Evaluation of Hypovirus Infection on the Vesicular Protein Expression Pattern of Cryphonectria parasitica by TMT-Based Proteomics Analysis
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Culture Conditions
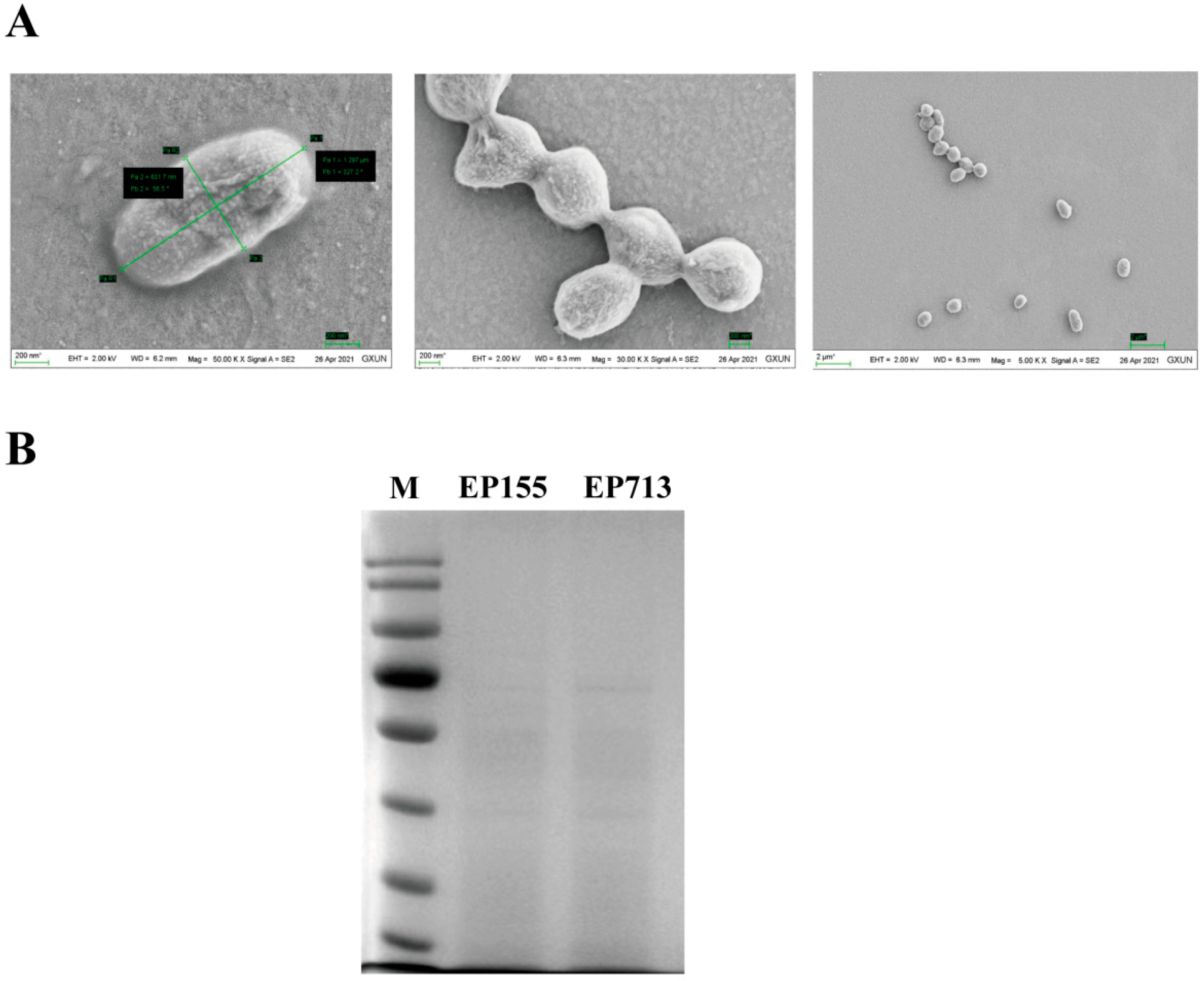
2.2. Vesicle Sample Preparation and Analysis
2.3. Protein Extraction and Digestion
2.4. Strong Cation Exchange (SCX) Fractionation
2.5. High pH Reversed-Phase Fractionation
2.6. LC-MS/MS Analysis
2.7. Identification and Quantitation of Proteins
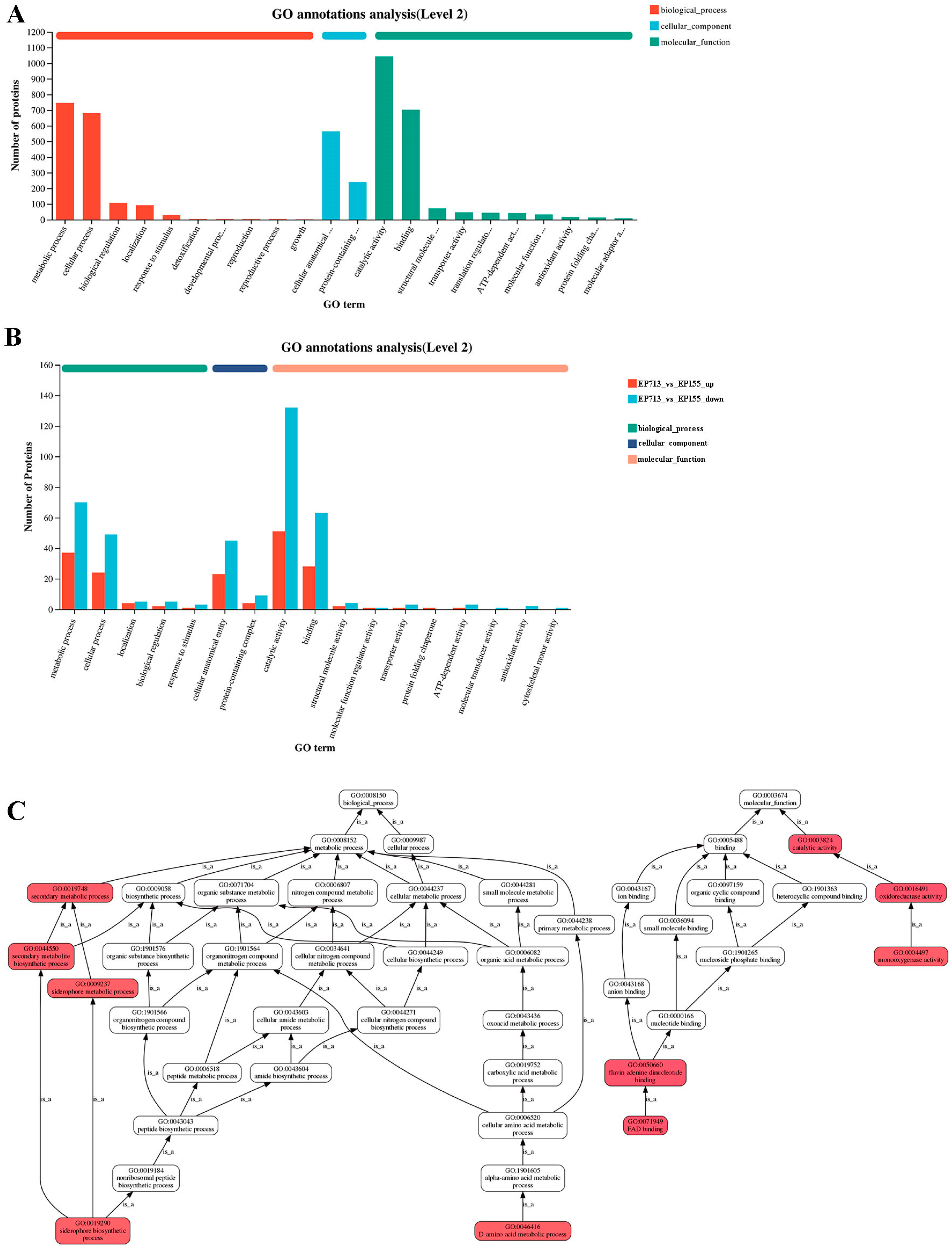
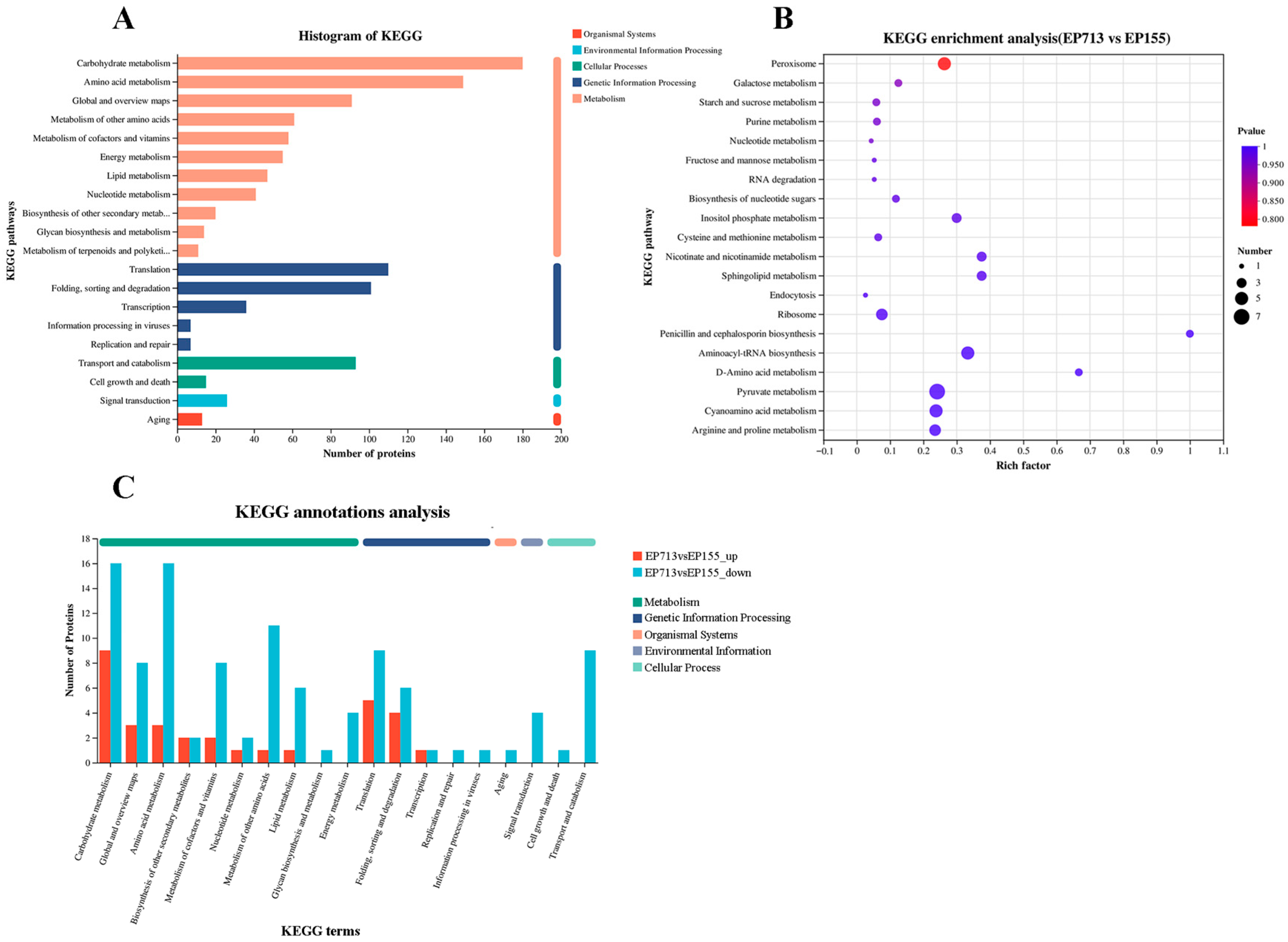
2.8. Bioinformatic Analysis
3. Results and Discussion
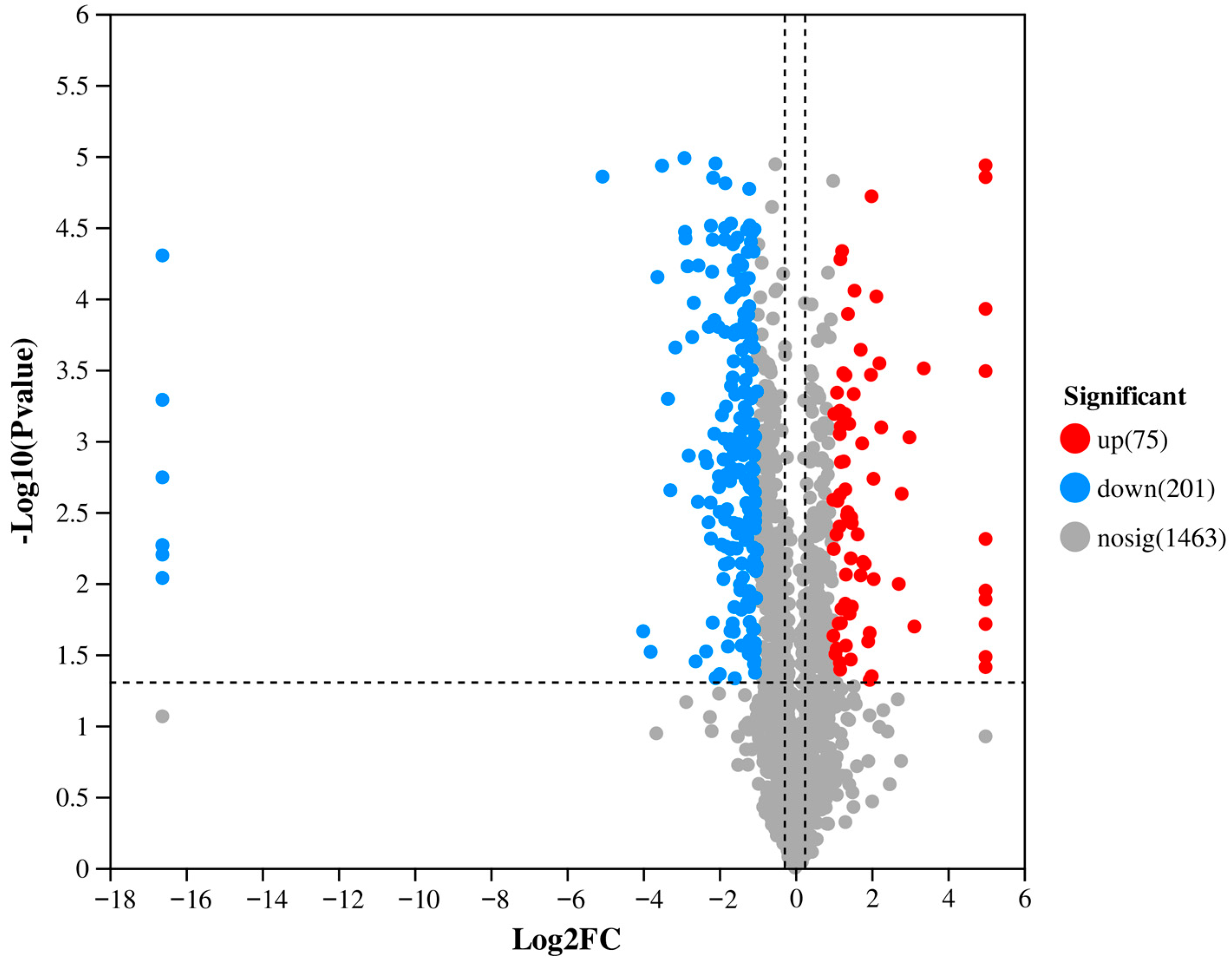
3.1. Differential Proteomic Analysis of Fungal Vesicle
3.2. Potential Impact of Hypovirus on Vesicle Trafficking and ER Homeostasis
3.3. Possible Stress Responses Associated with Vesicle Trafficking Changes
3.4. Altered Autophagy-Related Responses
3.5. Up-Regulation of t-SNARE Proteins and Potential Functional Shifts
3.6. Potential Impact of Down-Regulated Importin-β on Nuclear Transport and Pathogen Virulence
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rigling, D.; Prospero, S. Cryphonectria parasitica, the Causal Agent of Chestnut Blight: Invasion History, Population Biology and Disease Control. Mol. Plant Pathol. 2018, 19, 7–20. [Google Scholar] [CrossRef]
- Nuss, D.L. Hypovirulence: Mycoviruses at the Fungal–Plant Interface. Nat. Rev. Microbiol. 2005, 3, 632–642. [Google Scholar] [CrossRef]
- Hillman, B.I.; Suzuki, N. Viruses of the Chestnut Blight Fungus, Cryphonectria parasitica. Adv. Virus Res. 2004, 63, 423–472. [Google Scholar] [CrossRef] [PubMed]
- Turina, M.; Rostagno, L. Virus-Induced Hypovirulence in Cryphonectria parasitica: Still an Unresolved Conundrum. J. Plant Pathol. 2007, 89, 165–178. [Google Scholar]
- Heiniger, U.; Rigling, D. Biological Control of Chestnut Blight in Europe. Annu. Rev. Phytopathol. 1994, 32, 581–599. [Google Scholar] [CrossRef]
- Prospero, S.; Conedera, M.; Heiniger, U.; Rigling, D. Saprophytic Activity and Sporulation of Cryphonectria parasitica on Dead Chestnut Wood in Forests with Naturally Established Hypovirulence. Phytopathology 2006, 96, 1337–1344. [Google Scholar] [CrossRef]
- Allen, T.D.; Nuss, D.L. Specific and Common Alterations in Host Gene Transcript Accumulation Following Infection of the Chestnut Blight Fungus by Mild and Severe Hypoviruses. J. Virol. 2004, 78, 4145–4155. [Google Scholar] [CrossRef]
- Zheng, J.; Sugrue, R.J.; Tang, K. Mass Spectrometry Based Proteomic Studies on Viruses and Hosts–a Review. Anal. Chim. Acta 2011, 702, 149–159. [Google Scholar] [CrossRef]
- Powell, W.A.; Van Alfen, N.K. Two Nonhomologus Viruses of Cryphonectria (Endothia) parasitica Reduce Accumulation of Specific Virulence-Associated Polypeptides. J. Bacteriol. 1987, 169, 5324–5326. [Google Scholar] [CrossRef]
- Kim, J.-M.; Park, J.-A.; Kim, D.-H. Comparative Proteomic Analysis of Chestnut Blight Fungus, Cryphonectria parasitica, under Tannic-Acid-Inducing and Hypovirus-Regulating Conditions. Can. J. Microbiol. 2012, 58, 863–871. [Google Scholar] [CrossRef]
- Kazmierczak, P.; McCabe, P.; Turina, M.; Jacob-Wilk, D.; Van Alfen, N.K. The Mycovirus CHV1 Disrupts Secretion of a Developmentally Regulated Protein in Cryphonectria parasitica. J. Virol. 2012, 86, 6067–6074. [Google Scholar] [CrossRef]
- Wang, J.; Shi, L.; He, X.; Lu, L.; Li, X.; Chen, B. Comparative Secretome Analysis Reveals Perturbation of Host Secretion Pathways by a Hypovirus. Sci. Rep. 2016, 6, 34308. [Google Scholar] [CrossRef]
- Wang, J.; Wang, F.; Feng, Y.; Mi, K.; Chen, Q.; Shang, J.; Chen, B. Comparative Vesicle Proteomics Reveals Selective Regulation of Protein Expression in Chestnut Blight Fungus by a Hypovirus. J. Proteom. 2013, 78, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.; Ko, Y.-H.; Kim, D.-H. Transcriptome Analysis of Cryphonectria parasitica Infected with Cryphonectria Hypovirus 1 (CHV1) Reveals Distinct Genes Related to Fungal Metabolites, Virulence, Antiviral RNA-Silencing, and Their Regulation. Front. Microbiol. 2020, 11, 1711. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal Sample Preparation Method for Proteome Analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Choi, G.; Shapira, R.; Nuss, D. Cotranslational Autoproteolysis Involved in Gene Expression from a Double-Stranded RNA Genetic Element Associated with Hypovirulence of the Chestnut Blight Fungus. Proc. Natl. Acad. Sci. USA 1991, 88, 1167–1171. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Nuss, D.L.; Suzuki, N. Synergism between a Mycoreovirus and a Hypovirus Mediated by the Papain-like Protease P29 of the Prototypic Hypovirus CHV1-EP713. J. Gen. General. Virol. 2006, 87, 3703–3714. [Google Scholar] [CrossRef]
- Deng, F.; Nuss, D.L. Hypovirus Papain-Like Protease P48 Is Required for Initiation but Not for Maintenance of Virus RNA Propagation in the Chestnut Blight Fungus Cryphonectria parasitica. J. Virol. 2008, 82, 6369–6378. [Google Scholar] [CrossRef]
- Okazaki, Y.; Ohno, H.; Takase, K.; Ochiai, T.; Saito, T. Cell Surface Expression of Calnexin, a Molecular Chaperone in the Endoplasmic Reticulum. J. Biol. Chem. 2000, 275, 35751–35758. [Google Scholar] [CrossRef]
- Huang, T.; Ma, X.; Zhao, Z.; Qin, D.; Qin, W.; Wang, J.; Chen, B.; He, X. Homeostasis of Calnexin Is Essential for the Growth, Virulence, and Hypovirus RNA Accumulation in the Chestnut Blight Fungus. Mol. Microbiol. 2025, 123, 393–405. [Google Scholar] [CrossRef]
- Weimer, C.; Beck, R.; Eckert, P.; Reckmann, I.; Moelleken, J.; Brügger, B.; Wieland, F. Differential Roles of ArfGAP1, ArfGAP2, and ArfGAP3 in COPI Trafficking. J. Cell Biol. 2008, 183, 725–735. [Google Scholar] [CrossRef]
- Letourneur, F.; Gaynor, E.C.; Hennecke, S.; Demolliere, C.; Duden, R.; Emr, S.D.; Riezman, H.; Cosson, P. Coatomer Is Essential for Retrieval of Dilysine-Tagged Proteins to the Endoplasmic Reticulum. Cell 1994, 79, 1199–1207. [Google Scholar] [CrossRef]
- Spang, A.; Shiba, Y.; Randazzo, P.A. Arf GAPs: Gatekeepers of Vesicle Generation. FEBS Lett. 2010, 584, 2646–2651. [Google Scholar] [CrossRef]
- Saitoh, A.; Shin, H.-W.; Yamada, A.; Waguri, S.; Nakayama, K. Three Homologous ArfGAPs Participate in Coat Protein I-Mediated Transport *. J. Biol. Chem. 2009, 284, 13948–13957. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Li, X.; Ma, Y.; Zhang, K.; Yang, J. The Arf-GAP Proteins AoGcs1 and AoGts1 Regulate Mycelial Development, Endocytosis, and Pathogenicity in Arthrobotrys oligospora. J. Fungi 2022, 8, 463. [Google Scholar] [CrossRef] [PubMed]
- Musungu, B.M.; Bhatnagar, D.; Brown, R.L.; Payne, G.A.; OBrian, G.; Fakhoury, A.M.; Geisler, M. A Network Approach of Gene Co-Expression in the Zea Mays/Aspergillus Flavus Pathosystem to Map Host/Pathogen Interaction Pathways. Front. Genet. 2016, 7, 206. [Google Scholar] [CrossRef] [PubMed]
- Luo, P.M.; Boyce, M. Directing Traffic: Regulation of COPI Transport by Post-Translational Modifications. Front. Cell Dev. Biol. 2019, 7, 190. [Google Scholar] [CrossRef]
- Richie, D.L.; Feng, X.; Hartl, L.; Aimanianda, V.; Krishnan, K.; Powers-Fletcher, M.V.; Watson, D.S.; Galande, A.K.; White, S.M.; Willett, T.; et al. The Virulence of the Opportunistic Fungal Pathogen Aspergillus Fumigatus Requires Cooperation between the Endoplasmic Reticulum-Associated Degradation Pathway (ERAD) and the Unfolded Protein Response (UPR). Virulence 2011, 2, 12–21. [Google Scholar] [CrossRef]
- Yu, M. HacA, a Key Transcription Factor for the Unfolded Protein Response, Is Required for Fungal Development, Aflatoxin Biosynthesis and Pathogenicity of Aspergillus Flavus. Int. J. Food Microbiol. 2024, 417, 110693. [Google Scholar] [CrossRef]
- Kirchhausen, T. Clathrin. Annu. Rev. Biochem. 2000, 69, 699–727. [Google Scholar] [CrossRef]
- Jones, S.M.; Howell, K.E.; Henley, J.R.; Cao, H.; McNiven, M.A. Role of Dynamin in the Formation of Transport Vesicles from the Trans-Golgi Network. Science 1998, 279, 573–577. [Google Scholar] [CrossRef]
- Deloche, O.; Yeung, B.G.; Payne, G.S.; Schekman, R. Vps10p Transport from the Trans-Golgi Network to the Endosome Is Mediated by Clathrin-Coated Vesicles. Mol. Biol. Cell 2017, 12, 475–485. [Google Scholar] [CrossRef] [PubMed]
- Marcusson, E.G.; Horazdovsky, B.F.; Cereghino, J.L.; Gharakhanian, E.; Emr, S.D. The Sorting Receptor for Yeast Vacuolar Carboxypeptidase Y Is Encoded by the VPS10 Gene. Cell 1994, 77, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Ishii, A.; Kawai, M.; Noda, H.; Kato, H.; Takeda, K.; Asakawa, K.; Ichikawa, Y.; Sasanami, T.; Tanaka, K.; Kimura, Y. Accelerated Invagination of Vacuoles as a Stress Response in Chronically Heat-Stressed Yeasts. Sci. Rep. 2018, 8, 2644. [Google Scholar] [CrossRef] [PubMed]
- Rosenzweig, R.; Nillegoda, N.B.; Mayer, M.P.; Bukau, B. The Hsp70 Chaperone Network. Nat. Rev. Mol. Cell Biol. 2019, 20, 665–680. [Google Scholar] [CrossRef]
- Lubkowska, A.; Pluta, W.; Strońska, A.; Lalko, A. Role of Heat Shock Proteins (HSP70 and HSP90) in Viral Infection. Int. J. Mol. Sci. 2021, 22, 9366. [Google Scholar] [CrossRef]
- Sousa, R.; Lafer, E.M. The Role of Molecular Chaperones in Clathrin Mediated Vesicular Trafficking. Front. Mol. Biosci. 2015, 2, 26. [Google Scholar] [CrossRef]
- Hong, E.; Davidson, A.R.; Kaiser, C.A. A Pathway for Targeting Soluble Misfolded Proteins to the Yeast Vacuole. J. Cell Biol. 1996, 135, 623–633. [Google Scholar] [CrossRef]
- Jawed, A.; Ho, C.-T.; Grousl, T.; Shrivastava, A.; Ruppert, T.; Bukau, B.; Mogk, A. Balanced Activities of Hsp70 and the Ubiquitin Proteasome System Underlie Cellular Protein Homeostasis. Front. Mol. Biosci. 2023, 9, 1106477. [Google Scholar] [CrossRef]
- Wang, K.; Yu, W.; Yu, G.; Zhang, L.; Xian, L.; Wei, Y.; Perez-Sancho, J.; Xue, H.; Rufian, J.S.; Zhuang, H.; et al. A Bacterial Type III Effector Targets Plant Vesicle-associated Membrane Proteins. Mol. Plant Pathol. 2023, 24, 1154–1167. [Google Scholar] [CrossRef]
- Krishnan, K.; Askew, D.S. Endoplasmic Reticulum Stress and Fungal Pathogenesis. Fungal Biol. Rev. 2014, 28, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Nakatogawa, H. Mechanisms Governing Autophagosome Biogenesis. Nat. Rev. Mol. Cell Biol. 2020, 21, 439–458. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.-T.; Chang, A.-N.; Han, L.-T.; Guo, J.-S.; Li, Y.-H.; Liu, T.-B. Autophagy Regulates Fungal Virulence and Sexual Reproduction in Cryptococcus neoformans. Front. Cell Dev. Biol. 2020, 8, 374. [Google Scholar] [CrossRef]
- Shi, L.; Wang, J.; Quan, R.; Yang, F.; Shang, J.; Chen, B. CpATG8, a Homolog of Yeast Autophagy Protein ATG8, Is Required for Pathogenesis and Hypovirus Accumulation in the Chest Blight Fungus. Front. Cell. Infect. Microbiol. 2019, 9, 222. [Google Scholar] [CrossRef] [PubMed]
- Vogel, C.; Marcotte, E.M. Insights into the Regulation of Protein Abundance from Proteomic and Transcriptomic Analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef]
- Liu, Y.; Beyer, A.; Aebersold, R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef]
- Chen, H.; Charles, P.D.; Gu, Q.; Liberatori, S.; Robertson, D.L.; Palmarini, M.; Wilson, S.J.; Mohammed, S.; Castello, A. Omics Analyses Uncover Host Networks Defining Virus-Permissive and -Hostile Cellular States. Mol. Cell. Proteom. 2025, 24, 100966. [Google Scholar] [CrossRef]
- Werner, A.; Herzog, B.; Voigt, O.; Valerius, O.; Braus, G.H.; Pöggeler, S. NBR1 Is Involved in Selective Pexophagy in Filamentous Ascomycetes and Can Be Functionally Replaced by a Tagged Version of Its Human Homolog. Autophagy 2019, 15, 78–97. [Google Scholar] [CrossRef]
- Chen, Y.A.; Scheller, R.H. SNARE-Mediated Membrane Fusion. Nat. Rev. Mol. Cell Biol. 2001, 2, 98–106. [Google Scholar] [CrossRef]
- Bonifacino, J.S.; Glick, B.S. The Mechanisms of Vesicle Budding and Fusion. Cell 2004, 116, 153–166. [Google Scholar] [CrossRef]
- Buser, D.P.; Spang, A. Protein Sorting from Endosomes to the TGN. Front. Cell Dev. Biol. 2023, 11, 1140605. [Google Scholar] [CrossRef]
- Lu, Q.; Lu, Z.; Liu, Q.; Guo, L.; Ren, H.; Fu, J.; Jiang, Q.; Clarke, P.R.; Zhang, C. Chromatin-Bound NLS Proteins Recruit Membrane Vesicles and Nucleoporins for Nuclear Envelope Assembly via Importin-α/β. Cell Res. 2012, 22, 1562–1575. [Google Scholar] [CrossRef]
- Cingolani, G.; Petosa, C.; Weis, K.; Müller, C.W. Structure of Importin-β Bound to the IBB Domain of Importin-α. Nature 1999, 399, 221–229. [Google Scholar] [CrossRef]
- Roscioli, E.; Di Francesco, L.; Bolognesi, A.; Giubettini, M.; Orlando, S.; Harel, A.; Schininà, M.E.; Lavia, P. Importin-β Negatively Regulates Multiple Aspects of Mitosis Including RANGAP1 Recruitment to Kinetochores. J. Cell Biol. 2012, 196, 435–450. [Google Scholar] [CrossRef]
- Rotem, A.; Gruber, R.; Shorer, H.; Shaulov, L.; Klein, E.; Harel, A. Importin β Regulates the Seeding of Chromatin with Initiation Sites for Nuclear Pore Assembly. Mol. Biol. Cell 2009, 20, 4031–4042. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Li, H.; Xi, Y.; Hu, Q.; Liu, H.; Fan, J.; Xiang, Y.; Zhang, X.; Shui, W.; Lai, Y. Vesicle Trafficking and Vesicle Fusion: Mechanisms, Biological Functions, and Their Implications for Potential Disease Therapy. Mol. Biomed. 2022, 3, 29. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Sun, K.; Li, Y.; Ma, T.; Zhang, Y.; Yin, Y.; Zhang, S.; Shao, W. The Importin FgPse1 Is Required for Vegetative Development, Virulence, and Deoxynivalenol Production by Interacting with the Nuclear Polyadenylated RNA-Binding Protein FgNab2 in Fusarium Graminearum. Phytopathology 2022, 112, 1072–1080. [Google Scholar] [CrossRef] [PubMed]
- Shoji, J.; Kikuma, T.; Kitamoto, K. Vesicle Trafficking, Organelle Functions, and Unconventional Secretion in Fungal Physiology and Pathogenicity. Curr. Opin. Microbiol. 2014, 20, 1–9. [Google Scholar] [CrossRef]
| Accession | Description |
|---|---|
| Q04350 | ORFB polyprotein OS = Cryphonectria hypovirus 1 (strain EP713) OX = 12478 PE = 1 SV = 1 − [POLB_CHPVE] |
| A0A2H4UK75 | RNA-directed RNA polymerase OS = Cryphonectria hypovirus 1 OX = 40281 PE = 4 SV = 1 − [A0A2H4UK75_9VIRU] |
| A0A2H4UK88 | RNA-directed RNA polymerase OS = Cryphonectria hypovirus 1 OX = 40281 PE = 4 SV = 1 − [A0A2H4UK88_9VIRU] |
| A0A2H4UK77 | RNA-directed RNA polymerase OS = Cryphonectria hypovirus 1 OX = 40281 PE = 4 SV = 1 − [A0A2H4UK77_9VIRU] |
| A0A2H4UK87 | RNA-directed RNA polymerase OS = Cryphonectria hypovirus 1 OX = 40281 PE = 4 SV = 1 − [A0A2H4UK87_9VIRU] |
| A0A2H4UK85 | RNA-directed RNA polymerase OS = Cryphonectria hypovirus 1 OX = 40281 PE = 4 SV = 1 − [A0A2H4UK85_9VIRU] |
| P10941 | Polyprotein p69 (precursor to p29 and p40) OS = Cryphonectria hypovirus 1 (strain EP713) OX = 12478 PE = 1 SV = 2 − [POLA_CHPVE] |
| A0A2H4UK83 | ORF A OS = Cryphonectria hypovirus 1 OX = 40281 PE = 4 SV = 1 − [A0A2H4UK83_9VIRU] |
| E2GDE0 | Polyprotein OS = Cryphonectria hypovirus 1 OX = 40281 PE = 4 SV = 1 − [E2GDE0_9VIRU] |
| A0A2H4UK61 | ORF A OS = Cryphonectria hypovirus 1 OX = 40281 PE = 4 SV = 1 − [A0A2H4UK61_9VIRU] |
| W8EC18 | OrfA polyprotein (Fragment) OS = Cryphonectria hypovirus 1 OX = 40281 GN = OrfA PE = 4 SV = 1 − [W8EC18_9VIRU] |
| A0A2H4UK73 | ORF A OS = Cryphonectria hypovirus 1 OX = 40281 PE = 4 SV = 1 − [A0A2H4UK73_9VIRU] |
| Q9YME5 | Peptidase C7 domain-containing protein OS = Cryphonectria parasitica hypovirulence associated virus OX = 83190 PE = 4 SV = 1 − [Q9YME5_9VIRU] |
| Q7TDB9 | OrfA (Fragment) OS = Cryphonectria hypovirus 1 OX = 40281 PE = 4 SV = 1 − [Q7TDB9_9VIRU] |
| Q7T598 | ORF B (Fragment) OS = Cryphonectria hypovirus 1 OX = 40281 PE = 4 SV = 1 − [Q7T598_9VIRU] |
| Q7T594 | ORF B (Fragment) OS = Cryphonectria hypovirus 1 OX = 40281 PE = 4 SV = 1 − [Q7T594_9VIRU] |
| E2GDC9 | p48 (Fragment) OS = Cryphonectria hypovirus 1 OX = 40281 PE = 4 SV = 1 − [E2GDC9_9VIRU] |
| A0A7T0Q710 | ORF-A polyprotein (Fragment) OS = Cryphonectria hypovirus 1 OX = 40281 PE = 4 SV = 1 − [A0A7T0Q710_9VIRU] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, Z.; Lin, N.; Lu, T.; Xu, J.; Zhang, Z.; Wang, F.; Wang, J. Evaluation of Hypovirus Infection on the Vesicular Protein Expression Pattern of Cryphonectria parasitica by TMT-Based Proteomics Analysis. Biology 2025, 14, 1123. https://doi.org/10.3390/biology14091123
Zeng Z, Lin N, Lu T, Xu J, Zhang Z, Wang F, Wang J. Evaluation of Hypovirus Infection on the Vesicular Protein Expression Pattern of Cryphonectria parasitica by TMT-Based Proteomics Analysis. Biology. 2025; 14(9):1123. https://doi.org/10.3390/biology14091123
Chicago/Turabian StyleZeng, Zishan, Nanxin Lin, Tao Lu, Jian Xu, Zheng Zhang, Fang Wang, and Jinzi Wang. 2025. "Evaluation of Hypovirus Infection on the Vesicular Protein Expression Pattern of Cryphonectria parasitica by TMT-Based Proteomics Analysis" Biology 14, no. 9: 1123. https://doi.org/10.3390/biology14091123
APA StyleZeng, Z., Lin, N., Lu, T., Xu, J., Zhang, Z., Wang, F., & Wang, J. (2025). Evaluation of Hypovirus Infection on the Vesicular Protein Expression Pattern of Cryphonectria parasitica by TMT-Based Proteomics Analysis. Biology, 14(9), 1123. https://doi.org/10.3390/biology14091123







