | 4566 ± 321 | ne | ne | na |
| 14 | NT5 | S | 27.2 ± 0.32 | ne | ne | na | 2340 ± 234 | ne | ne | na |
| 15 | SC10 | S | 31.5 ± 0.58 | ne | ne | na | 176.4 ± 67 | ne | ne | na |
| 16 | S-y5/2 | S | ne | 29.9 ± 0.54 | ne | na | ne | 898 ± 89 | ne | na |
| 17 | S-y2/11 | S | ne | − | ne | na | ne | 98 ± 7.2 | ne | na |
| 18 | S-y3/2 | S | ne | − | ne | na | ne | − | ne | na |
| 19 | S-y4/1 | S | ne | − | ne | na | ne | − | ne | na |
| 20 | S-y4/3 | S | ne | − | ne | na | ne | − | ne | na |
| 21 | S-y4/5 | S | ne | 29.8 ± 0.81 | ne | na | ne | 452 ± 54 | ne | na |
| 22 | S-y4/9 | S | 29.8 ± 0.51 | − | ne | na | 818 ± 56 | 48 ± 5.3 | ne | na |
| 23 | S-y5/4 | S | 29.7 ± 0.51 | 29.1 ± 0.53 | ne | na | 1452 ± 121 | 846.8 ± 89 | ne | na |
| 24 | S-y5/5 | S | 30.2 ± 0.51 | 30.2 ± 0.82 | ne | na | 2899 ± 99 | 368.4 ± 23 | ne | na |
| 25 | S-y5/6 | S | 30.1 ± 0.51 | − | ne | na | 786 ± 43 | 35 ± 4.3 | ne | na |
| 26 | S-y5/7 | S | 27.2 ± 0.51 | − | ne | na | 2404 ± 188 | 192.8 ± 22 | ne | na |
| 27 | S-y5/12 | S | 29.8 ± 0.51 | 29.1 ± 0.55 | ne | na | 1320 ± 201 | 543 ± 88 | ne | na |
| 28 | R-y5/1 | R | na | na | − | ne | na | na | 13 ± 3.3 | ne |
| 29 | R-y5/2 | R | na | na | − | ne | na | na | 9.8 ± 2.2 | ne |
| 30 | R-y5/3 | R | na | na | − | ne | na | na | 0 | ne |
| 31 | R-y5/6 | R | na | na | − | ne | na | na | 0 | ne |
| 32 | R-y5/8 | R | na | na | − | ne | na | na | 0 | ne |
| 33 | R-y1/9 | R | na | na | ne | 31.4 ± 0.43 | na | na | ne | 54 ± 7 |
| 34 | R-y1/11 | R | na | na | ne | − | na | na | ne | 16 ± 3 |
| 35 | R-y2/1 | R | na | na | ne | 31.2 ± 0.29 | na | na | ne | 88 ± 21 |
| 36 | R-y2/2 | R | na | na | ne | − | na | na | ne | − |
| 37 | R-y2/4 | R | na | na | ne | 30.9 ± 0.48 | na | na | ne | 97 ± 12 |
| 38 | R-y2/7 | R | na | na | ne | − | na | na | ne | 58 ± 3.5 |
| 39 | R-y2/12 | R | na | na | ne | − | na | na | ne | 30 ± 4.1 |
| 40 | R-y3/6 | R | na | na | ne | − | na | na | ne | − |
| 41 | R-y4/4 | R | na | na | ne | − | na | na | ne | 34 ± 6.1 |
| 42 | R-y4/5 | R | na | na | ne | − | na | na | ne | − |
| 43 | R-y4/6 | R | na | na | ne | − | na | na | ne | − |
| 44 | R-y5/4 | R | na | na | ne | − | na | na | ne | − |
| 45 | AS2 | A | na | na | − | − | na | na | − | 13 ± 2.3 |
| 46 | AS10 | A | na | na | − | − | na | na | − | 24 ± 3.3 |
| 47 | AS9 | A | na | na | − | − | na | na | − | − |
| 48 | AS-DBL | A | na | na | − | − | na | na | − | − |
| 49 | AS-K3 | A | na | na | − | − | na | na | − | − |
| 50 | AS-NT5 | A | na | na | − | − | na | na | − | − |
| Total positive samples | 21 | 5 | 1 | 3 | 21 | 9 | 5 | 9 | ||
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Quaglino, F.; Zhao, Y.; Casati, P.; Bulgari, D.; Bianco, P.A.; Wei, W.; Davis, R.E. ‘Candidatus Phytoplasma solani’, a novel taxon associated with stolbur- and Bois noir-related diseases of plants. Int. J. Syst. Evol. Microbiol. 2013, 63, 2879–2894. [Google Scholar] [CrossRef]
- Belli, G.; Bianco, P.A.; Conti, M. Grapevine yellows in Italy: Past, present and future. J. Plant Pathol. 2010, 92, 303–326. [Google Scholar]
- Pierro, R.; Moussa, A.; Mori, N.; Marcone, C.; Quaglino, F.; Romanazzi, G. Bois noir management in vineyard: A review on effective and promising control strategies. Front. Plant. Sci. 2024, 15, 1364241. [Google Scholar] [CrossRef]
- Zahavi, T.; Sharon, R.; Sapir, G.; Mawassi, M.; Dafny-Yelin, M.; Naor, V. The long-term effect of stolbur phytoplasma on grapevines in the Golan Heights Aust. J. Grape Wine Res. 2013, 19, 277–284. [Google Scholar] [CrossRef]
- Aryan, A.; Brader, G.; Mörtel, J.; Pastar, M.; Riedle-Bauer, M. An abundant ‘Candidatus Phytoplasma solani’ tuf b strain is associated with grapevine, stinging nettle and Hyalesthes obsoletus. Eur. J. Plant Pathol. 2014, 140, 213–227. [Google Scholar] [CrossRef]
- Bertaccini, A.; Duduk, B.; Paltrinieri, S.; Contaldo, N. Phytoplasmas and phytoplasma diseases: A severe threat to agriculture. Am. J. Plant Sci. 2014, 5, 1763–1788. [Google Scholar] [CrossRef]
- Mehle, N.; Kavčič, S.; Mermal, S.; Vidmar, S.; Pompe Novak, M.; Riedle-Bauer, M.; Brader, G.; Kladnik, A.; Dermastia, M. Geographical and temporal diversity of ‘Candidatus Phytoplasma solani’ in wine-growing regions in Slovenia and Austria. Front. Plant Sci. 2022, 13, 889675. [Google Scholar] [CrossRef]
- EPPO (European and Mediterranean Plant Protection Organization). ‘Candidatus Phytoplasma solani’ (PHYPSO). EPPO Datasheet: ‘Candidatus Phytoplasma solani’. Available online: https://gd.eppo.int/taxon/PHYPSO/datasheet (accessed on 8 January 2025).
- Contaldo, N.; Stepanović, J.; Pacini, F.; Bertaccini, A.; Duduk, B. Molecular variability and host distribution of “Candidatus Phytoplasma solani” strains from different geographic origins. Microorganisms 2021, 9, 2530. [Google Scholar] [CrossRef] [PubMed]
- Quaglino, F.; Maghradze, D.; Casati, P.; Chkhaidze, N.; Lobjanidze, M.; Ravasio, A.; Passera, A.; Venturini, G.; Failla, O.; Bianco, P.A. Identification and characterization of new ‘Candidatus Phytoplasma solani’ strains associated with bois noir disease in Vitis vinifera L. cultivars showing a range of symptom severity in Georgia, the Caucasus Region. Plant Dis. 2016, 100, 904–915. [Google Scholar] [CrossRef] [PubMed]
- Landi, L.; Riolo, P.; Murolo, S.; Romanazzi, G.; Nardi, S.; Isidoro, N. Genetic variability of stolbur phytoplasma in Hyalesthes obsoletus (Hemiptera: Cixiidae) and its main host plants in vineyard agroecosystems. J. Econ. Entomol. 2015, 108, 1506–1515. [Google Scholar] [CrossRef]
- Murolo, S.; Romanazzi, G. In-vineyard population structure of ‘Candidatus Phytoplasma solani’ using multilocus sequence typing analysis. Infect. Genet. Evol. 2015, 31, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Langer, M.; Maixner, M. Molecular characterization of grapevine yellows associated phytoplasmas of the stolbur group based on RFLP analysis of non-ribosomal DNA. Vitis 2004, 43, 191–199. [Google Scholar]
- Plavec, J.; Ivančan, G.; Škorić, D.; Foissac, X.; Šeruga Musić, M. Genetically divergent ‘Candidatus Phytoplasma solani’ isolates in Croatian vineyard pathosystems suggest complex epidemiological networks. Phytopathol. Res. 2024, 6, 46. [Google Scholar]
- Chuche, J.; Danet, J.-L.; Salar, P.; Foissac, X.; Thiéry, D. Transmission of ‘Candidatus Phytoplasma solani’ by Reptalus quinquecostatus (Hemiptera: Cixiidae). Ann. Appl. Biol. 2016, 169, 214–223. [Google Scholar] [CrossRef]
- Fialova, R.; Valova, P.; Balakishiyeva, G.; Danet, J.L.; Safarova, D.; Foissac, X.; Navratil, M. Genetic variability of stolbur phytoplasma in annual crop and wild plant species in South Moravia. J. Plant Pathol. 2009, 91, 411–416. [Google Scholar]
- Kosovac, A.; Radonjić, S.; Hrnčić, S.; Krstić, O.; Toševski, I.; Jović, J. Molecular tracing of the transmission routes of bois noir in Mediterranean vineyards of Montenegro and experimental evidence for the epidemiological role of Vitex agnus-castus (Lamiaceae) and associated Hyalesthes obsoletus (Cixiidae). Plant Pathol. 2016, 65, 285–298. [Google Scholar] [CrossRef]
- Johannesen, J.; Foissac, X.; Kehrli, P.; Maixner, M. Impact of vector dispersal and host-plant fidelity on the dissemination of an emerging plant pathogen. PLoS ONE 2012, 7, e51809. [Google Scholar] [CrossRef] [PubMed]
- Conigliaro, G.; Jamshidi, E.; Lo Verde, G.; Bella, P.; Mondello, V.; Giambra, S.; D’Urso, V.; Tsolakis, H.; Murolo, S.; Burruano, S.; et al. Epidemiological investigations and molecular characterization of ‘Candidatus Phytoplasma solani’ in grapevines, weeds, vectors and putative vectors in western Sicily, (Southern Italy). Pathogens 2020, 9, 918. [Google Scholar] [CrossRef]
- Mori, N.; Cargnus, E.; Martini, M.; Pavan, F. Relationships between Hyalesthes obsoletus, its herbaceous hosts and Bois noir epidemiology in Northern Italian vineyards. Insects 2020, 11, 606. [Google Scholar] [CrossRef]
- Terlizzi, F.; Credi, R. Uneven distribution of stolbur phytoplasma in Italian grapevines as revealed by nested-PCR. Bull. Insectol. 2007, 60, 365–366. [Google Scholar]
- Constable, F.E.; Gibb, K.S.; Symons, R.H. Seasonal distribution of phytoplasmas in Australian grapevines. Plant Pathol. 2003, 52, 267–276. [Google Scholar] [CrossRef]
- Carraro, L.; Ermacora, P.; Loi, N.; Osler, R. The recovery phenomenon in apple proliferation infected apple trees. J. Plant Pathol. 2004, 86, 141–146. [Google Scholar]
- Hren, M.; Nikolić, P.; Rotter, A.; Blejec, A.; Terrier, N.; Ravnikar, M.; Dermastia, M.; Gruden, K. ‘Bois Noir’ phytoplasma induces significant reprogramming of the leaf transcriptome in the field grown grapevine. BMC Genom. 2009, 10, 460. [Google Scholar] [CrossRef]
- Landi, L.; Murolo, S.; Romanazzi, G. Detection of ‘Candidatus Phytoplasma solani’ in roots from Bois noir symptomatic and recovered grapevines. Sci. Rep. 2019, 9, 2013. [Google Scholar] [CrossRef]
- Cimerman, A.; Pacifico, D.; Salar, P.; Marzachi, C.; Foissac, X. Striking diversity of vmp1, a variable gene encoding a putative membrane protein of the stolbur phytoplasma. Appl. Environ. Microbiol. 2009, 75, 2951–2957. [Google Scholar] [CrossRef]
- Lee, I.M.; Davis, R.; Gundesen-Rindal, D.E. Phytoplasma: Phytopathogenic mollicutes. Annu. Rev. Microbiol. 2000, 54, 221–255. [Google Scholar] [CrossRef] [PubMed]
- Pacifico, D.; Alma, A.; Bagnoli, B.; Foissac, X.; Pasquini, G.; Tessitori, M.; Marzachì, C. Characterization of Bois noir isolates by restriction fragment length polymorphism of a Stolbur-specific putative membrane protein gene. Phytopathology 2009, 99, 711–715. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Angelini, E.; Bianchi, G.L.; Filippin, L.; Motassutti, C.; Borgo, M. A new TaqMan method for the identification of phytoplasmas associated with grapevine yellows by real-time PCR assay. J. Microbiol. Meth. 2007, 68, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Hren, M.; Boben, J.; Rotter, A.; Kralj, P.; Gruden, K.; Ravnikar, M. Real-time PCR detection systems for Flavescence doree and Bois noir phytoplasmas in grapevine: Comparison with conventional PCR detection and application in diagnostics. Plant Pathol. 2007, 56, 785–796. [Google Scholar] [CrossRef]
- Pelletier, C.; Salar, P.; Gillet, J.; Cloquemin, G.; Very, P.; Foissac, X.; Malembic-Maher, S. Triplex real-time PCR assay for sensitive and simultaneous detection of grapevine phytoplasmas of the 16SrV and 16SrXII-A groups with an endogenous analytical control. Vitis 2009, 48, 87–95. [Google Scholar]
- Pinheiro, L.B.; Coleman, V.A.; Hindson, C.M.; Herrmann, J.; Hindson, B.J.; Bhat, S.; Emslie, K.R. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal. Chem. 2012, 84, 1003–1011. [Google Scholar] [CrossRef] [PubMed]
- Landi, L.; Foglia, R.; Murolo, S.; Romanazzi, G. The mycorrizal status in vineyards affected by esca. J. Fungi. 2021, 7, 869. [Google Scholar] [CrossRef]
- Whale, A.S.; Huggett, J.F.; Cowen, S.; Speirs, V.; Shaw, J.; Ellison, S.; Foy, C.A.; Scott, D.J. Comparison of microfluidic digital PCR and conventional quantitative PCR for measuring copy number variation. Nucleic Acids Res. 2012, 40, e82. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef]
- Yang, X.; Yu, F.; Yan, C.; Li, M.; Liang, X.; Lao, X.; Sun, R.; Lv, W.; Zhang, H.; Zhang, F. Global trends in the application of droplet digital PCR technology in the field of infectious disease pathogen diagnosis: A bibliometric analysis from 2012 to 2023. Diagn. Microbiol. Infect. Dis. 2025, 111, 116623. [Google Scholar] [CrossRef]
- Zhang, Y.; Wei, Z.; Zhang, J.; Chen, C.; Liu, F. Application of PCR and PCR-derived technologies for the detection of pathogens infecting crops. Physiol. Mol. Plant Pathol. 2025, 136, 102589. [Google Scholar] [CrossRef]
- Ratti, C.; Minguzzi, S.; Turina, M. A rapid protocol of crude RNA/DNA extraction for RT-qPCR detection and quantification. Methods Mol. Biol. 2019, 1875, 159–169. [Google Scholar] [PubMed]
- Bahder, B.W.; Soto, N.; Komondy, L.; Mou, D.F.; Humphries, A.R.; Helmick, E.E. Detection and quantification of the 16SrIV-D phytoplasma in leaf tissue of common ornamental palm species in Florida using qPCR and dPCR. Plant Dis. 2019, 103, 1918–1922. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Zhang, X.; Song, W.; Qin, W. Accurate and sensitive detection of areca palm yellow leaf phytoplasma in China by droplet digital PCR targeting tuf gene sequence. Ann. Appl. Biol. 2022, 181, 152–159. [Google Scholar] [CrossRef]
- BioRad Bullettin. Droplet Digital PCR Applications Guide. 2025. Available online: http://www.bio-rad.com/webroot/web/pdf/lsr/literature/Bulletin_6407.pdf (accessed on 16 August 2025).
- Whale, A.S.; De Spiegelaere, W.; Trypsteen, W.; Nour, A.A.; Bae, Y.-K.; Benes, V.; Burke, D.; Cleveland, M.; Corbisier, P.; Devonshire, A.S.; et al. The Digital MIQE Guidelines Update: Minimum Information for Publication of Quantitative Digital PCR Experiments for 2020. Clin. Chem. 2020, 66, 1012–1029. [Google Scholar] [PubMed]
- Huggett, J.F.; Foy, C.A.; Benes, V.; Emslie, K.; Garson, J.A.; Haynes, R. The digital MIQE guidelines: Minimum information for publication of quantitative digital PCR experiments. Clin. Chem. 2013, 59, 892–902. [Google Scholar] [CrossRef] [PubMed]
- Technology Networks. DNA Copy Number Calculator. 2025. Available online: https://www.technologynetworks.com/tn/tools/copynumbercalculator (accessed on 11 February 2025).
- Mehle, N.; Dreo, T. Quantitative analysis with droplet digital PCR. Methods Mol. Biol. 2019, 1875, 171–186. [Google Scholar]
- Pilar Martínez-Diz, M.; Andrés-Sodupe, M.; Berbegal, M.; Bujanda, R.; Díaz-Losada, E.; Gramaje, D. Droplet digital PCR technology for detection of Ilyonectria liriodendri from grapevine environmental samples. Plant Dis. 2020, 104, 1114–1150. [Google Scholar] [CrossRef]
- Wang, L.; Tian, Q.; Zhou, P.; Zhao, W.; Sun, X. Evaluation of Droplet Digital PCR for the detection of black canker disease in tomato. Plant Dis. 2022, 106, 395–495. [Google Scholar] [CrossRef] [PubMed]
- Murolo, S.; Moumni, M.; Mancini, V.; Allagui, M.B.; Landi, L.; Romanazzi, G. Detection and quantification of Stagonosporopsis cucurbitacearum in seeds of Cucurbita maxima using droplet digital polymerase chain reaction. Front. Microbiol. 2022, 12, 764447. [Google Scholar] [CrossRef]
- Wang, D.; Liu, E.; Liu, H.; Jin, X.; Niu, C.; Gao, Y.; Su, X.A. A droplet digital PCR assay for detection and quantification of Verticillium nonalfalfae and V. albo-atrum. Front. Cell Infect. Microbiol. 2023, 12, 1110684. [Google Scholar] [CrossRef] [PubMed]
- Dreo, T.; Pirc, M.; Ramšak, Ž.; Pavšič, J.; Milavec, M.; Zel, J.; Gruden, K. Optimising droplet digital PCR analysis approaches for detection and quantification of bacteria: A case study of fire blight and potato brown rot. Anal. Bioanal. Chem. 2014, 406, 6513–6528. [Google Scholar] [CrossRef]
- Amoia, S.S.; Minafra, A.; Ligorio, A.; Cavalieri, V.; Boscia, D.; Saponari, M.; Loconsole, G. Detection of Xylella fastidiosa in host plants and insect vectors by droplet digital PCR. Agriculture 2023, 13, 716. [Google Scholar] [CrossRef]
- Dupas, E.; Legendre, B.; Olivier, V.; Poliakoff, F.; Manceau, C.; Cunty, A. Comparison of real-time PCR and droplet digital PCR for the detection of Xylella fastidiosa in plants. J. Microbiol. Method 2019, 162, 86–95. [Google Scholar] [CrossRef]
- Ramírez, J.D.; Herrera, G.; Muskus, C.; Mendez, C.; Duque, M.C.; Butcher, R. Development of a digital droplet Polymerase Chain Reaction (ddPCR) assay to detect Leishmania DNA in samples from Cutaneous Leishmaniasis patients. Int. J. Infect. Dis. 2019, 79, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Sidstedt, M.; Rådström, P.; Hedman, J. PCR inhibition in qPCR, dPCR and MPS-mechanisms and solutions. Anal. Bioanal. Chem. 2020, 412, 2009–2023. [Google Scholar] [CrossRef]
- Wnuk, E.; Waśko, A.; Walkiewicz, A.; Bartmiński, P.; Bejger, R.; Mielnik, L.; Bieganowski, A. The effects of humic substances on DNA isolation from soils. PeerJ. 2020, 8, e9378. [Google Scholar] [CrossRef]
- Svec, D.; Tichopad, A.; Novosadova, V.; Pfaffl, M.W.; Kubista, M. How good is a PCR efficiency estimate: Recommendations for precise and robust qPCR efficiency assessments. Biomol. Detect. Quantif. 2015, 3, 9–16. [Google Scholar] [CrossRef]
- Zhao, Y.; Xia, Q.; Yin, Y.; Wang, Z. Comparison of droplet digital PCR and quantitative PCR assays for quantitative detection of Xanthomonas citri subsp. citri. PLoS ONE 2016, 11, e0159004. [Google Scholar] [CrossRef]
- Kralik, P.; Ricchi, M. A Basic Guide to Real Time PCR in Microbial diagnostics: Definitions, parameters, and everything. Front. Microbiol. 2017, 8, 108. [Google Scholar] [CrossRef]
- Lal, P.; Tiwari, R.K.; Kumar, A.; Altaf, M.A.; Alsahli, A.A.; Lal, M.K.; Kumar, R. Bibliometric analysis of real-time PCR-based pathogen detection in plant protection research: A comprehensive study. Front. Plant Sci. 2023, 14, 1129714. [Google Scholar] [CrossRef]
- Landi, L.; Romanazzi, G. Seasonal variation of defense-related gene expression in leaves from Bois noir affected and recovered grapevines. J. Agric. Food. Chem. 2011, 59, 6628–6637. [Google Scholar] [CrossRef] [PubMed]
- Pacifico, D.; Margaria, P.; Galetto, L.; Legovich, M.; Abbà, S.; Veratti, F.; Marzachì, C.; Palmano, S. Differential gene expression in two grapevine cultivars recovered from “Flavescence dorée”. Microbiol. Res. 2019, 220, 72–82. [Google Scholar] [CrossRef]
- Sanders, R.; Mason, D.J.; Foy, C.A.; Huggett, J.F. Considerations for accurate gene expression measurement by reverse transcription quantitative PCR when analysing clinical samples. Anal. Bioanal. Chem. 2014, 406, 6471–6483. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Nagendran, K.; Rai, A.B.; Singh, B.; Rao, G.P.; Bertaccini, A. Global status of phytoplasma diseases in vegetable crops. Front. Microbiol. 2019, 10, 1349. [Google Scholar] [CrossRef] [PubMed]
- Wright, A.A.; Shires, M.; Molnar, C.; Bishop, G.; Johnson, A.; Frias, C.; Harper, S.J. Titer and distribution of ‘Candidatus Phytoplasma pruni’ in Prunus avium. Phytopathology 2022, 112, 1406–1412. [Google Scholar] [CrossRef] [PubMed]
- Quaglino, F.; Sanna, F.; Moussa, A.; Faccincani, M.; Passera, A.; Casati, P.; Bianco, P.A.; Mori, N. Identification and ecology of alternative insect vectors of ‘Candidatus Phytoplasma solani’ to grapevine. Sci. Rep. 2019, 9, 19522. [Google Scholar] [CrossRef] [PubMed]
- Oropeza, C.; Cordova, I.; Chumba, A.; Narvaez, M.; Saenz, L.; Ashburner, R.; Harrison, N. Phytoplasma distribution in coconut palms affected by lethal yellow disease. Ann. Appl. Biol. 2011, 159, 109–117. [Google Scholar] [CrossRef]
- Prezelj, N.; Nikolic, P.; Gruden, K.; Ravnikar, M.; Dermastia, M. Spatiotemporal distribution of Flavescence dorée phytoplasma in grapevine. Plant Pathol. 2012, 62, 760–766. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Landi, L.; Murolo, S.; Romanazzi, G. Droplet Digital PCR Assay for Detection and Quantification of ‘Candidatus Phytoplasma solani’ in Grapevine Samples. Biology 2025, 14, 1251. https://doi.org/10.3390/biology14091251
Landi L, Murolo S, Romanazzi G. Droplet Digital PCR Assay for Detection and Quantification of ‘Candidatus Phytoplasma solani’ in Grapevine Samples. Biology. 2025; 14(9):1251. https://doi.org/10.3390/biology14091251
Chicago/Turabian StyleLandi, Lucia, Sergio Murolo, and Gianfranco Romanazzi. 2025. "Droplet Digital PCR Assay for Detection and Quantification of ‘Candidatus Phytoplasma solani’ in Grapevine Samples" Biology 14, no. 9: 1251. https://doi.org/10.3390/biology14091251
APA StyleLandi, L., Murolo, S., & Romanazzi, G. (2025). Droplet Digital PCR Assay for Detection and Quantification of ‘Candidatus Phytoplasma solani’ in Grapevine Samples. Biology, 14(9), 1251. https://doi.org/10.3390/biology14091251








