Organelle Genome Characteristics and Phylogenetic Analysis of a Warm-Season Turfgrass Eremochloa ophiuroides (Poaceae)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Sequencing
2.2. Organelle Genome Assembly and Annotation
2.3. Codon Usage Bias Analysis
2.4. Repeat Sequences Identification
2.5. Gene Transfer Events Between Chloroplast and Mitochondrial Genomes
2.6. Phylogenetic Analyses in the Gramineae Family
2.7. Structural Variation Among Mitochondrial Genomes
2.8. Organelle Genome Marker Development and Utilization
3. Results
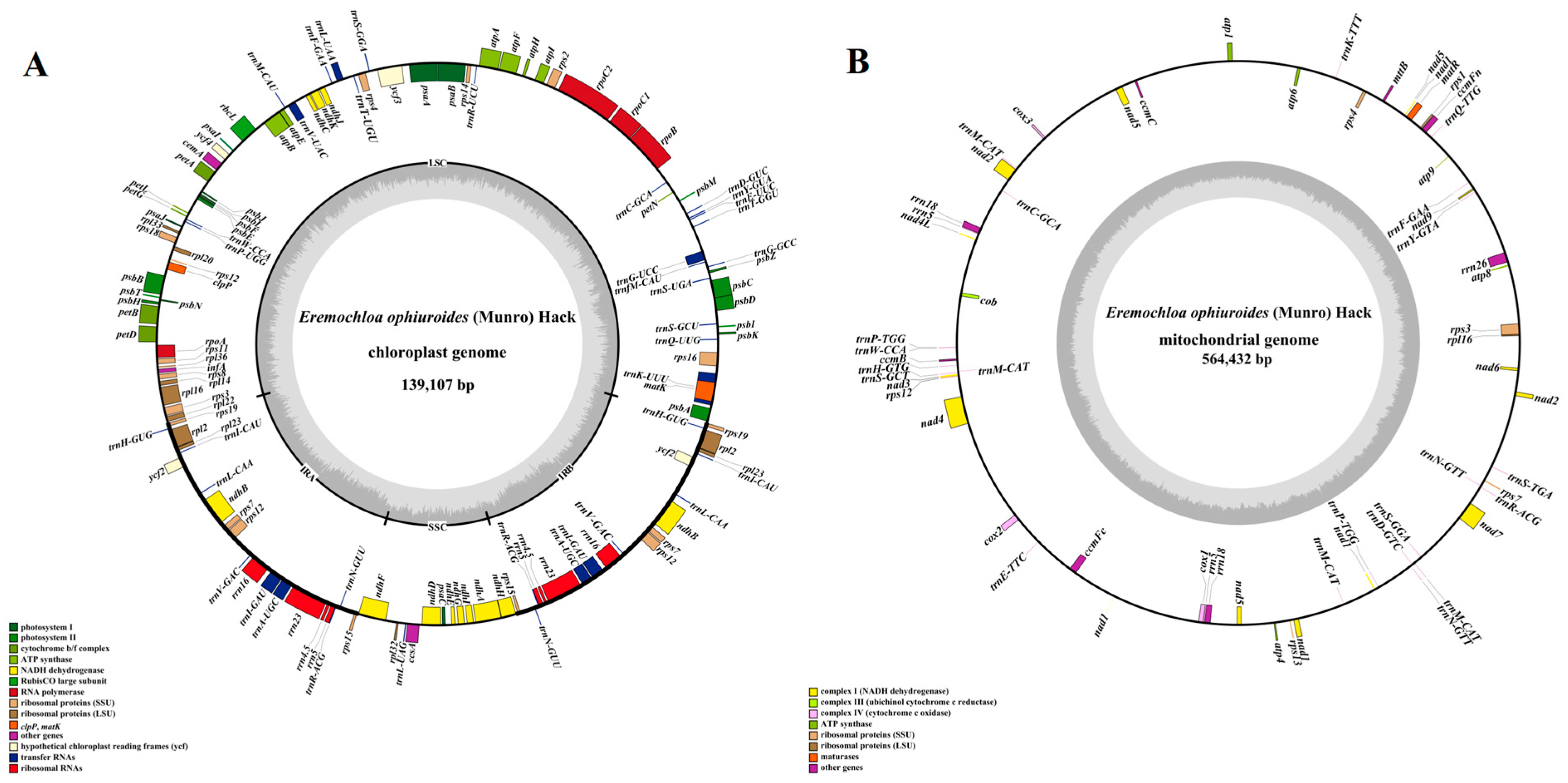
3.1. The Assembly and Annotation of the Organelle Genomes of Centipedegrass
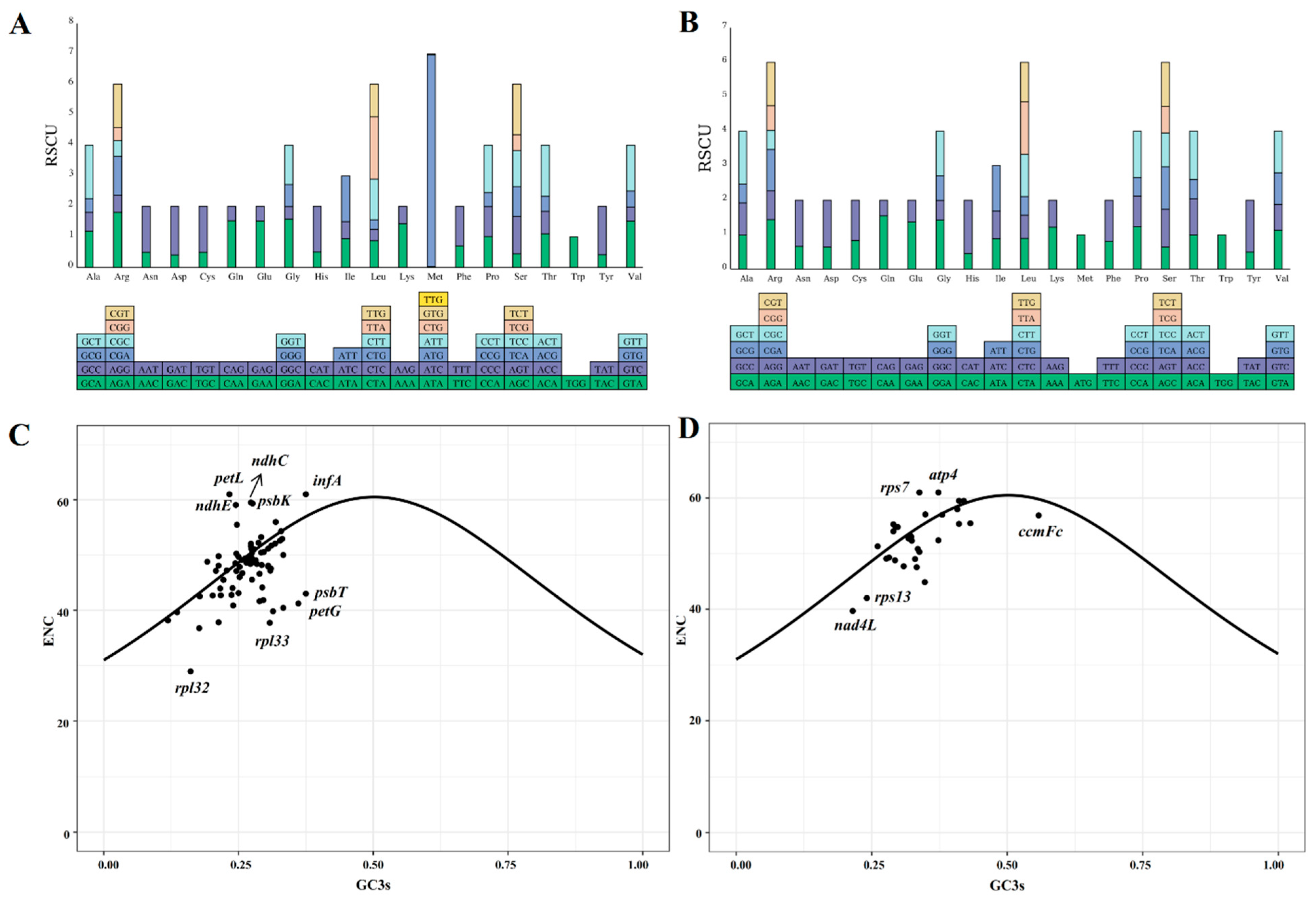
3.2. Codon Bias Analysis
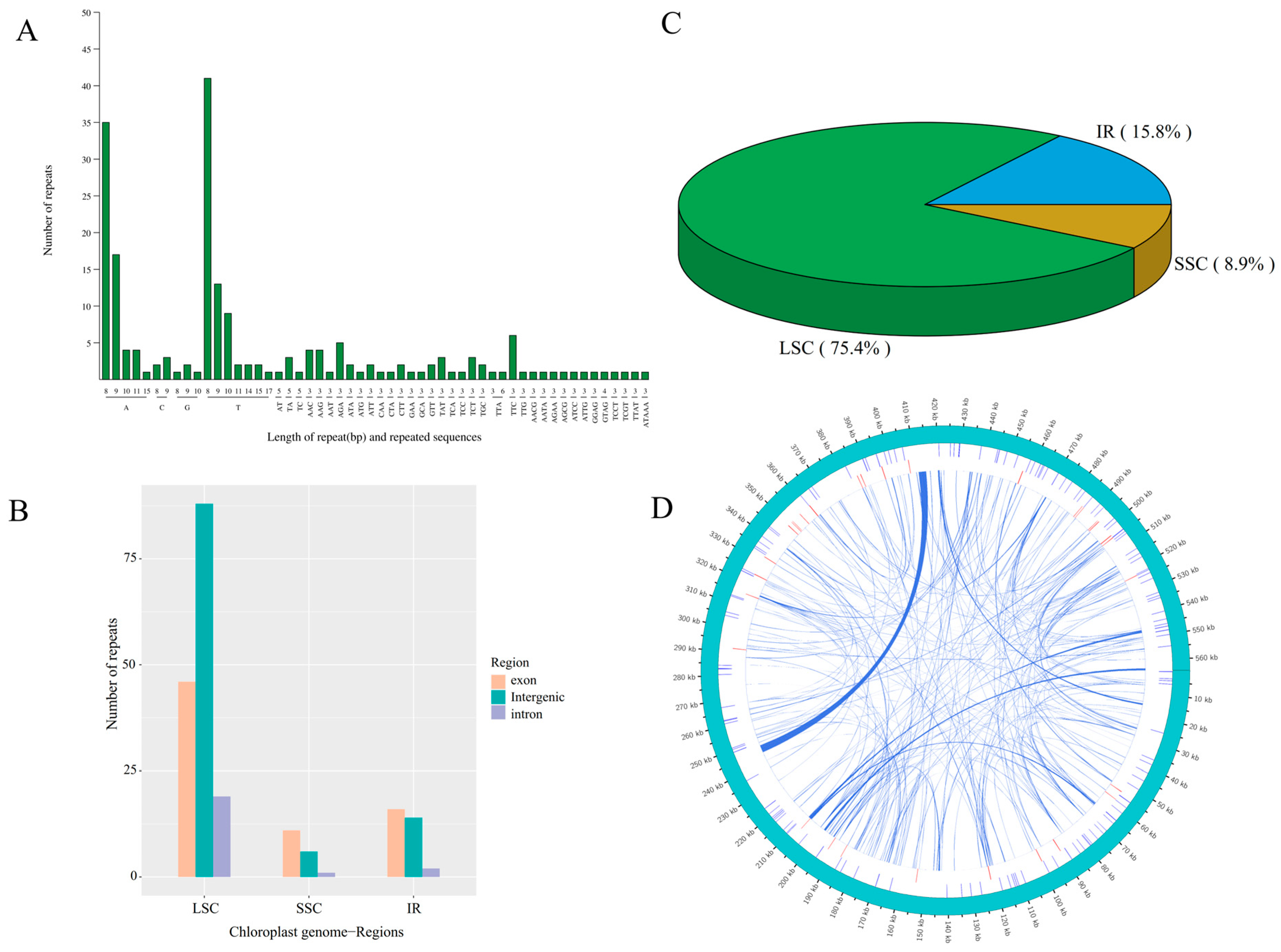
3.3. Repeat Sequence Analysis
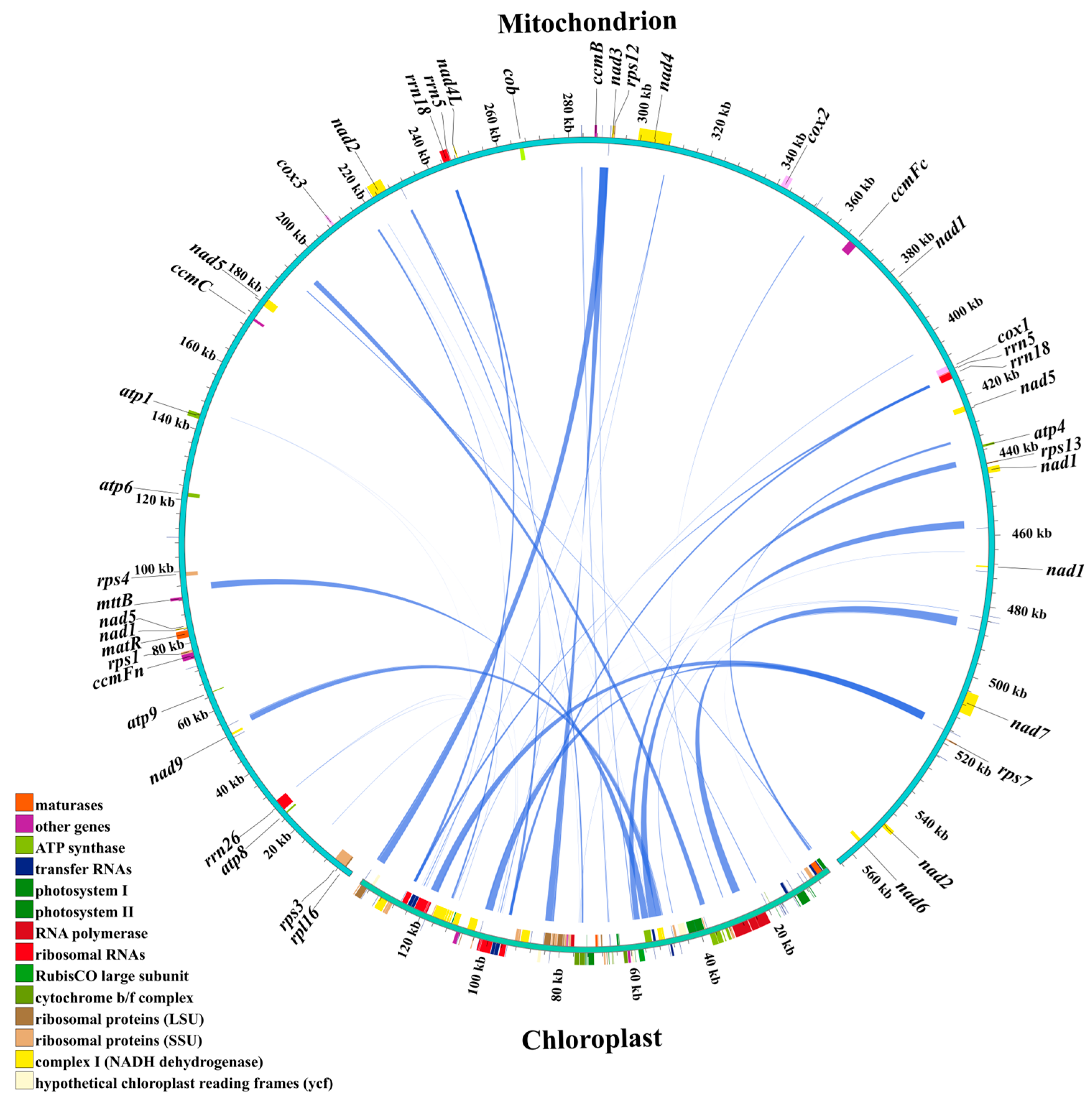
3.4. Homology Sequences Between Chloroplast and Mitochondrial Genomes of Centipedegrass
3.5. Phylogenetic Analyses and Divergence Times in the Family Gramineae
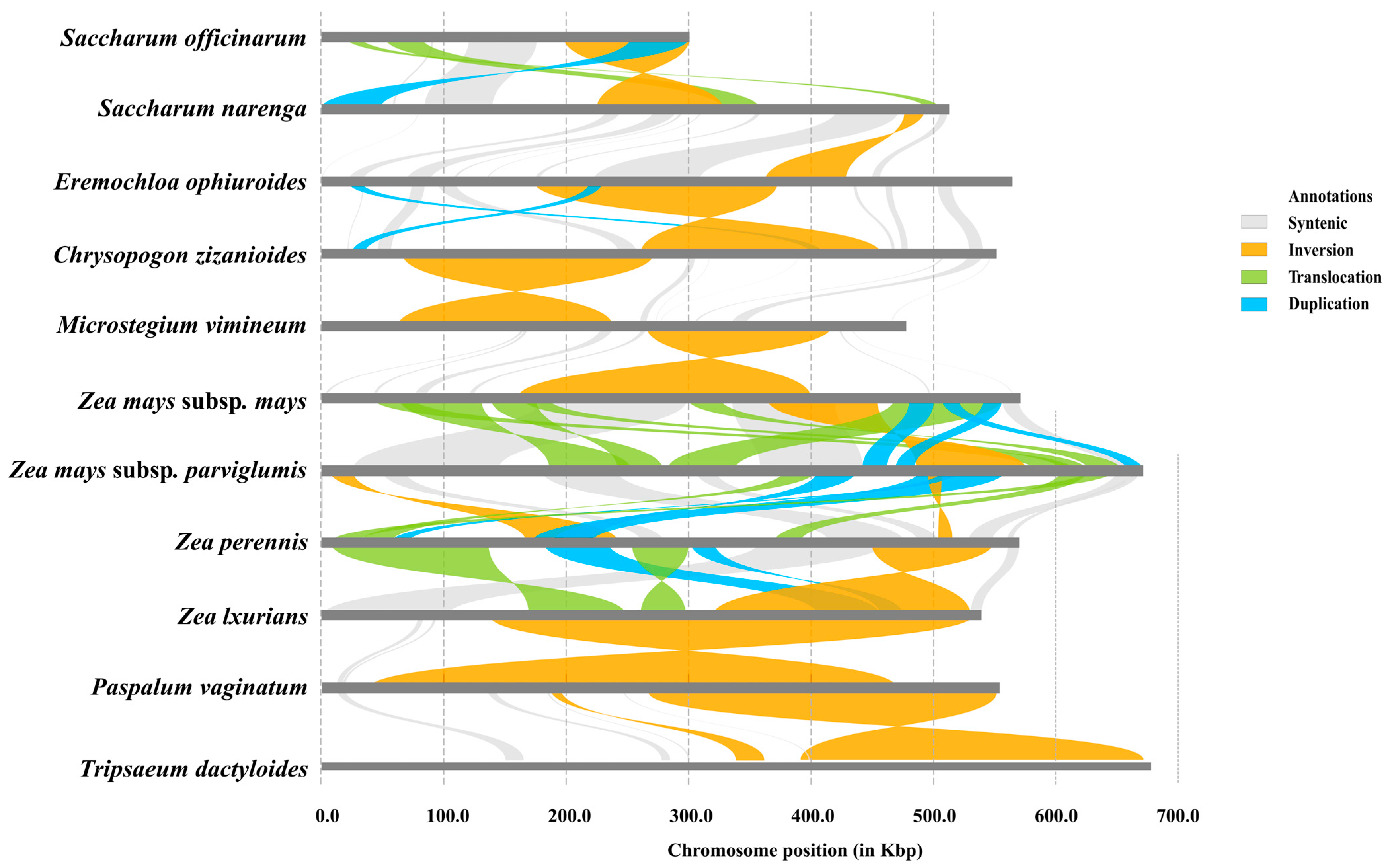
3.6. Structural Variations Among Mitochondrial Genomes
3.7. Genetic Diversity Indices Based on Organelle Genome Markers
4. Discussions
4.1. Centipedegrass Mitogenome Reveals Evolutionary and Breeding Clues
4.2. Repeated Sequences Identified in the Centipedegrass Mitogenome
4.3. Plastid DNA Integration in Centipedegrass Mitogenome
4.4. Codon Usage Patterns in Centipedegrass Organelle Genomes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ENC | Effective number of codons |
| GC3s | 3rd synonymous codon |
| IGT | Intracellular gene transfer |
| LSC | Large single copy |
| NGS | Next-generation sequencing |
| NJ | Neighbor-joining |
| PCGs | Protein-coding genes |
| RSCU | Relative synonymous codon usage |
| SSRs | Simple sequence repeats |
| SVs | Structural variations |
References
- Li, J.; Guo, H.; Zong, J.; Chen, J.; Li, D.; Liu, J. Genetic diversity in centipedegrass [Eremochloa ophiuroides (Munro) Hack.]. Hortic. Res. 2020, 7, 4. [Google Scholar] [CrossRef]
- Milla-Lewis, S.R.; Kimball, J.A.; Zuleta, M.C.; Harris-Shultz, K.R.; Schwartz, B.M.; Hanna, W.W. Use of sequence-related amplified polymorphism (SRAP) markers for comparing levels of genetic diversity in centipedegrass (Eremochloa ophiuroides (Munro) Hack.) germplasm. Genet. Resour. Crop Evol. 2012, 59, 1517–1526. [Google Scholar] [CrossRef]
- Shen, X.X.; Mei, X.D.; Wang, Z.A.; Jiang, J.M. Chinese She Medicine Plant Atlas; Zhejiang Science and Technology Press: Hangzhou, China, 2019; Volume 2. [Google Scholar]
- Zimorski, V.; Ku, C.; Martin, W.F.; Gould, S.B. Endosymbiotic theory for organelle origins. Curr. Opin. Microbiol. 2014, 22, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.X.; Chen, X.D.; Long, L.J.; Jost, M.; Zhao, R.; Liu, L.M.; Mower, J.P.; dePamphilis, C.W.; Wanke, S.; Jiao, Y.N. De novo assembly and comparative analyses of mitochondrial genomes in Piperales. Genome Biol. Evol. 2023, 15, evad041. [Google Scholar] [CrossRef]
- Hiesel, R.; von Haeseler, A.; Brennicke, A. Plant mitochondrial nucleic acid sequences as a tool for phylogenetic analysis. Proc. Natl. Acad. Sci. USA 1994, 91, 634–638. [Google Scholar] [CrossRef]
- Duminil, J.; Besnard, G. Utility of the mitochondrial genome in plant taxonomic studies. In Molecular Plant Taxonomy: Methods and Protocols; Humana: New York, NY, USA, 2021; pp. 107–118. [Google Scholar] [CrossRef]
- Wang, X.L.; Cheng, F.; Rohlsen, D.; Bi, C.W.; Wang, C.Y.; Xu, Y.Q.; Wei, S.Y.; Ye, Q.L.; Yin, T.M.; Ye, N. Organellar genome assembly methods and comparative analysis of horticultural plants. Hortic. Res. 2018, 5, 3. [Google Scholar] [CrossRef]
- Govindarajulu, R.; Parks, M.; Tennessen, J.A.; Liston, A.; Ashman, T.L. Comparison of nuclear, plastid, and mitochondrial phylogenies and the origin of wild octoploid strawberry species. Am. J. Bot. 2015, 102, 544–554. [Google Scholar] [CrossRef]
- Rydin, C.; Wikström, N.; Bremer, B. Conflicting results from mitochondrial genomic data challenge current views of Rubiaceae phylogeny. Am. J. Bot. 2017, 104, 1522–1532. [Google Scholar] [CrossRef]
- Hebert, P.D.; Cywinska, A.; Ball, S.L.; DeWaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. B 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Chen, Y.; Gibson, K.; Liu, S.; Yu, Z.; Chen, N. High genetic diversity of the harmful algal bloom species Phaeocystis globosa revealed using the molecular marker COX1. Harmful Algae 2021, 107, 102065. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes de novo assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, 29–37. [Google Scholar] [CrossRef]
- Laslett, D.; Canback, B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L. Holt KE.Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef]
- McGinnis, S.; Madden, T.L. BLAST: At the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004, 32, 20–25. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Julie, D.T.; Desmond, G.H.; Toby, J.G. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Lam-Tung, N.; Heiko, A.S.; Arndt von, H.; Bui, Q.M. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Alexei, J.D.; Marc, A.S.; Dong, X.; Andrew, R. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [PubMed]
- Goel, M.; Sun, H.; Jiao, W.B.; Schneeberger, K. SyRI: Finding genomic rearrangements and local sequence differences from whole-genome assemblies. Genome Biol. 2019, 20, 277. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P. Variant annotation and functional prediction: SnpEff. Methods Mol. Biol. 2022, 2493, 289–314. [Google Scholar] [CrossRef]
- Goel, M.; Schneeberger, K. Plotsr: Visualizing structural similarities and rearrangements between multiple genomes. Bioinformatics 2022, 38, 2922–2928. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Xiong, Y.L.; Shu, X.; Yu, Q.Q.; Lei, X.; Li, D.X.; Yan, L.J.; Bai, S.Q.; Ma, X. Molecular phylogeography and intraspecific divergences in siberian wildrye (Elymus sibiricus L.) wild populations in China, inferred from chloroplast DNA sequence and cpSSR markers. Front. Plant Sci. 2022, 13, 862759. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Chevigny, N.; Schatz-Daas, D.; Lotfi, F.; Gualberto, J.M. DNA repair and the stability of the plant mitochondrial genome. Int. J. Mol. Sci. 2020, 21, 328. [Google Scholar] [CrossRef]
- Chen, W.T.; Li, M.; Hu, S.Y.; Wang, S.H.; Yuan, M.L. Comparative mitogenomic and evolutionary analysis of Lycaenidae (Insecta: Lepidoptera): Potential association with high-altitude adaptation. Front. Genet. 2023, 14, 1137588. [Google Scholar] [CrossRef]
- Dong, S.S.; Zhao, C.X.; Chen, F.; Liu, Y.H.; Zhang, S.Z.; Wu, H.; Zhang, L.S.; Liu, Y. The complete mitochondrial genome of the early flowering plant Nymphaea colorata is highly repetitive with low recombination. BMC Genom. 2018, 19, 614. [Google Scholar] [CrossRef]
- Donnelly, K.; Cottrell, J.; Ennos, R.A.; Vendramin, G.G.; A’Hara, S.; King, S.; Perry, A.; Wachowiak, W.; Cavers, S. Reconstructing the plant mitochondrial genome for marker discovery: A case study using Pinus. Mol. Ecol. Resour. 2017, 17, 943–954. [Google Scholar] [CrossRef]
- Liu, Q.; Yuan, H.Y.; Xu, J.X.; Cui, D.L.; Xiong, G.; Schwarzacher, T.; Heslop-Harrison, J.S. The mitochondrial genome of the diploid oat Avena longiglumis. BMC Plant Biol. 2023, 23, 218. [Google Scholar] [CrossRef]
- Asaf, S.; Khan, A.L.; Khan, A.R.; Waqas, M.; Kang, S.M.; Khan, M.A.; Shahzad, R.; Seo, C.W.; Shin, J.H.; Lee, I.J. Mitochondrial genome analysis of wild rice (Oryza minuta) and its comparison with other related species. PLoS ONE 2016, 11, e0152937. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, G.; Wei, J. Assembly and comparative analysis of the first complete mitochondrial genome of Setaria italica. Planta 2024, 260, 23. [Google Scholar] [CrossRef]
- Park, S.; Grewe, F.; Zhu, A.; Ruhlman, T.A.; Sabir, J.; Mower, J.P.; Jansen, R.K. Dynamic evolution of Geranium mitochondrial genomes through multiple horizontal and intracellular gene transfers. New Phytol. 2015, 208, 570–583. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J. Promiscuous DNA—Chloroplast genes inside plant mitochondria. Nature 1982, 299, 678–679. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.S.; Park, S. Complete plastid and mitochondrial genomes of Aeginetia indica reveal intracellular gene transfer (IGT), horizontal gene transfer (HGT), and cytoplasmic male sterility (CMS). Int. J. Mol. Sci. 2021, 22, 6143. [Google Scholar] [CrossRef]
- Nakazono, M.; Nishiwaki, S.; Tsutsumi, N.; Hirai, A. A chloroplast-derived sequence is utilized as a source of promoter sequences for the gene for subunit 9 of NADH dehydrogenase (nad9) in rice mitochondria. Mol. Genet. Genom. 1996, 252, 371–378. [Google Scholar] [CrossRef]
- Miyata, S.I.; Nakazono, M.; Hirai, A. Transcription of plastid-derived tRNA genes in rice mitochondria. Curr. Genet. 1998, 34, 216–220. [Google Scholar] [CrossRef] [PubMed]
- Hao, W.; Palmer, J.D. Fine-scale mergers of chloroplast and mitochondrial genes create functional, transcompartmentally chimeric mitochondrial genes. Proc. Natl. Acad. Sci. USA 2009, 106, 16728–16733. [Google Scholar] [CrossRef]
- Cerutti, H.; Jagendorf, A. Movement of DNA across the chloroplast envelope: Implications for the transfer of promiscuous DNA. Photosynth. Res. 1995, 46, 329–337. [Google Scholar] [CrossRef]
- Bock, R. Witnessing genome evolution: Experimental reconstruction of endosymbiotic and horizontal gene transfer. Annu. Rev. Genet. 2017, 51, 1–22. [Google Scholar] [CrossRef]
- Tang, P.; Ni, Y.; Li, J.L.; Lu, Q.Q.; Liu, C.; Guo, J.L. The complete mitochondrial genome of Paeonia lactiflora Pall. (Saxifragales: Paeoniaceae): Evidence of gene transfer from chloroplast to mitochondrial genome. Genes 2024, 15, 239. [Google Scholar] [CrossRef]
- Bömmer, D.; Haberhausen, G.; Zetsche, K. A large deletion in the plastid DNA of the holoparasitic flowering plant Cuscuta reflexa concerning two ribosomal proteins (rpl2, rpl23), one transfer RNA (trnI) and an ORF 2280 homologue. Curr. Genet. 1993, 24, 171–176. [Google Scholar] [CrossRef]
- Lin, C.S.; Chen, J.J.; Huang, Y.T.; Chan, M.T.; Daniell, H.; Chang, W.J.; Hsu, C.T.; Liao, D.C.; Wu, F.H.; Lin, S.Y.; et al. The location and translocation of ndh genes of chloroplast origin in the Orchidaceae family. Sci. Rep. 2015, 5, 9040. [Google Scholar] [CrossRef]
- Gao, C.W.; Wu, C.H.; Zhang, Q.; Zhao, X.; Wu, M.X.; Chen, R.R.; Zhao, Y.L.; Li, Z. Characterization of chloroplast genomes from two Salvia medicinal plants and gene transfer among their mitochondrial and chloroplast genomes. Front. Genet. 2020, 11, 574962. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Luo, Y.Y.; Sha, A.J.; Xiao, W.Q.; Xiong, Z.; Chen, X.D.; He, J.; Peng, L.X.; Zou, L. Analysis of synonymous codon usage patterns in mitochondrial genomes of nine Amanita species. Front. Micro 2023, 14, 1134228. [Google Scholar] [CrossRef] [PubMed]
- Mazumder, G.A.; Uddin, A.; Chakraborty, S. Analysis of codon usage bias in mitochondrial CO gene among platyhelminthes. Mol. Biochem. Parasit. 2021, 245, 111410. [Google Scholar] [CrossRef] [PubMed]
- Pepe, D. Codon bias analyses on thyroid carcinoma genes. Minerva Endocrinol. 2020, 45, 295–305. [Google Scholar] [CrossRef]

| Genes | Sample | Haplotype Number | Haplotype Diversity | Nucleotide Diversity | Tajima’s D |
|---|---|---|---|---|---|
| cox1 | 24 | 15 | 0.936 | 0.00305 | −1.24332 |
| atp6 | 24 | 2 | 0.359 | 0.00039 | −0.66674 |
| psbA-trnH | 21 | 5 | 0.424 | 0.01287 | −2.56233 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, J.; Xiong, Y.; Xu, M.; Gou, W.; Yang, T.; Xiong, Y.; Dong, Z.; Pan, L.; Sha, L.; Luo, H.; et al. Organelle Genome Characteristics and Phylogenetic Analysis of a Warm-Season Turfgrass Eremochloa ophiuroides (Poaceae). Biology 2025, 14, 975. https://doi.org/10.3390/biology14080975
Zhao J, Xiong Y, Xu M, Gou W, Yang T, Xiong Y, Dong Z, Pan L, Sha L, Luo H, et al. Organelle Genome Characteristics and Phylogenetic Analysis of a Warm-Season Turfgrass Eremochloa ophiuroides (Poaceae). Biology. 2025; 14(8):975. https://doi.org/10.3390/biology14080975
Chicago/Turabian StyleZhao, Junming, Yanli Xiong, Maotao Xu, Wenlong Gou, Tingyong Yang, Yi Xiong, Zhixiao Dong, Ling Pan, Lina Sha, Hong Luo, and et al. 2025. "Organelle Genome Characteristics and Phylogenetic Analysis of a Warm-Season Turfgrass Eremochloa ophiuroides (Poaceae)" Biology 14, no. 8: 975. https://doi.org/10.3390/biology14080975
APA StyleZhao, J., Xiong, Y., Xu, M., Gou, W., Yang, T., Xiong, Y., Dong, Z., Pan, L., Sha, L., Luo, H., & Ma, X. (2025). Organelle Genome Characteristics and Phylogenetic Analysis of a Warm-Season Turfgrass Eremochloa ophiuroides (Poaceae). Biology, 14(8), 975. https://doi.org/10.3390/biology14080975






