Eimeria ovinoidalis Infection Reshapes Gut Microbial Communities and Metabolic Profiles in Tan Sheep
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Parasitological Methods
2.3. Molecular Methods
2.4. Microbiome Sequencing and Bioinformatics Analysis
2.5. Taxonomic and Functional Analysis
2.6. Untargeted Metabolomic Analysis
2.7. Statistical Analysis
3. Result
3.1. Metagenome Profiling
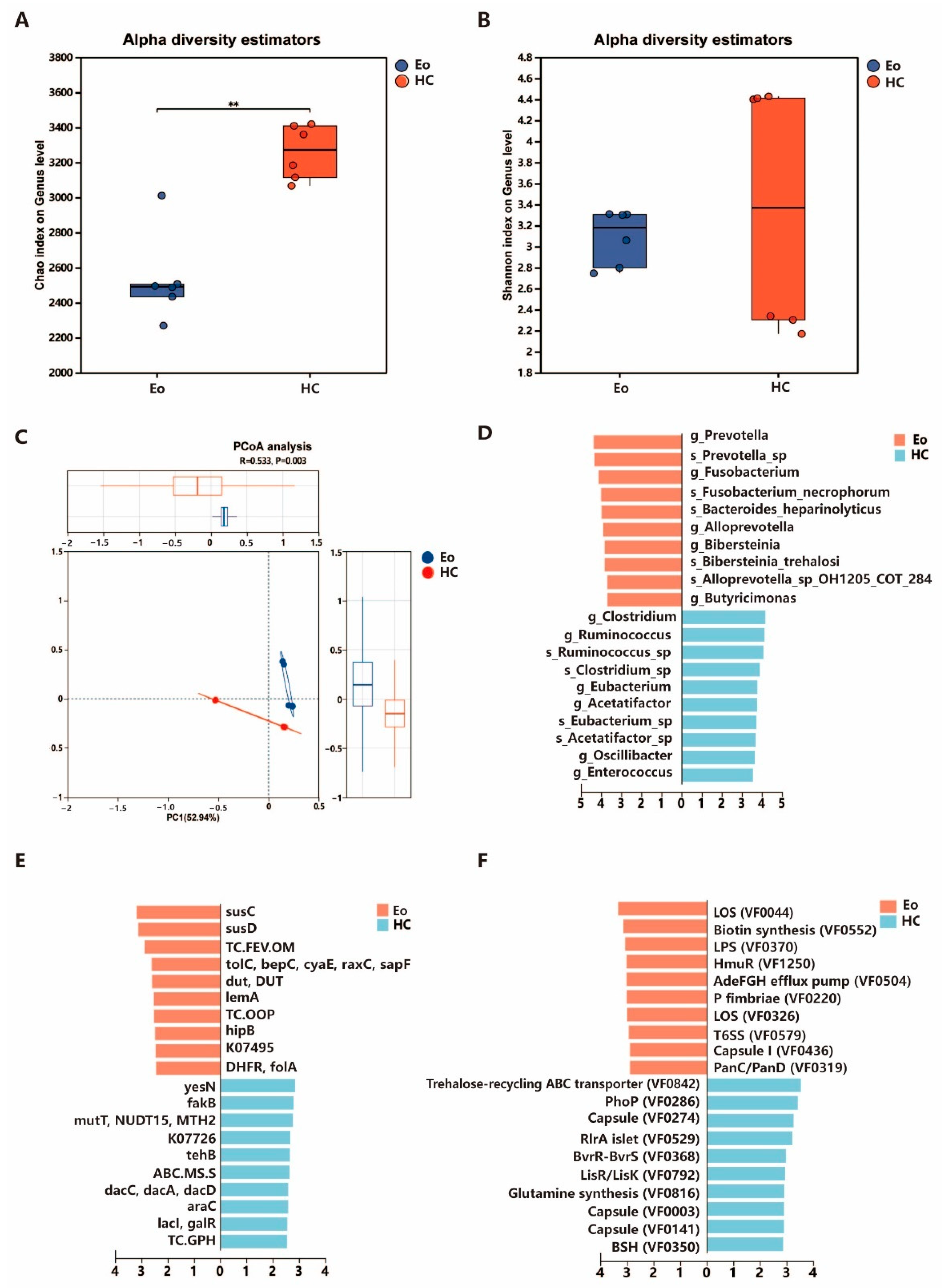
3.2. Alpha and Beta Diversity Analysis
3.3. Abundance and Composition of Different Bacteria Between Groups
3.4. Functions Different Between the Eo and HC Sheep
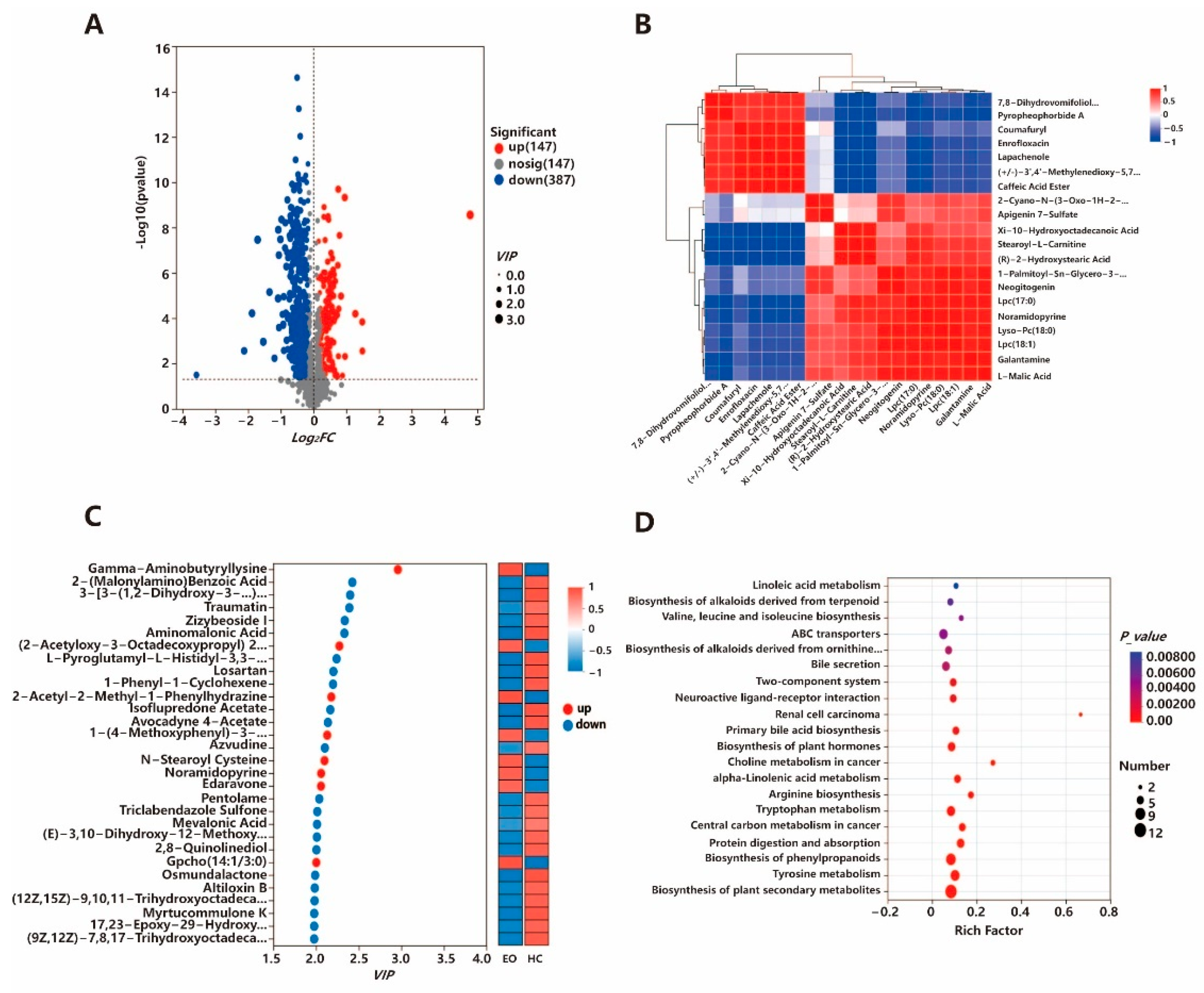
3.5. Non-Target Metabolomics Analysis
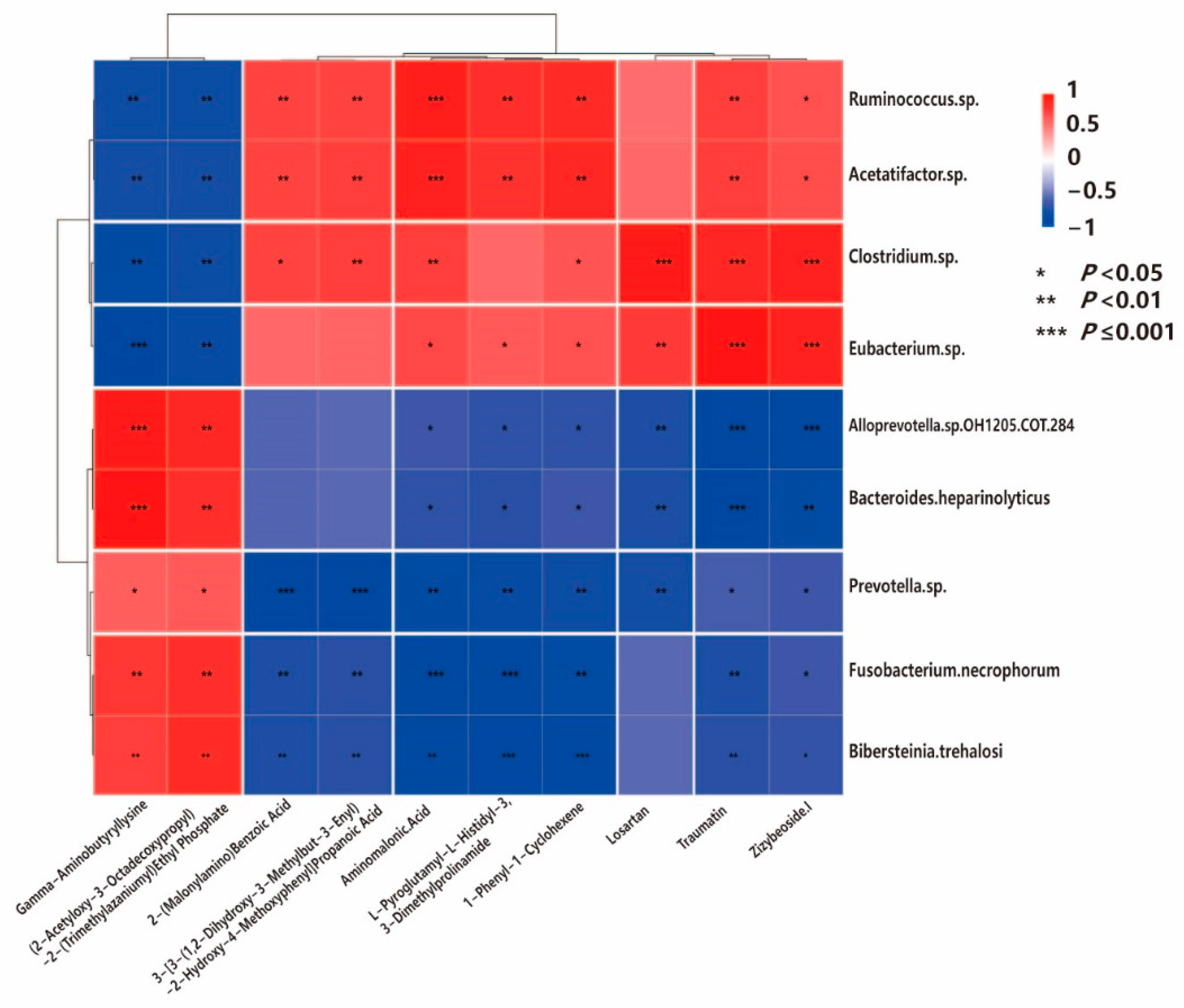
3.6. Integrated Metagenome–Metabolome Analysis
4. Discussion
4.1. Coccidian Infection Is Associated with a Decrease in Intestinal Microbiota Diversity
4.2. Coccidiosis-Associated Microbial Imbalance: Depletion of Beneficial Bacteria and Enrichment of Opportunistic Pathogens
4.3. Functional Reprogramming of Gut Microbiota: From Homeostatic to Stress-Adapted Communities in Coccidiosis
4.4. Coccidiosis Infection Shifts the Gut Microbiome from Commensal to Competitive Virulence Profiles
4.5. Gut Microbiota–Metabolite Associations Observed in E. ovinoidalis Infection and Stress-Induced Metabolic Imbalance
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- de Macedo, L.O.; Bezerra-Santos, M.A.; de Mendonça, C.L.; Alves, L.C.; Ramos, R.A.N.; de Carvalho, G.A. Prevalence and risk factors associated with infection by Eimeria spp. in goats and sheep in Northeastern Brazil. J. Parasit. Dis. 2020, 44, 607–612. [Google Scholar] [CrossRef]
- Mohammed, N.H.; Alobaidii, W.A.; Hasan, M.H. Coccidiosis In Sheep And Goats (Review). Assiut Vet. Med. J. 2021, 67, 33–39. [Google Scholar] [CrossRef]
- Chartier, C.; Paraud, C. Coccidiosis due to Eimeria in sheep and goats, a review. Small Rumin. Res. 2012, 103, 84–92. [Google Scholar] [CrossRef]
- Gül, A.; Değer, S. The Prevalance and Distribution of Eimeria Species Found in Sheep in Van. Turk. J. Vet. Anim. Sci. 2002, 26, 859–864. [Google Scholar]
- Mohamaden, W.I.; Sallam, N.H.; Abouelhassan, E.M. Prevalence of Eimeria species among sheep and goats in Suez Governorate, Egypt. Int. J. Vet. Sci. Med. 2018, 6, 65–72. [Google Scholar] [CrossRef]
- Olmos, L.; Caro, L.C.; Avellaneda-Cáceres, A.; Medina, D.; Sandoval, V.; Aguirre, D.; Micheloud, J. First record of clinical coccidiosis (Eimeria ovinoidalis) in adult sheep from northwestern Argentina. Vet. Parasitol. Reg. Stud. Rep. 2020, 21, 100429. [Google Scholar] [CrossRef]
- Cheng, S.; Wang, N.; Wang, C.; Liu, S.; Li, S.; Li, D.; Zhang, S.; Xu, H.; Zhang, L.; Jian, F. Impacts of a highly pathogenic ovine Eimeria ovinoidalis on the growth of Hu lambs. Vet. Parasitol. 2024, 330, 110250. [Google Scholar] [CrossRef]
- Einarsson, E.; Ma’ayeh, S.; Svärd, S.G. An up-date on Giardia and giardiasis. Current Opinion in Microbiology 2016, 34, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhu, N.; Su, X.; Gao, Y.; Yang, R. Gut-Microbiota-Derived Metabolites Maintain Gut and Systemic Immune Homeostasis. Cells 2023, 12, 793. [Google Scholar] [CrossRef] [PubMed]
- Wiertsema, S.P.; van Bergenhenegouwen, J.; Garssen, J.; Knippels, L.M.J. The Interplay between the Gut Microbiome and the Immune System in the Context of Infectious Diseases throughout Life and the Role of Nutrition in Optimizing Treatment Strategies. Nutrients 2021, 13, 886. [Google Scholar] [CrossRef]
- Ding, G.; Yang, X.; Li, Y.; Wang, Y.; Du, Y.; Wang, M.; Ye, R.; Wang, J.; Zhang, Y.; Chen, Y.; et al. Gut microbiota regulates gut homeostasis, mucosal immunity and influences immune-related diseases. Mol. Cell. Biochem. 2024, 480, 1969–1981. [Google Scholar] [CrossRef]
- Zhang, R.; Ding, N.; Feng, X.; Liao, W. The gut microbiome, immune modulation, and cognitive decline: Insights on the gut-brain axis. Front. Immunol. 2025, 16, 1529958. [Google Scholar] [CrossRef]
- Huang, S.-C.; Liu, K.-L.; Chen, P.; Xu, B.-W.; Ding, W.-L.; Yue, T.-J.; Lu, Y.-N.; Li, S.-Y.; Li, J.-K.; Jian, F.-C. New insights into the combined effects of aflatoxin B1 and Eimeria ovinoidalis on uterine function by disrupting the gut-blood-reproductive axis in sheep. Microbiome 2024, 12, 269. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, K.; Wu, W.; Giannoulatou, E.; Ho, J.W.; Li, L. Host and microbiome multi-omics integration: Applications and methodologies. Biophys. Rev. 2019, 11, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Singleton, S.S.; Bhuiyan, U.; Krammer, L.; Mazumder, R. Multi-omics approaches to studying gastrointestinal microbiome in the context of precision medicine and machine learning. Front. Mol. Biosci. 2024, 10, 1337373. [Google Scholar] [CrossRef] [PubMed]
- Chetty, A.; Blekhman, R. Multi-omic approaches for host-microbiome data integration. Gut Microbes 2024, 16, 2297860. [Google Scholar] [CrossRef] [PubMed]
- Duan, D.; Wang, M.; Han, J.; Li, M.; Wang, Z.; Zhou, S.; Xin, W.; Li, X. Advances in multi-omics integrated analysis methods based on the gut microbiome and their applications. Front. Microbiol. 2025, 15, 1509117. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, M.; Stražar, M.; Avila-Pacheco, J.; Rojas-Tapias, D.F.; Brown, E.M.; Temple, E.; Deik, A.; Bullock, K.; Jeanfavre, S.; Pierce, K.; et al. Linking microbial genes to plasma and stool metabolites uncovers host-microbial interactions underlying ulcerative colitis disease course. Cell Host Microbe 2024, 32, 209–226.e7. [Google Scholar] [CrossRef]
- Li, R.W.; Liu, F.; Solano-Aguilar, G.; Urban, J.F. 40 The gut microbiota modifies host-parasite interactions. J. Anim. Sci. 2024, 102, 65–66. [Google Scholar] [CrossRef]
- Su, F.; Su, M.; Wei, W.; Wu, J.; Chen, L.; Sun, X.; Liu, M.; Sun, S.; Mao, R.; Bourgonje, A.R.; et al. Integrating multi-omics data to reveal the host-microbiota interactome in inflammatory bowel disease. Gut Microbes 2025, 17, 2476570. [Google Scholar] [CrossRef]
- Sloss, M.W.; Kemp, R.L. Veterinary Clinical Parasitology; Iowa State College Press: Ames, IA, USA, 1994. [Google Scholar]
- Xiao, L.H.; Morgan, U.M.; Limor, J.; Escalante, A.; Lal, A.A. Genetic Diversity within Cryptosporidium parvum and Related Cryptosporidium Species. Appl. Environ. Microbiol. 1999, 65, 3386–3391. [Google Scholar] [CrossRef]
- Appelbee, A.J.; Frederick, L.M.; Heitman, T.L.; Olson, M.E. Prevalence and genotyping of Giardia duodenalis from beef calves in Alberta, Canada. Vet. Parasitol. 2003, 112, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Soares, V.M.; dos Santos, E.A.R.; Tadielo, L.E.; Cerqueira-Cézar, C.K.; da Cruz Encide Sampaio, A.N.; Eisen, A.K.A.; de Oliveira, K.G.; Padilha, M.B.; de Moraes Guerra, M.E.; Gasparetto, R.; et al. Detection of adenovirus, rotavirus, and hepatitis E virus in meat cuts marketed in Uruguaiana, Rio Grande do Sul, Brazil. One Health 2022, 14, 100377. [Google Scholar] [CrossRef]
- Ursula, C.O.; Jane, S.F.; Dominique, L.; Michal, P.; Arthur, G. Development of molecular assays for the identification of the 11 Eimeria species of the domestic rabbit (Oryctolagus cuniculus). Vet. Parasitol. 2010, 176, 275–280. [Google Scholar] [CrossRef]
- Gomes-dos-Santos, A.; Fonseca, E.; Riccardi, N.; Hinzmann, M.; Lopes-Lima, M.; Froufe, E. The transcriptome assembly of the European freshwater mussel Unio elongatulus C. Pfeiffer, 1825. Sci. Data 2024, 11, 377. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Buchfink, B. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar]
- Coordinators, N.R. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2015, 44, D7–D19. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Minoru, K.; Susumu, G.; Yoko, S.; Masayuki, K.; Miho, F.; Mao, T. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2013, 42, D199–D205. [Google Scholar] [CrossRef]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2004, 33, D325–D328. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Smith, A.D.; Solano-Aguilar, G.; Wang, T.T.Y.; Li, R.W. Mechanistic insights into the attenuation of intestinal inflammation and modulation of the gut microbiome by krill oil using in vitro and in vivo models. Microbiome 2020, 8, 83. [Google Scholar] [CrossRef]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Ruben Abagyan, A.; Siuzdak, G. XCMS: Processing Mass Spectrometry Data for Metabolite Profiling Using Nonlinear Peak Alignment, Matching, and Identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Wang, X.; Li, L. Human gut microbiome: The second genome of human body. Protein Cell 2010, 1, 718–725. [Google Scholar] [CrossRef]
- Alegado, R.A.; King, N. Bacterial Influences on Animal Origins. Cold Spring Harb. Perspect. Biol. 2014, 6, a016162. [Google Scholar] [CrossRef] [PubMed]
- Kostic, A.D.; Howitt, M.R.; Garrett, W.S. Exploring host–microbiota interactions in animal models and humans. Genes Dev. 2013, 27, 701–718. [Google Scholar] [CrossRef]
- McFall-Ngai, M.; Hadfield, M.G.; Bosch, T.C.G.; Carey, H.V.; Domazet-Lošo, T.; Douglas, A.E.; Dubilier, N.; Eberl, G.; Fukami, T.; Gilbert, S.F.; et al. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. USA 2013, 110, 3229–3236. [Google Scholar] [CrossRef] [PubMed]
- Minamoto, Y.; Otoni, C.C.; Steelman, S.M.; Büyükleblebici, O.; Steiner, J.M.; Jergens, A.E.; Suchodolski, J.S. Alteration of the fecal microbiota and serum metabolite profiles in dogs with idiopathic inflammatory bowel disease. Gut Microbes 2014, 6, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Plaizier, J.C.; Krause, D.O.; Gozho, G.N.; McBride, B.W. Subacute ruminal acidosis in dairy cows: The physiological causes, incidence and consequences. Vet. J. 2008, 176, 21–31. [Google Scholar] [CrossRef]
- Malmuthuge, N.; Griebel, P.J.; Guan, L.L. The Gut Microbiome and Its Potential Role in the Development and Function of Newborn Calf Gastrointestinal Tract. Front. Vet. Sci. 2015, 2, 36. [Google Scholar] [CrossRef]
- Gresse, R.; Chaucheyras-Durand, F.; Fleury, M.A.; Van de Wiele, T.; Forano, E.; Blanquet-Diot, S. Gut Microbiota Dysbiosis in Postweaning Piglets: Understanding the Keys to Health. Trends Microbiol. 2017, 25, 851–873. [Google Scholar] [CrossRef]
- Lu, C.; Yan, Y.; Jian, F.; Ning, C. Coccidia-Microbiota Interactions and Their Effects on the Host. Front. Cell. Infect. Microbiol. 2021, 11, 751481. [Google Scholar] [CrossRef]
- Thabile, M.; Moses, O.; Matthew Adekunle, A. Understanding the interactions between Eimeria infection and gut microbiota, towards the control of chicken coccidiosis: A review. Parasite 2021, 28, 48. [Google Scholar] [CrossRef]
- Huang, G.; Tang, X.; Bi, F.; Hao, Z.; Han, Z.; Suo, J.; Zhang, S.; Wang, S.; Duan, C.; Yu, Z.; et al. Eimeria tenella infection perturbs the chicken gut microbiota from the onset of oocyst shedding. Vet. Parasitol. 2018, 258, 30–37. [Google Scholar] [CrossRef]
- Amachawadi, R.G.; Tom, W.A.; Hays, M.P.; Fernando, S.C.; Hardwidge, P.R.; Nagaraja, T.G. Bacterial community analysis of purulent material from liver abscesses of crossbred cattle and Holstein steers fed finishing diets with or without tylosin. J. Anim. Sci. 2021, 99, skab076. [Google Scholar] [CrossRef]
- Huang, Y.; Tang, J.; Cai, Z.; Zhou, K.; Chang, L.; Bai, Y.; Ma, Y. Prevotella Induces the Production of Th17 Cells in the Colon of Mice. J. Immunol. Res. 2020, 2020, 9607328. [Google Scholar] [CrossRef]
- Glover, J.S.; Ticer, T.D.; Engevik, M.A. Characterizing the mucin-degrading capacity of the human gut microbiota. Sci. Rep. 2022, 12, 8456. [Google Scholar] [CrossRef] [PubMed]
- Wright, D.P.; Rosendale, D.I.; Roberton, A.M. Prevotella enzymes involved in mucin oligosaccharide degradation and evidence for a small operon of genes expressed during growth on mucin. FEMS Microbiol. Lett. 2000, 190, 73–79. [Google Scholar] [CrossRef]
- Sun, J.; Xu, X.; Gao, S.; Pan, Q.; Liu, Z.; Huang, Y.; Lian, Y. Refractory pneumonia caused by Prevotella heparinolytica: A case report. J. Med. Case Rep. 2024, 18, 213. [Google Scholar] [CrossRef]
- Wood, M.E.; Fox, K.A.; Jennings-Gaines, J.; Killion, H.J.; Amundson, S.; Miller, M.W.; Edwards, W.H. How Respiratory Pathogens Contribute to Lamb Mortality in a Poorly Performing Bighorn Sheep (Ovis canadensis) Herd. J. Wildl. Dis. 2017, 53, 126. [Google Scholar] [CrossRef]
- Singh, V.; Lee, G.; Son, H.; Koh, H.; Kim, E.S.; Unno, T.; Shin, J.-H. Butyrate producers,”The Sentinel of Gut”: Their intestinal significance with and beyond butyrate, and prospective use as microbial therapeutics. Front. Microbiol. 2023, 13, 1103836. [Google Scholar] [CrossRef]
- Zhang, D.; Jian, Y.-P.; Zhang, Y.-N.; Li, Y.; Gu, L.-T.; Sun, H.-H.; Liu, M.-D.; Zhou, H.-L.; Wang, Y.-S.; Xu, Z.-X. Short-chain fatty acids in diseases. Cell Commun. Signal. 2023, 21, 212. [Google Scholar] [CrossRef]
- Narushima, S.; Sugiura, Y.; Oshima, K.; Atarashi, K.; Hattori, M.; Suematsu, M.; Honda, K. Characterization of the 17 strains of regulatory T cell-inducing human-derived Clostridia. Gut Microbes 2014, 5, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef]
- Martens, E.C.; Koropatkin, N.M.; Smith, T.J.; Gordon, J.I. Complex glycan catabolism by the human gut microbiota: The Bacteroidetes Sus-like paradigm. J. Biol. Chem. 2009, 284, 24673–24677. [Google Scholar] [CrossRef]
- Piddock, L.J. Multidrug-resistance efflux pumps—not just for resistance. Nat. Rev. Microbiol. 2006, 4, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Yadav, M.; Ghosh, C.; Rathore, J.S. Bacterial toxin-antitoxin modules: Classification, functions, and association with persistence. Curr. Res. Microb. Sci. 2021, 2, 100047. [Google Scholar] [CrossRef]
- Wen, Y.; Behiels, E.; Devreese, B. Toxin-Antitoxin systems: Their role in persistence, biofilm formation, and pathogenicity. Pathog. Dis. 2014, 70, 240–249. [Google Scholar] [CrossRef]
- Dawan, J.; Ahn, J. Bacterial Stress Responses as Potential Targets in Overcoming Antibiotic Resistance. Microorganisms 2022, 10, 1385. [Google Scholar] [CrossRef] [PubMed]
- Sonika, S.; Singh, S.; Mishra, S.; Verma, S. Toxin-antitoxin systems in bacterial pathogenesis. Heliyon 2023, 9, e14220. [Google Scholar] [CrossRef]
- Karimi, S.; Ghafourian, S.; Taheri Kalani, M.; Azizi Jalilian, F.; Hemati, S.; Sadeghifard, N. Association Between Toxin-Antitoxin Systems and Biofilm Formation. Jundishapur J. Microbiol. 2014, 8, e14540. [Google Scholar] [CrossRef] [PubMed]
- Ostyn, E.; Augagneur, Y.; Pinel-Marie, M.L. Insight into the environmental cues modulating the expression of bacterial toxin-antitoxin systems. FEMS Microbiol. Rev. 2025, 49, fuaf007. [Google Scholar] [CrossRef] [PubMed]
- Qu, Q.; Peng, H.; Chen, M.; Liu, X.; Che, R.; Bello-Onaghise, G.; Zhang, Z.; Chen, X.; Li, Y. The relationship between resistance evolution and carbon metabolism in Staphylococcus xylosus under ceftiofur sodium stress. Arch. Microbiol. 2024, 206, 370. [Google Scholar] [CrossRef]
- Poetsch, A.R. The genomics of oxidative DNA damage, repair, and resulting mutagenesis. Comput. Struct. Biotechnol. J. 2020, 18, 207–219. [Google Scholar] [CrossRef]
- Xiao, Q.; Xia, M.; Tang, W.; Zhao, H.; Chen, Y.; Zhong, J. The lipid metabolism remodeling: A hurdle in breast cancer therapy. Cancer Lett. 2024, 582, 216512. [Google Scholar] [CrossRef]
- Arczewska, K.D.; Kuśmierek, J.T. Bacterial DNA repair genes and their eukaryotic homologues: 2. Role of bacterial mutator gene homologues in human disease. Overview of nucleotide pool sanitization and mismatch repair systems. Acta Biochim. Pol. 2007, 54, 435–457. [Google Scholar] [CrossRef]
- Lu, A.L.; Li, X.; Gu, Y.; Wright, P.M.; Chang, D.-Y. Repair of Oxidative DNA Damage: Mechanisms and Functions. Cell Biochem. Biophys. 2001, 35, 141–170. [Google Scholar] [CrossRef]
- Typas, A.; Banzhaf, M.; Gross, C.A.; Vollmer, W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 2012, 10, 123–136. [Google Scholar] [CrossRef]
- Robert, S. AraC protein, regulation of the l-arabinose operon in Escherichia coli, and the light switch mechanism of AraC action. FEMS Microbiol. Rev. 2010, 34, 779–796. [Google Scholar]
- Christie, P.J.; Cascales, E. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 2003, 1, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Russell, A.B.; Peterson, S.B.; Mougous, J.D. Type VI secretion system effectors: Poisons with a purpose. Nat. Rev. Microbiol. 2014, 12, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Bachman, M.A.; Oyler, J.E.; Burns, S.H.; Caza, M.; Lépine, F.; Dozois, C.M.; Weiser, J.N.; Bäumler, A.J. Klebsiella pneumoniae yersiniabactin promotes respiratory tract infection through evasion of lipocalin 2. Infect. Immun. 2011, 79, 3309–3316. [Google Scholar] [CrossRef]
- Zelante, T.; Iannitti, R.G.; Cunha, C.; De Luca, A.; Giovannini, G.; Pieraccini, G.; Zecchi, R.; D’Angelo, C.; Massi-Benedetti, C.; Fallarino, F.; et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 2013, 39, 372–385. [Google Scholar] [CrossRef]
- Avonce, N.; Mendoza-Vargas, A.; Morett, E.; Iturriaga, G. Insights on the evolution of trehalose biosynthesis. BMC Evol. Biol. 2006, 6, 109. [Google Scholar] [CrossRef]
- Groisman, E.A. The Pleiotropic Two-Component Regulatory System PhoP-PhoQ. J. Bacteriol. 2001, 183, 1835–1842. [Google Scholar] [CrossRef] [PubMed]
- Rendueles, O.; Ghigo, J.M. Mechanisms of Competition in Biofilm Communities. Microbiol. Spectr. 2015, 3, 319–342. [Google Scholar] [CrossRef]
- Tettelin, H.; Masignani, V.; Cieslewicz, M.; Donati, C.; Medini, D.; Ward, N.; Angiuoli, S.; Crabtree, J.; Jones, A.; Durkin, A. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. USA 2005, 102, 13950–13955. [Google Scholar] [CrossRef] [PubMed]
- Joyce, S.A.; Shanahan, F.; Hill, C.; Gahan, C.G.M. Bacterial bile salt hydrolase in host metabolism: Potential for influencing gastrointestinal microbe-host crosstalk. Gut Microbes 2014, 5, 669–674. [Google Scholar] [CrossRef] [PubMed]
- Agolino, G.; Cristofolini, M.; Vaccalluzzo, A.; Tagliazucchi, D.; Cattivelli, A.; Pino, A.; Caggia, C.; Solieri, L.; Randazzo, C.L. Genome Mining and Characterization of Two Novel Lacticaseibacillus rhamnosus Probiotic Candidates with Bile Salt Hydrolase Activity. Biomolecules 2025, 15, 86. [Google Scholar] [CrossRef]
- Chwals, W. Regulation of the Cellular and Physiological Effects of Glutamine. Mini-Rev. Med. Chem. 2004, 4, 833–838. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Guo, J.; Whitmore Melanie, A.; Tobin, I.; Kim Dohyung, M.; Zhao, Z.; Zhang, G. Dynamic response of the intestinal microbiome to Eimeria maxima-induced coccidiosis in chickens. Microbiol. Spectr. 2024, 12, e00823–e00824. [Google Scholar] [CrossRef]
- Tang, J.; Wang, Q.; Yu, H.; Dong, L.; Tang, M.; Arif, A.; Zhang, G.; Zhang, T.; Xie, K.; Su, S.; et al. A Comparison of the Cecal Microbiota between the Infection and Recovery Periods in Chickens with Different Susceptibilities to Eimeria tenella. Animals 2024, 14, 2709. [Google Scholar] [CrossRef]
- Taranto, D.; Kloosterman, D.J.; Akkari, L. Macrophages and T cells in metabolic disorder-associated cancers. Nat. Rev. Cancer 2024, 24, 744–767. [Google Scholar] [CrossRef]
- Liu, S.; Li, S.; Cheng, S.; Liu, M.; Li, J.; Li, S.; Li, X.; Zhang, L.; Jian, F. Effect of Artemisia annua on anticoccidial action, intestinal microbiota and metabolites of Hu lambs. BMC Vet. Res. 2025, 21, 41. [Google Scholar] [CrossRef]
- Kallenbach, M.; Gilardoni, P.A.; Allmann, S.; Baldwin, I.T.; Bonaventure, G. C12 derivatives of the hydroperoxide lyase pathway are produced by product recycling through lipoxygenase-2 in Nicotiana attenuata leaves. New Phytol. 2011, 191, 1054–1068. [Google Scholar] [CrossRef]
- Ravanan, P.; Singh, S.K.; Rao, G.S.R.S.; Kondaiah, P. Growth inhibitory, apoptotic and anti-inflammatory activities displayed by a novel modified triterpenoid, cyano enone of methyl boswellates. J. Biosci. 2011, 36, 297–307. [Google Scholar] [CrossRef]
- Braga, J.D.; Thongngam, M.; Kumrungsee, T. Gamma-aminobutyric acid as a potential postbiotic mediator in the gut–brain axis. npj Sci. Food 2024, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Holmes, E.; Wilson, I.D. Gut microorganisms, mammalian metabolism and personalized health care. Nat. Rev. Microbiol. 2005, 3, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Zhao, Y.; Wang, P.; Yu, Y.; Xu, Y.; Yang, Y. Eimeria ovinoidalis Infection Reshapes Gut Microbial Communities and Metabolic Profiles in Tan Sheep. Biology 2025, 14, 1190. https://doi.org/10.3390/biology14091190
Wang J, Zhao Y, Wang P, Yu Y, Xu Y, Yang Y. Eimeria ovinoidalis Infection Reshapes Gut Microbial Communities and Metabolic Profiles in Tan Sheep. Biology. 2025; 14(9):1190. https://doi.org/10.3390/biology14091190
Chicago/Turabian StyleWang, Jiandong, Yuxi Zhao, Pan Wang, Youli Yu, Yarong Xu, and Yuqiu Yang. 2025. "Eimeria ovinoidalis Infection Reshapes Gut Microbial Communities and Metabolic Profiles in Tan Sheep" Biology 14, no. 9: 1190. https://doi.org/10.3390/biology14091190
APA StyleWang, J., Zhao, Y., Wang, P., Yu, Y., Xu, Y., & Yang, Y. (2025). Eimeria ovinoidalis Infection Reshapes Gut Microbial Communities and Metabolic Profiles in Tan Sheep. Biology, 14(9), 1190. https://doi.org/10.3390/biology14091190





