Global Profiling of Protein β-hydroxybutyrylome in Porcine Liver
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Cell Culture
2.3. Protein Extraction and Western Blot
2.4. Trypsin Digestion
2.5. Affinity Enrichment of Kbhb Peptides
2.6. LC-MS/MS Analysis
2.7. Database Search
2.8. Functional Annotation
2.9. Targeted Metabolome
3. Results
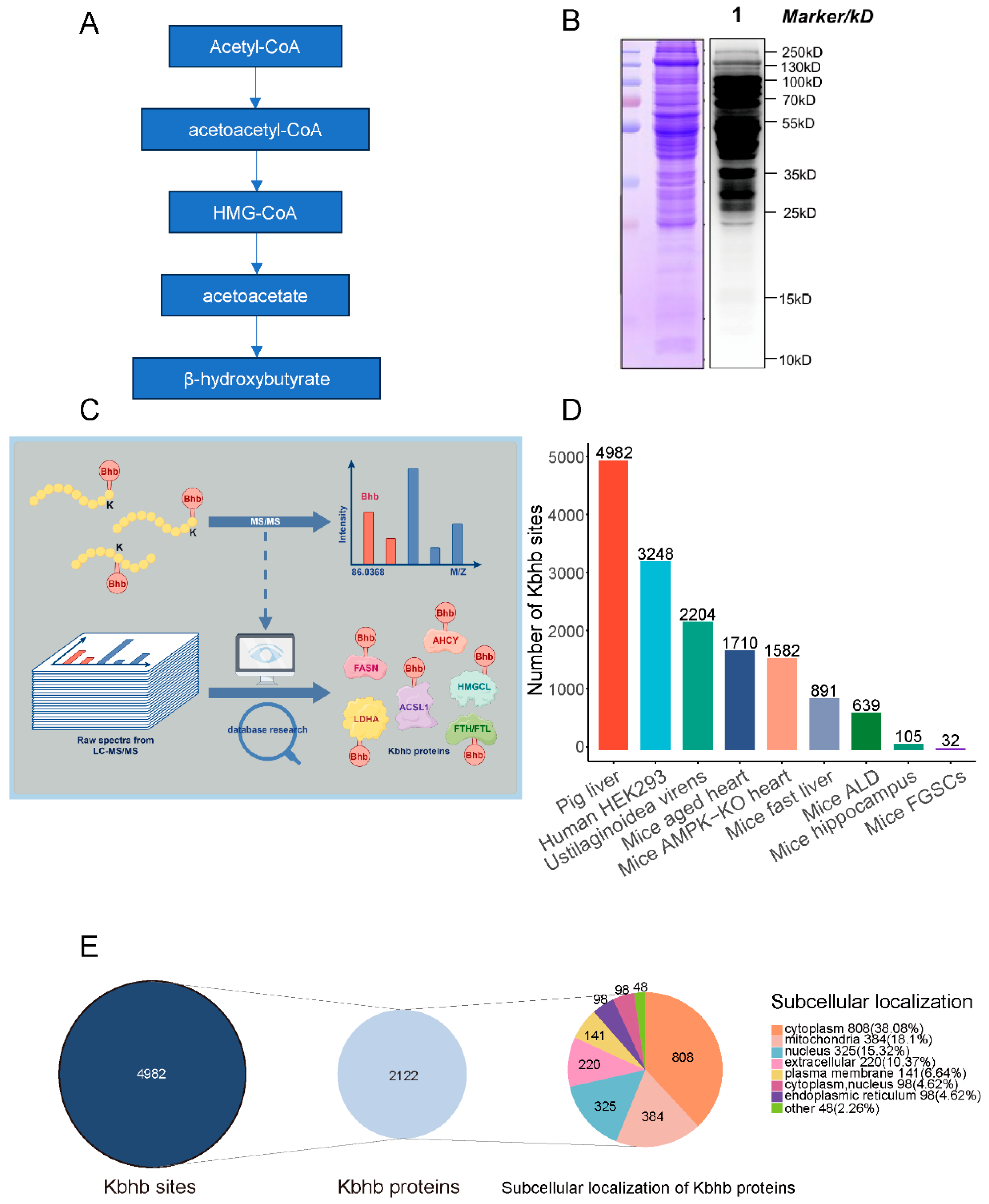
3.1. Global Profiling of Kbhb in Porcine Liver
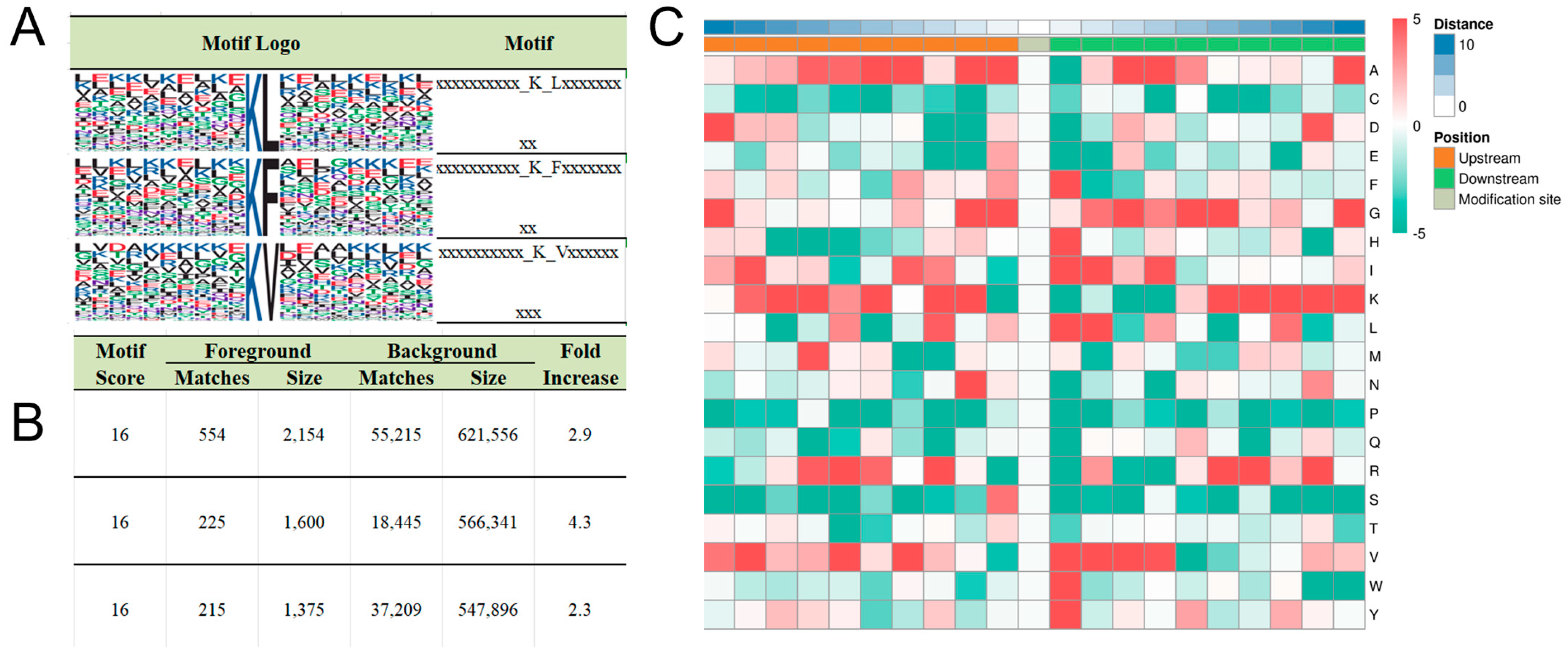
3.2. Motif Analysis of Kbhb Sites
3.3. Functional Enrichment of Kbhb Proteins
3.4. Histone Kbhb in Porcine Liver
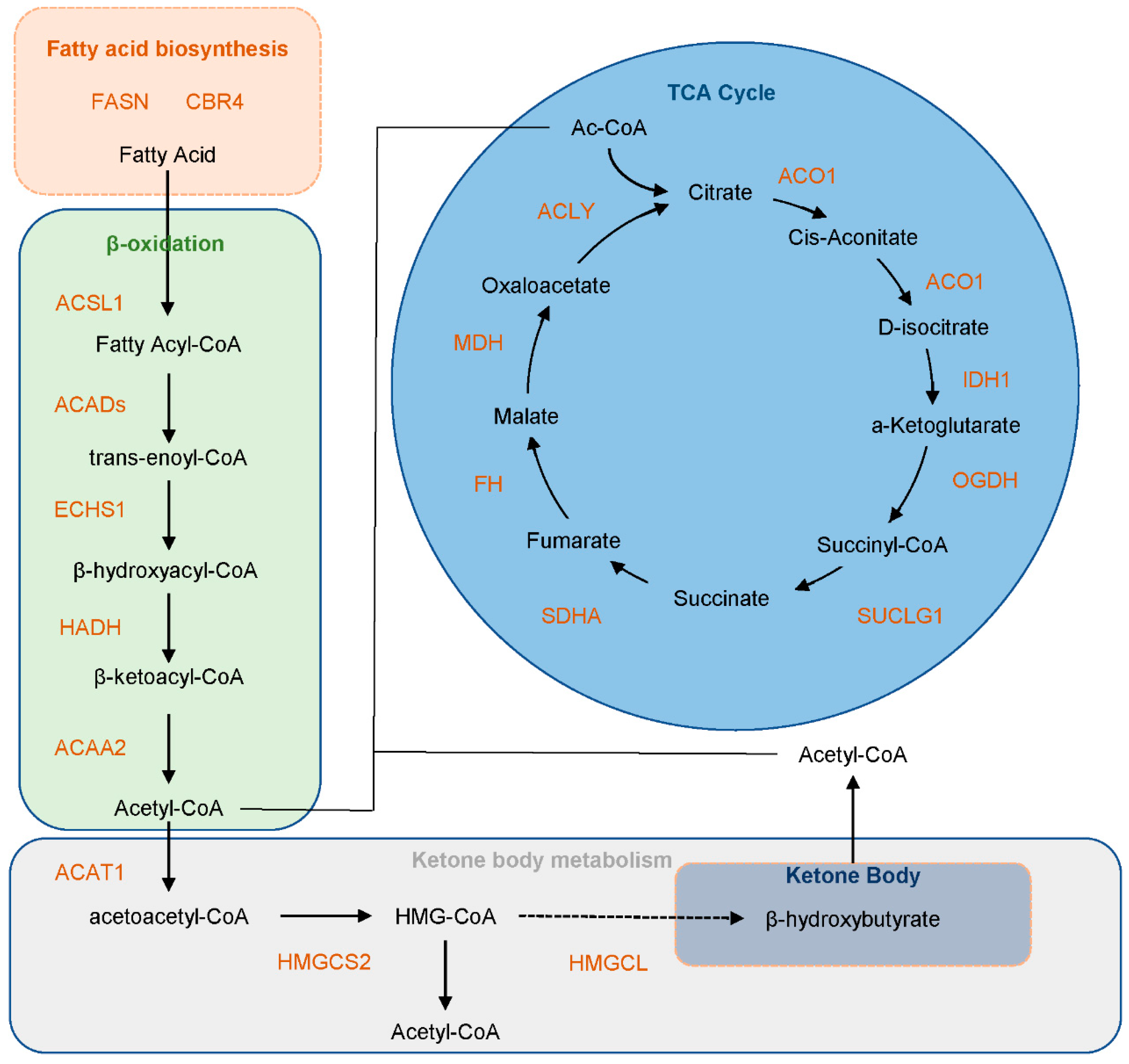
3.5. Functional Insights into Kbhb-Regulated Energy Metabolism
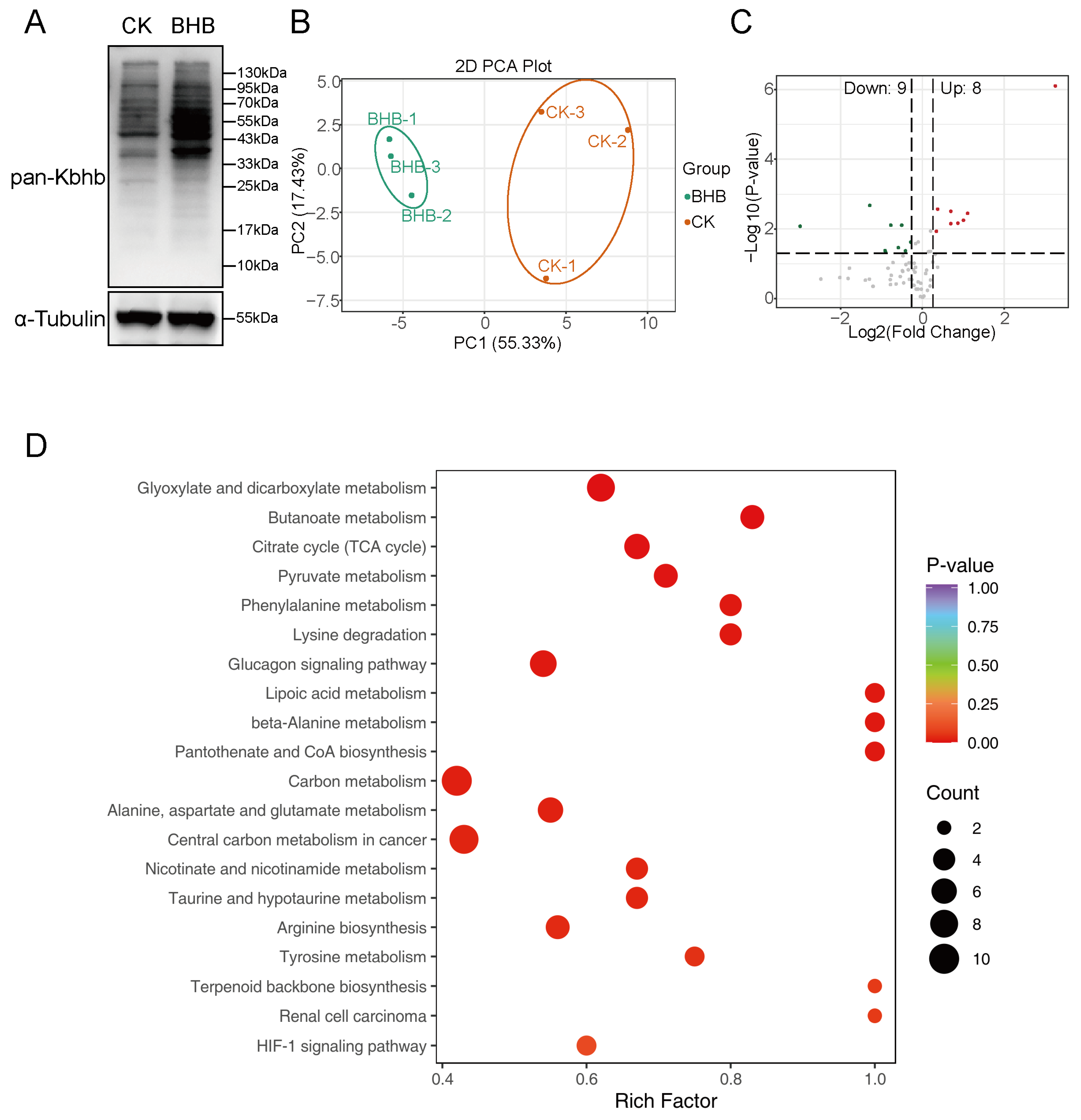
3.6. Kbhb Regulates Hepatic Energy Metabolism
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Lee, J.M.; Hammarén, H.M.; Savitski, M.M.; Baek, S.H. Control of protein stability by post-translational modifications. Nat. Commun. 2023, 14, 201. [Google Scholar] [CrossRef]
- Wang, S.; Osgood, A.O.; Chatterjee, A. Uncovering post-translational modification-associated protein-protein interactions. Curr. Opin. Struct. Biol. 2022, 74, 102352. [Google Scholar] [CrossRef] [PubMed]
- Diskin, C.; Ryan, T.A.J.; O’Neill, L.A.J. Modification of Proteins by Metabolites in Immunity. Immunity 2021, 54, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Sabari, B.R.; Zhang, D.; Allis, C.D.; Zhao, Y. Metabolic regulation of gene expression through histone acylations. Nat. Rev. Mol. Cell Biol. 2017, 18, 90–101. [Google Scholar] [CrossRef]
- Sabari, B.R.; Tang, Z.; Huang, H.; Yong-Gonzalez, V.; Molina, H.; Kong, H.E.; Dai, L.; Shimada, M.; Cross, J.R.; Zhao, Y.; et al. Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol. Cell 2015, 58, 203–215. [Google Scholar] [CrossRef]
- Simithy, J.; Sidoli, S.; Yuan, Z.-F.; Coradin, M.; Bhanu, N.V.; Marchione, D.M.; Klein, B.J.; Bazilevsky, G.A.; McCullough, C.E.; Magin, R.S.; et al. Characterization of histone acylations links chromatin modifications with metabolism. Nat. Commun. 2017, 8, 1141. [Google Scholar] [CrossRef]
- Chen, Y.; Sprung, R.; Tang, Y.; Ball, H.; Sangras, B.; Kim, S.C.; Falck, J.R.; Peng, J.; Gu, W.; Zhao, Y. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell Proteom. 2007, 6, 812–819. [Google Scholar] [CrossRef]
- Tan, M.; Luo, H.; Lee, S.; Jin, F.; Yang, J.S.; Montellier, E.; Buchou, T.; Cheng, Z.; Rousseaux, S.; Rajagopal, N.; et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 2011, 146, 1016–1028. [Google Scholar] [CrossRef]
- Peng, C.; Lu, Z.; Xie, Z.; Cheng, Z.; Chen, Y.; Tan, M.; Luo, H.; Zhang, Y.; He, W.; Yang, K.; et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol. Cell Proteom. 2011, 10, M111.012658. [Google Scholar] [CrossRef]
- Zhang, Z.; Tan, M.; Xie, Z.; Dai, L.; Chen, Y.; Zhao, Y. Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 2011, 7, 58–63. [Google Scholar] [CrossRef]
- Tan, M.; Peng, C.; Anderson, K.A.; Chhoy, P.; Xie, Z.; Dai, L.; Park, J.; Chen, Y.; Huang, H.; Zhang, Y.; et al. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 2014, 19, 605–617. [Google Scholar] [CrossRef]
- Dai, L.; Peng, C.; Montellier, E.; Lu, Z.; Chen, Y.; Ishii, H.; Debernardi, A.; Buchou, T.; Rousseaux, S.; Jin, F.; et al. Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nat. Chem. Biol. 2014, 10, 365–370. [Google Scholar] [CrossRef]
- Xie, Z.; Zhang, D.; Chung, D.; Tang, Z.; Huang, H.; Dai, L.; Qi, S.; Li, J.; Colak, G.; Chen, Y.; et al. Metabolic Regulation of Gene Expression by Histone Lysine β-Hydroxybutyrylation. Mol. Cell 2016, 62, 194–206. [Google Scholar] [CrossRef]
- Zhang, D.; Tang, Z.; Huang, H.; Zhou, G.; Cui, C.; Weng, Y.; Liu, W.; Kim, S.; Lee, S.; Perez-Neut, M.; et al. Metabolic regulation of gene expression by histone lactylation. Nature 2019, 574, 575–580. [Google Scholar] [CrossRef]
- Huang, H.; Zhang, D.; Wang, Y.; Perez-Neut, M.; Han, Z.; Zheng, Y.G.; Hao, Q.; Zhao, Y. Lysine benzoylation is a histone mark regulated by SIRT2. Nat. Commun. 2018, 9, 3374. [Google Scholar] [CrossRef]
- Gao, Y.; Sheng, X.; Tan, D.; Kim, S.; Choi, S.; Paudel, S.; Lee, T.; Yan, C.; Tan, M.; Kim, K.M.; et al. Identification of Histone Lysine Acetoacetylation as a Dynamic Post-Translational Modification Regulated by HBO1. Adv. Sci. 2023, 10, e2300032. [Google Scholar] [CrossRef] [PubMed]
- Newman, J.C.; Verdin, E. β-Hydroxybutyrate: A Signaling Metabolite. Annu. Rev. Nutr. 2017, 37, 51–76. [Google Scholar] [CrossRef] [PubMed]
- Shimazu, T.; Hirschey, M.D.; Newman, J.; He, W.; Shirakawa, K.; Le Moan, N.; Grueter, C.A.; Lim, H.; Saunders, L.R.; Stevens, R.D.; et al. Suppression of oxidative stress by β-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science 2013, 339, 211–214. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.-M.; Ramprasath, T.; Zou, M.-H. β-hydroxybutyrate and its metabolic effects on age-associated pathology. Exp. Mol. Med. 2020, 52, 548–555. [Google Scholar] [CrossRef]
- Li, K.; Wang, W.-H.; Wu, J.-B.; Xiao, W.-H. β-hydroxybutyrate: A crucial therapeutic target for diverse liver diseases. Biomed. Pharmacother. 2023, 165, 115191. [Google Scholar] [CrossRef]
- Gregoretti, I.V.; Lee, Y.-M.; Goodson, H.V. Molecular evolution of the histone deacetylase family: Functional implications of phylogenetic analysis. J. Mol. Biol. 2004, 338, 17–31. [Google Scholar] [CrossRef]
- Xu, K.; Zhang, K.; Wang, Y.; Gu, Y. Comprehensive review of histone lactylation: Structure, function, and therapeutic targets. Biochem. Pharmacol. 2024, 225, 116331. [Google Scholar] [CrossRef]
- Koronowski, K.B.; Greco, C.M.; Huang, H.; Kim, J.-K.; Fribourgh, J.L.; Crosby, P.; Mathur, L.; Ren, X.; Partch, C.L.; Jang, C.; et al. Ketogenesis impact on liver metabolism revealed by proteomics of lysine β-hydroxybutyrylation. Cell Rep. 2021, 36, 109487. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wan, X.; Wu, Z.; Zhou, Y.; Chen, R.; Xu, W.; Zhang, J.; Yang, Z.; Bai, L.; Zhang, J.; et al. β-hydroxybutyrate reduces reinstatement of cocaine conditioned place preference through hippocampal CaMKII-α β-hydroxybutyrylation. Cell Rep. 2022, 41, 111724. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Li, F.; Sun, Q.; Lin, N.; Han, H.; You, K.; Tian, F.; Mao, Z.; Li, T.; Tong, T.; et al. p53 β-hydroxybutyrylation attenuates p53 activity. Cell Death Dis. 2019, 10, 243. [Google Scholar] [CrossRef] [PubMed]
- González-Valero, L.; Rodríguez-López, J.M.; Lachica, M.; Fernández-Fígares, I. Metabolic differences in hepatocytes of obese and lean pigs. Animal 2014, 8, 1873–1880. [Google Scholar] [CrossRef]
- Lu, J.F.; Zhu, M.Q.; Xia, B.; Zhang, N.N.; Liu, X.P.; Liu, H.; Zhang, R.X.; Xiao, J.Y.; Yang, H.; Zhang, Y.Q.; et al. GDF15 is a major determinant of ketogenic diet-induced weight loss. Cell Metab. 2024, 36, 454–456. [Google Scholar] [CrossRef]
- Meng, X.; Baine, J.M.; Yan, T.; Wang, S. Comprehensive Analysis of Lysine Lactylation in Rice (Oryza sativa) Grains. J. Agric. Food Chem. 2021, 69, 8287–8297. [Google Scholar] [CrossRef]
- Wang, X.; Tian, G.G.; Cheng, W.; Yu, X.; Li, X.; Wu, J. Metformin promotes female germline stem cell proliferation by upregulating Gata-binding protein 2 with histone β-hydroxybutyrylation. Stem Cell Res. Ther. 2023, 14, 144. [Google Scholar] [CrossRef]
- Luo, W.; He, M.; Luo, Q.; Li, Y. Proteome-wide analysis of lysine β-hydroxybutyrylation in the myocardium of diabetic rat model with cardiomyopathy. Front. Cardiovasc. Med. 2022, 9, 1066822. [Google Scholar] [CrossRef]
- Huang, H.; Zhang, D.; Weng, Y.; Delaney, K.; Tang, Z.; Yan, C.; Qi, S.; Peng, C.; Cole, P.A.; Roeder, R.G.; et al. The regulatory enzymes and protein substrates for the lysine β-hydroxybutyrylation pathway. Sci. Adv. 2021, 7, eabe2771. [Google Scholar] [CrossRef]
- Ding, W.-J.; Li, X.-H.; Tang, C.-M.; Yang, X.-C.; Sun, Y.; Song, Y.-P.; Ling, M.-Y.; Yan, R.; Gao, H.-Q.; Zhang, W.-H.; et al. Quantification and Proteomic Characterization of β-Hydroxybutyrylation Modification in the Hearts of AMPKα2 Knockout Mice. Mol. Cell Proteom. 2023, 22, 100494. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Duan, Y.; Ren, Z.; Niu, T.; Xu, Q.; Wang, Z.; Zheng, L.; Wang, Y.; Chen, X.; Huang, J.; et al. Post-Translational Modification β-Hydroxybutyrylation Regulates Ustilaginoidea virens Virulence. Mol. Cell Proteom. 2023, 22, 100616. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, X.; Yu, N.; Yan, R.; Sun, Y.; Tang, C.; Ding, W.; Ling, M.; Song, Y.; Gao, H.; et al. Proteomics and β-hydroxybutyrylation Modification Characterization in the Hearts of Naturally Senescent Mice. Mol. Cell Proteom. 2023, 22, 100659. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Tang, K.; Ma, J.; Zhou, L.; Liu, J.; Zeng, L.; Zhu, L.; Xu, P.; Chen, J.; Wei, K.; et al. Ketogenesis-generated β-hydroxybutyrate is an epigenetic regulator of CD8+ T-cell memory development. Nat. Cell Biol. 2020, 22, 18–25. [Google Scholar] [CrossRef]
- Zhang, X.-J.; She, Z.-G.; Wang, J.; Sun, D.; Shen, L.-J.; Xiang, H.; Cheng, X.; Ji, Y.-X.; Huang, Y.-P.; Li, P.-L.; et al. Multiple omics study identifies an interspecies conserved driver for nonalcoholic steatohepatitis. Sci. Transl. Med. 2021, 13, eabg8117. [Google Scholar] [CrossRef]
- Sahar, S.; Masubuchi, S.; Eckel-Mahan, K.; Vollmer, S.; Galla, L.; Ceglia, N.; Masri, S.; Barth, T.K.; Grimaldi, B.; Oluyemi, O.; et al. Circadian control of fatty acid elongation by SIRT1 protein-mediated deacetylation of acetyl-coenzyme A synthetase 1. J. Biol. Chem. 2014, 289, 6091–6097. [Google Scholar] [CrossRef]
- Zhu, R.; Ye, X.; Lu, X.; Xiao, L.; Yuan, M.; Zhao, H.; Guo, D.; Meng, Y.; Han, H.; Luo, S.; et al. ACSS2 acts as a lactyl-CoA synthetase and couples KAT2A to function as a lactyltransferase for histone lactylation and tumor immune evasion. Cell Metab. 2024, 37, 361–376. [Google Scholar] [CrossRef]
- Ringel, A.E.; Tucker, S.A.; Haigis, M.C. Chemical and Physiological Features of Mitochondrial Acylation. Mol. Cell 2018, 72, 610–624. [Google Scholar] [CrossRef]
- Zheng, Y.; Sun, W.; Shan, C.; Li, B.; Liu, J.; Xing, H.; Xu, Q.; Cui, B.; Zhu, W.; Chen, J.; et al. β-hydroxybutyrate inhibits ferroptosis-mediated pancreatic damage in acute liver failure through the increase of H3K9bhb. Cell Rep. 2022, 41, 111847. [Google Scholar] [CrossRef]
- Zhang, H.; Chang, Z.; Qin, L.-N.; Liang, B.; Han, J.-X.; Qiao, K.-L.; Yang, C.; Liu, Y.-R.; Zhou, H.-G.; Sun, T. MTA2 triggered R-loop trans-regulates BDH1-mediated β-hydroxybutyrylation and potentiates propagation of hepatocellular carcinoma stem cells. Signal Transduct. Target. Ther. 2021, 6, 135. [Google Scholar] [CrossRef]
- Qin, J.; Huang, X.; Gou, S.; Zhang, S.; Gou, Y.; Zhang, Q.; Chen, H.; Sun, L.; Chen, M.; Liu, D.; et al. Ketogenic diet reshapes cancer metabolism through lysine β-hydroxybutyrylation. Nat. Metab. 2024, 6, 1505–1528. [Google Scholar] [CrossRef]
- Zuo, Y.; Wang, Q.; Tian, W.; Zheng, Z.; He, W.; Zhang, R.; Zhao, Q.; Miao, Y.; Yuan, Y.; Wang, J.; et al. β-hydroxybutyrylation and O-GlcNAc modifications of STAT1 modulate antiviral defense in aging. Cell Mol. Immunol. 2025, 22, 403–417. [Google Scholar] [CrossRef]
- Jiang, W.; Wang, M.; Wang, J.; Hao, Q.; Li, Y.; Liu, L.; Zhou, T.; Song, W.; Liu, J.; Liu, M.; et al. β-Hydroxybutyrate promotes cancer metastasis through β-hydroxybutyrylation-dependent stabilization of Snail. Nat. Commun. 2025, 16, 6592. [Google Scholar] [CrossRef]
- Weinert, B.T.; Schölz, C.; Wagner, S.A.; Iesmantavicius, V.; Su, D.; Daniel, J.A.; Choudhary, C. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 2013, 4, 842–851. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, S.; Guan, J.; Tian, F.; Ye, H.; Wang, Q.; Lv, L.; Liu, Y.; Zheng, X.; Yin, Z.; Zhang, X. Global Profiling of Protein β-hydroxybutyrylome in Porcine Liver. Biology 2025, 14, 1183. https://doi.org/10.3390/biology14091183
Fan S, Guan J, Tian F, Ye H, Wang Q, Lv L, Liu Y, Zheng X, Yin Z, Zhang X. Global Profiling of Protein β-hydroxybutyrylome in Porcine Liver. Biology. 2025; 14(9):1183. https://doi.org/10.3390/biology14091183
Chicago/Turabian StyleFan, Shuhao, Jinyu Guan, Fang Tian, Haibo Ye, Qianqian Wang, Lei Lv, Yuanyuan Liu, Xianrui Zheng, Zongjun Yin, and Xiaodong Zhang. 2025. "Global Profiling of Protein β-hydroxybutyrylome in Porcine Liver" Biology 14, no. 9: 1183. https://doi.org/10.3390/biology14091183
APA StyleFan, S., Guan, J., Tian, F., Ye, H., Wang, Q., Lv, L., Liu, Y., Zheng, X., Yin, Z., & Zhang, X. (2025). Global Profiling of Protein β-hydroxybutyrylome in Porcine Liver. Biology, 14(9), 1183. https://doi.org/10.3390/biology14091183






