Genome-Wide Identification, Characterization, and Expression Analysis of NRT Gene Family in Suaeda glauca
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Samples and NaCl Concentration Treatments
2.2. Identification of NRT Genes in Suaeda glauca
2.3. Phylogenetic Analysis of NRT Families
2.4. Motif, Conserved Domain, and Gene Structure Analysis of SgNRTs
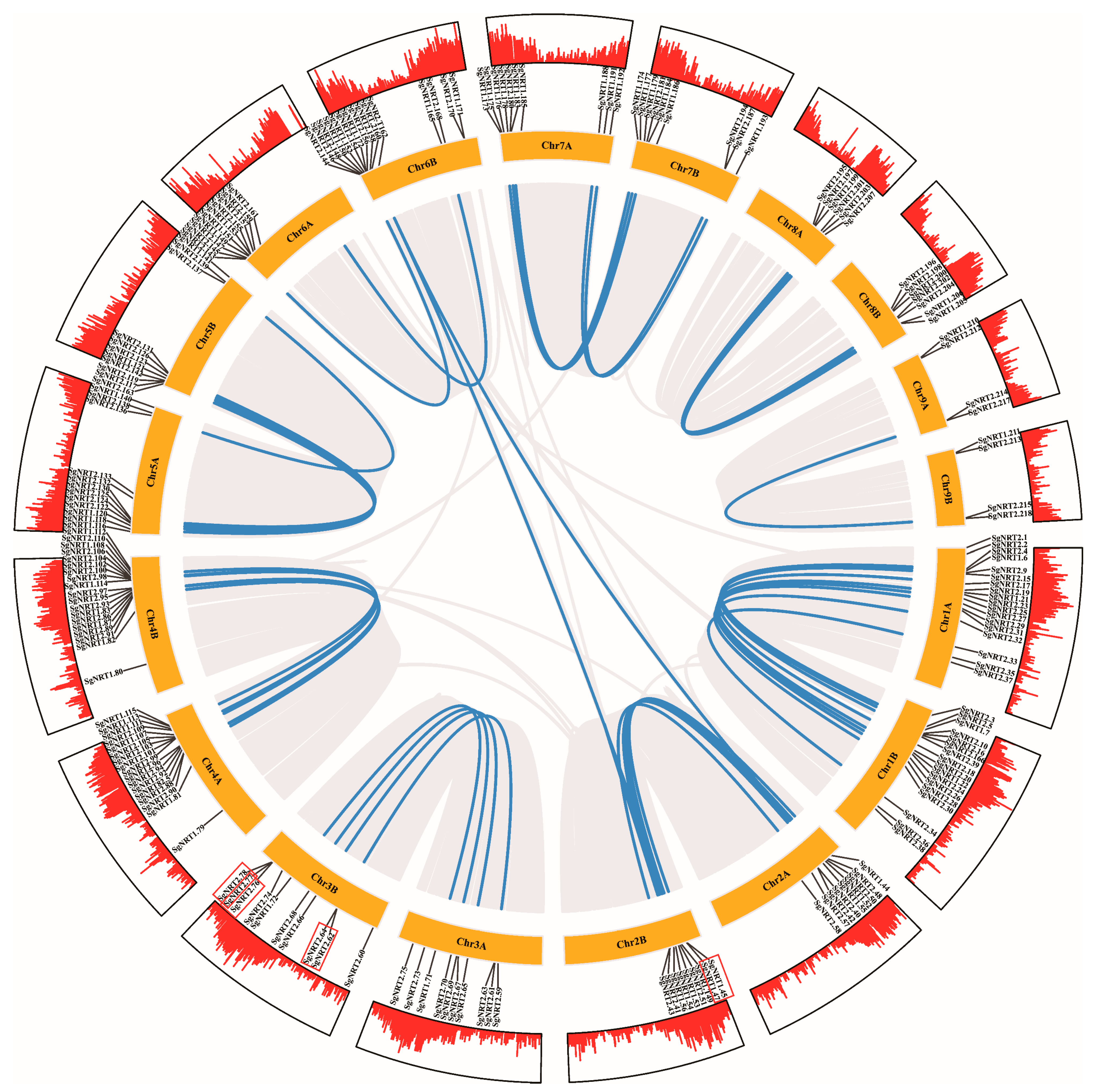
2.5. Chromosomal Localization and Synteny Correlation of NRT Family
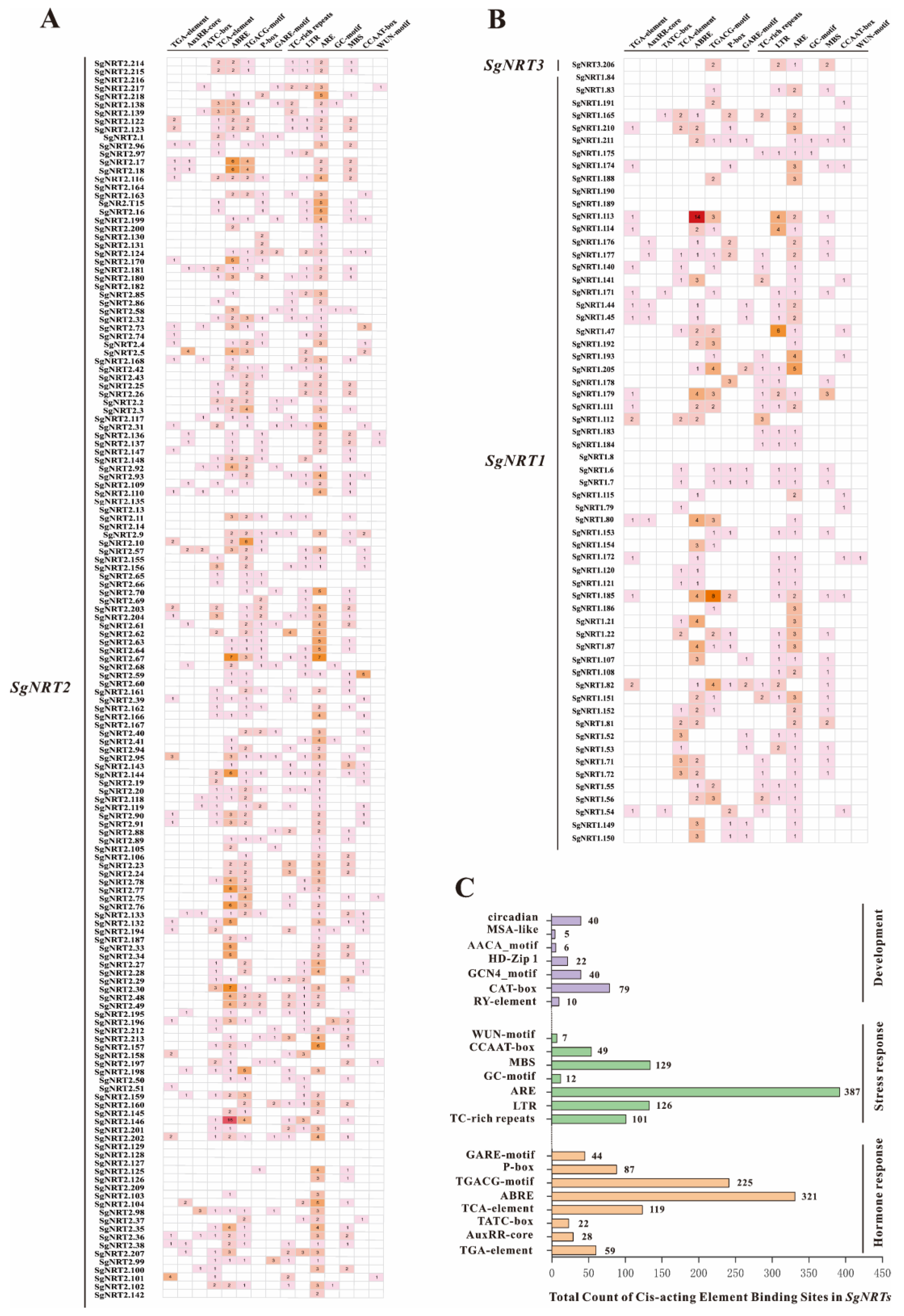
2.6. Cis-Element Analysis of SgNRT Gene Promoters
2.7. Gene Expression Profiling of the NRT Genes
3. Result
3.1. Genome-Wide Identification and Characterization of NRT Members in Suaeda glauca
3.2. Evolutionary Relationships of NRT Members Between S. glauca and Other Species
3.3. Motif, Conserved Domain, and Gene Structure Distribution of SgNRT
3.4. Amplification and Evolutionary Patterns of SgNRT Genes
3.5. Characterization of Cis-Acting in the SgNRT Genes Promoter Region
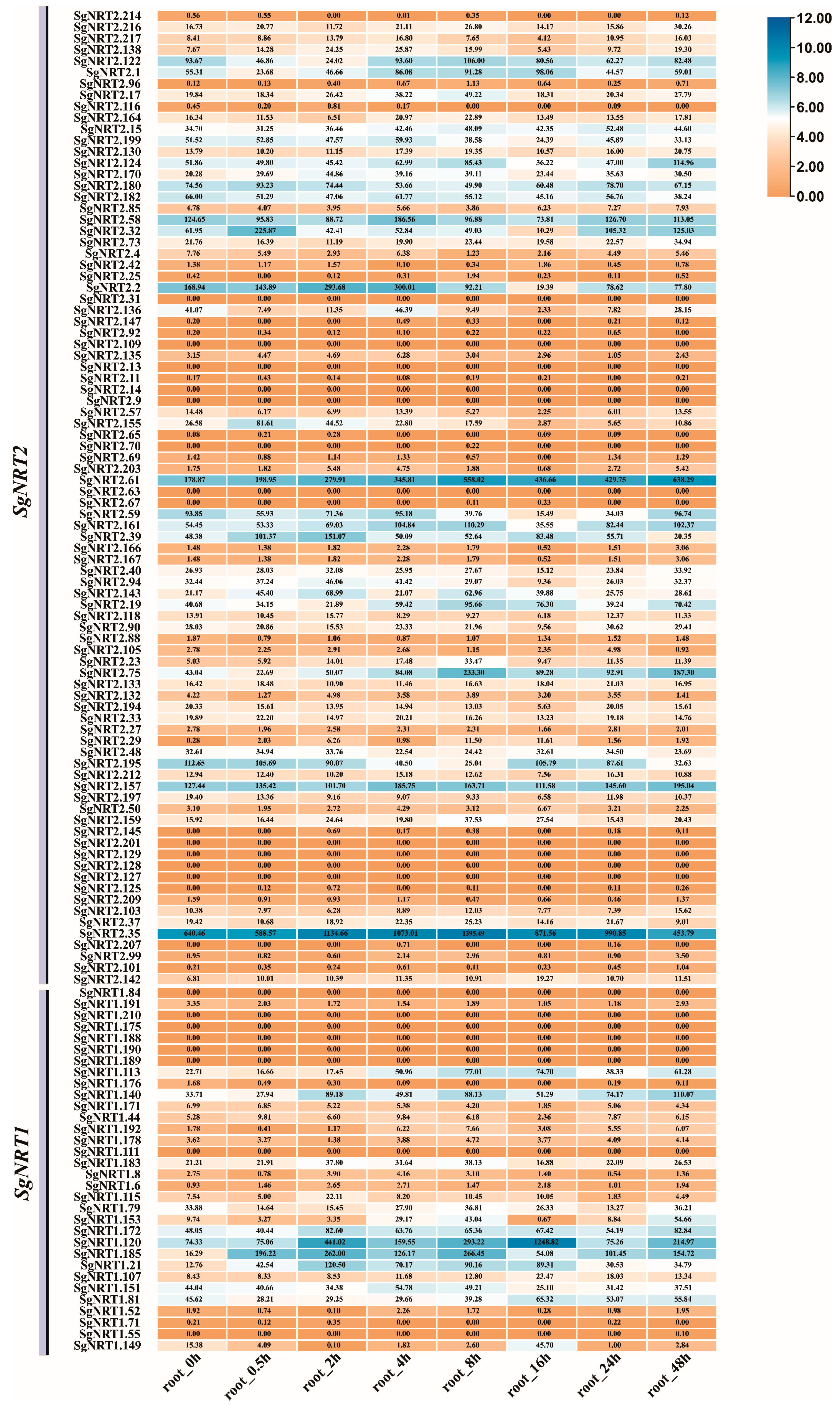
3.6. The Specific Expression of SgNRT Genes in 13 Different Organs or Tissues
3.7. The Dynamic Expression of SgNRT Genes Within 48 h After Salt Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- M’hamdi, M.; Abid, G.; Chikh-Rouhou, H.; Razgallah, N.; Hassen, A. Effect of genotype and growing season on nitrate accumulation and expression patterns of nitrate transporter genes in potato (Solanum tuberosum L.). Arch. Agron. Soil Sci. 2016, 62, 1508–1520. [Google Scholar] [CrossRef]
- Fan, X.; Naz, M.; Fan, X.; Xuan, W.; Miller, A.J.; Xu, G. Plant nitrate transporters: From gene function to application. J. Exp. Bot. 2017, 68, 2463–2475. [Google Scholar] [CrossRef] [PubMed]
- Li, J.F.; Cao, H.R.; Li, S.X.; Dong, X.N.; Zhao, Z.; Jia, Z.T.; Yuan, L.X. Genetic and molecular mechanisms underlying nitrogen use efficiency in maize. J. Genet. Genom. 2025, 52, 276–286. [Google Scholar] [CrossRef] [PubMed]
- Asim, M.; Ullah, Z.; Oluwaseun, A.; Wang, Q.; Liu, H.B. Signalling Overlaps between Nitrate and Auxin in Regulation of The Root System Architecture: Insights from the Arabidopsis thaliana. Int. J. Mol. Sci. 2020, 21, 2880. [Google Scholar] [CrossRef]
- Tsay, Y.F.; Chiu, C.C.; Tsai, C.B.; Ho, C.H.; Hsu, P.K. Nitrate transporters and peptide transporters. FEBS Lett. 2007, 581, 2290–2300. [Google Scholar] [CrossRef]
- Dechorgnat, J.; Nguyen, C.T.; Armengaud, P.; Jossier, M.; Diatloff, E.; Filleur, S.; Daniel-Vedele, F. From the soil to the seeds: The long journey of nitrate in plants. J. Exp. Bot. 2011, 62, 1349–1359. [Google Scholar] [CrossRef]
- Liu, K.H.; Huang, C.Y.; Tsay, Y.F. CHL1 is a dual-affinity nitrate transporter of arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 1999, 11, 865–874. [Google Scholar] [CrossRef][Green Version]
- Ho, C.H.; Lin, S.H.; Hu, H.C.; Tsay, Y.F. CHL1 Functions as a Nitrate Sensor in Plants. Cell 2009, 138, 1184–1194. [Google Scholar] [CrossRef]
- Corratgé-Faillie, C.; Lacombe, B. Substrate (un)specificity of Arabidopsis NRT1/PTR FAMILY (NPF) proteins. J. Exp. Bot. 2017, 68, 3107–3113. [Google Scholar] [CrossRef] [PubMed]
- Krapp, A.; David, L.C.; Chardin, C.; Girin, T.; Marmagne, A.; Leprince, A.S.; Chaillou, S.; Ferrario-Méry, S.; Meyer, C.; Daniel-Vedele, F. Nitrate transport and signalling in Arabidopsis. J. Exp. Bot. 2014, 65, 789–798. [Google Scholar] [CrossRef]
- Okamoto, M.; Kumar, A.; Li, W.B.; Wang, Y.; Siddiqi, M.Y.; Crawford, N.M.; Glass, A.D.M. High-affinity nitrate transport in roots of Arabidopsis depends on expression of the NAR2-like gene AtNRT3.1. Plant Physiol. 2006, 140, 1036–1046. [Google Scholar] [CrossRef]
- Bai, H.; Euring, D.; Volmer, K.; Janz, D.; Polle, A. The Nitrate Transporter (NRT) Gene Family in Poplar. PLoS ONE 2013, 8, e72126. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.A.; Vega, A.; Bouguyon, E.; Krouk, G.; Gojon, A.; Coruzzi, G.; Gutierrez, R.A. Nitrate Transport, Sensing, and Responses in Plants. Mol. Plant 2016, 9, 837–856. [Google Scholar] [CrossRef]
- Kong, W.W.; Tian, L.B.; Li, R.C.; Xiao, L.N.; Cui, F.; Liu, Y.Y.; Li, G.W.; Wan, S.B. Characterization of NRT1 Family and Their Response to Salt Stress in Peanut. J. Peanut Sci. 2019, 48, 1–7. [Google Scholar] [CrossRef]
- Wang, Y.B.; Zhang, Z.X.; Zhang, C.M.; Liu, W.; Zhang, Y.W.; Zhang, L.F.; Xu, R. Identification of Soybean Salt Stress Responsive NRT Genes and Cloning and Expression Analysis of GmNRT1.5A. Soybean Sci. 2023, 42, 674–682. [Google Scholar]
- Liu, G.D.; Rui, L.; Yang, Y.Y.; Liu, R.X.; Li, H.L.; Ye, F.; You, C.X.; Zhang, S. Identification and Functional Characterization of MdNRT1.1 in Nitrogen Utilization and Abiotic Stress Tolerance in Malus domestica. Int. J. Mol. Sci. 2023, 24, 9291. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, J.; Deng, C.; Liu, Z.; Yin, K.X.; Zhang, Y.; Zhao, Z.Y.; Zhao, R.; Zhao, N.; Zhou, X.Y.; et al. Effect of NaCl on ammonium and nitrate uptake and transport in salt-tolerant and salt-sensitive poplars. Tree Physiol. 2024, 44, tpae020. [Google Scholar] [CrossRef]
- Cheng, Y.; Yang, P.; Zhao, L.H.; Priyadarshani, S.; Zhou, Q.; Li, Z.Y.; Li, W.M.; Xiong, J.J.; Lin, Z.B.; Li, L.; et al. Studies on genome size estimation, chromosome number, gametophyte development and plant morphology of salttolerant halophyte Suaeda salsa. BMC Plant Biol. 2019, 19, 473. [Google Scholar] [CrossRef]
- Flowers, T.J.; Colmer, T.D. Salinity tolerance in halophytes. New Phytol. 2008, 179, 945–963. [Google Scholar] [CrossRef]
- Guo, J.; Li, Y.; Han, G.; Song, J.; Wang, B. NaCl markedly improved the reproductive capacity of the euhalophyte Suaeda salsa. Funct. Plant Biol. 2018, 45, 350–361. [Google Scholar] [CrossRef]
- Yu, W.C.; Wu, W.W.; Zhang, N.; Wang, L.P.; Wang, Y.H.; Wang, B.; Lan, Q.K.; Wang, Y. Research Advances on Molecular Mechanism of Salt Tolerance in Suaeda. Biology 2022, 11, 1273. [Google Scholar] [CrossRef]
- Salazar, O.R.; Chen, K.; Melino, V.J.; Reddy, M.P.; Hribová, E.; Cízková, J.; Beránková, D.; Vega, J.P.A.; Leal, L.M.C.; Aranda, M.; et al. SOS1 tonoplast neo-localization and the RGG protein SALTY are important in the extreme salinity tolerance of Salicornia bigelovii. Nat. Commun. 2024, 15, 4279. [Google Scholar] [CrossRef]
- Cheng, Y.; Sun, J.; Jiang, M.W.; Luo, Z.G.; Wang, Y.; Liu, Y.H.; Li, W.M.; Hu, B.; Dong, C.X.; Ye, K.Z.; et al. Chromosome-scale genome sequence of Suaeda glauca sheds light on salt stress tolerance in halophytes. Hortic. Res. 2023, 10, uhad161. [Google Scholar] [CrossRef]
- Qian, Y.; Rao, L. Effects of saline-alkali stress on the growth and chlorophyll fluorescence characteristics of Lycium barbarum seedlings. J. For. Environ. 2022, 42, 271–278. [Google Scholar]
- Zhang, K.Y.; Chang, L.; Li, G.H.; Li, Y.F. Advances and future research in ecological stoichiometry under saline-alkali stress. Environ. Sci. Pollut. Res. 2023, 30, 5475–5486. [Google Scholar] [CrossRef]
- Xiong, Y.; Wang, S.S.; Wu, X.Y.; Cui, C.J.; Zhu, J.B. Overexpression of SsNRT1.1D gene from Suaeda salsa improves salt tolerance in transgenic tomato plants. Vitr. Cell. Dev. Biol.-Plant 2024, 60, 168–175. [Google Scholar] [CrossRef]
- Muhammad, M.T.; Hui, W.; Bilal, A.; Yu, L.; Sheng, F.; Ke, L.; Chao, L.; Kamran, S.; Shaohuan, L.; Dong, Z. Identiffcation and characterization of NRT gene family reveals their critical response to nitrate regulation during adventitious root formation and development in apple rootstock. Sci. Hortic. 2021, 275, 109642. [Google Scholar]
- Choi, M.G.; Kim, E.J.; Song, J.Y.; Choi, S.B.; Cho, S.W.; Park, C.S.; Kang, C.S.; Park, Y.I. Peptide transporter2 (PTR2) enhances water uptake during early seed germination in Arabidopsis thaliana. Plant Mol. Biol. 2020, 102, 615–624. [Google Scholar] [CrossRef]
- Feng, H.; Fan, X.; Fan, X.; Liu, X.; Miller, A.J.; Xu, G. Multiple roles of nitrate transport accessory protein NAR2 in plants. Plant Signal. Behav. 2011, 6, 1286–1289. [Google Scholar] [CrossRef]
- Chen, C.J.; Wu, Y.; Li, J.W.; Wang, X.; Zeng, Z.H.; Xu, J.; Liu, Y.L.; Feng, J.T.; Chen, H.; He, Y.H.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Cai, X.F.; Xu, C.X.; Wang, Q.H. Identification and characterization of the NPF, NRT2 and NRT3 in spinach. Plant Physiol. Biochem. 2021, 158, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Deng, D.; Yan, N. Structural basis and transport mechanism of the major facility superfamily (MFS) transporter. Chin. Sci. Bull. 2015, 60, 720–728. [Google Scholar] [CrossRef]
- Zhao, Z.; Li, M.; Xu, W.; Liu, J.-H.; Li, C. Genome-Wide Identification of NRT Gene Family and Expression Analysis of Nitrate Transporters in Response to Salt Stress in Poncirus trifoliata. Genes 2022, 13, 1115. [Google Scholar] [CrossRef]
- Feng, J.; Zhu, C.; Cao, J.; Liu, C.; Zhang, J.; Cao, F.; Zhou, X. Genome-wide identification and expression analysis of the NRT genes in Ginkgo biloba under nitrate treatment reveal the potential roles during calluses browning. BMC Genom. 2023, 24, 633. [Google Scholar] [CrossRef]
- Plett, D.; Toubia, J.; Garnett, T.; Tester, M.; Kaiser, B.N.; Baumann, U. Dichotomy in the NRT Gene Families of Dicots and Grass Species. PLoS ONE 2010, 5, e15289. [Google Scholar] [CrossRef]
- Chen, M.; Zhu, K.; Xie, J.; Liu, J.; Tan, P.; Peng, F. Genome-Wide Identification and Expression Analysis of AMT and NRT Gene Family in Pecan (Carya illinoinensis) Seedlings Revealed a Preference for NH4+-N. Int. J. Mol. Sci. 2022, 23, 13314. [Google Scholar] [CrossRef]
- Santos, T.B.d.; Lima, J.E.; Felicio, M.S.; Soares, J.D.M.; Domingues, D.S. Genome-wide identification, classification and transcriptional analysis of nitrate and ammonium transporters in Coffea. Genet. Mol. Biol. 2017, 40, 346–359. [Google Scholar] [CrossRef]
- You, H.; Liu, Y.; Minh, T.N.; Lu, H.; Zhang, P.; Li, W.; Xiao, J.; Ding, X.; Li, Q. Genome-wide identification and expression analyses of nitrate transporter family genes in wild soybean (Glycine soja). J. Appl. Genet. 2020, 61, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Guo, B.; Li, Y.; Wang, S.; Li, D.; Lv, C.; Xu, R. Characterization of the Nitrate Transporter gene family and functional identification of HvNRT2.1 in barley (Hordeum vulgare L.). PLoS ONE 2020, 15, e0232056. [Google Scholar] [CrossRef]
- Zhao, X.Y.; Wu, H.B.; Li, B.Y.; Wang, P.Y.; Zhang, P.; Shen, H.L.; Yang, J.F. Genome-Wide Identification and Expression Analysis of PkNRT Gene Family in Korean Pine (Pinus koraiensis). Plants 2025, 14, 238. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.Y.; Mei, J.J.; Zheng, W.; Khan, F.S.; Bhuiyan, M.N.; Wang, K.J.; Rhaman, M.S.; Abe-Kanoh, N.; Ji, W. Identification of Grape NRT Gene Family and Analysis of Its Expression in Leaves Under Nitrogen-Deficiency Stress. Horticulturae 2025, 11, 252. [Google Scholar] [CrossRef]
- Wang, W.; Hu, B.; Li, A.F.; Chu, C.C. NRT1.1s in plants: Functions beyond nitrate transport. J. Exp. Bot. 2020, 71, 4373–4379. [Google Scholar] [CrossRef]
- Qian, Y.; Cha, Q.; Kong, M.; Liu, Z.; Li, Y.; Hou, X.; Liu, T. Molecular evolution of NRT2 gene family in plant. Jiangsu J. Agric. Sci. 2015, 31, 45–54. [Google Scholar]
- Hu, B.; Wang, W.; Ou, S.J.; Tang, J.Y.; Li, H.; Che, R.H.; Zhang, Z.H.; Chai, X.Y.; Wang, H.R.; Wang, Y.Q.; et al. Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 2015, 47, 834. [Google Scholar] [CrossRef]
- Saier, M.H. Genome archeology leading to the characterization and classification of transport proteins. Curr. Opin. Microbiol. 1999, 2, 555–561. [Google Scholar] [CrossRef]
- Kanstrup, C.; Nour-Eldin, H.H. The emerging role of the nitrate and peptide transporter family: NPF in plant specialized metabolism. Curr. Opin. Plant Biol. 2022, 68, 102243. [Google Scholar] [CrossRef]
- Haj-Amor, Z.; Araya, T.; Kim, D.G.; Bouri, S.; Lee, J.; Ghilou, W.; Yang, Y.; Kang, H.; Jhariya, M.K.; Banerjee, A.; et al. Soil salinity and its associated effects on soil microorganisms, greenhouse gas emissions, crop yield, biodiversity and desertification: A review. Sci. Total Environ. 2022, 843, 156946. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Zheng, Y.B. Suaeda glauca Can Be Produced Hydroponically at Moderate NaCl Salinity. Hortscience 2015, 50, 847–850. [Google Scholar] [CrossRef]
- Song, X.Q.; Yang, N.; Su, Y.H.; Lu, X.Y.; Liu, J.; Liu, Y.; Zhang, Z.H.; Tang, Z.H. Suaeda glauca and Suaeda salsa Employ Different Adaptive Strategies to Cope with Saline-Alkali Environments. Agronomy 2022, 12, 2496. [Google Scholar] [CrossRef]
- Tegeder, M.; Masclaux-Daubresse, C. Source and sink mechanisms of nitrogen transport and use. New Phytol. 2018, 217, 35–53. [Google Scholar] [CrossRef]
- Aslam, M.T.; Khan, I.; Chattha, M.U.; Maqbool, R.; Ziaulhaq, M.; Lihong, W.; Usman, S.; Rasheed, A.; Hassan, M.U.; Hashem, M.; et al. The critical role of nitrogen in plants facing the salinity stress: Review and future prospective. Not. Bot. Horti Agrobot. Cluj-Napoca 2023, 51, 13347. [Google Scholar] [CrossRef]
- Yang, Z.; Tan, S.; Yang, Q.L.; Chen, S.M.; Qi, C.M.; Liu, X.G.; Liang, J.P.; Wang, H.D. Nitrogen Application Alleviates Impairments for Jatropha curcas L. Seedling Growth under Salinity Stress by Regulating Photosynthesis and Antioxidant Enzyme Activity. Agronomy 2023, 13, 1749. [Google Scholar] [CrossRef]
- Singh, M.; Singh, V.P.; Prasad, S.M. Responses of photosynthesis, nitrogen and proline metabolism to salinity stress in Solanum lycopersicum under different levels of nitrogen supplementation. Plant Physiol. Biochem. 2016, 109, 72–83. [Google Scholar] [CrossRef]
- Kotur, Z.; Glass, A.D.M. A 150kDa plasma membrane complex of AtNRT2.5 and AtNAR2.1 is the major contributor to constitutive high-affinity nitrate influx in Arabidopsis thaliana. Plant Cell Environ. 2015, 38, 1490–1502. [Google Scholar] [CrossRef] [PubMed]
- Drechsler, N.; Courty, P.E.; Brulé, D.; Kunze, R. Identification of arbuscular mycorrhiza-inducible Nitrate Transporter 1/Peptide Transporter Family (NPF) genes in rice. Mycorrhiza 2018, 28, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, C.; Tian, C.; Kong, C.; Xu, C.; Liu, X. Identification of nitrate transporter protein 1/peptide transporter protein family 6.4 gene (ScNPF6.4) and functional analysis of its regulation of tillering in sugarcane. Acta Agron. Sin. 2024, 50, 2131–2142. [Google Scholar]

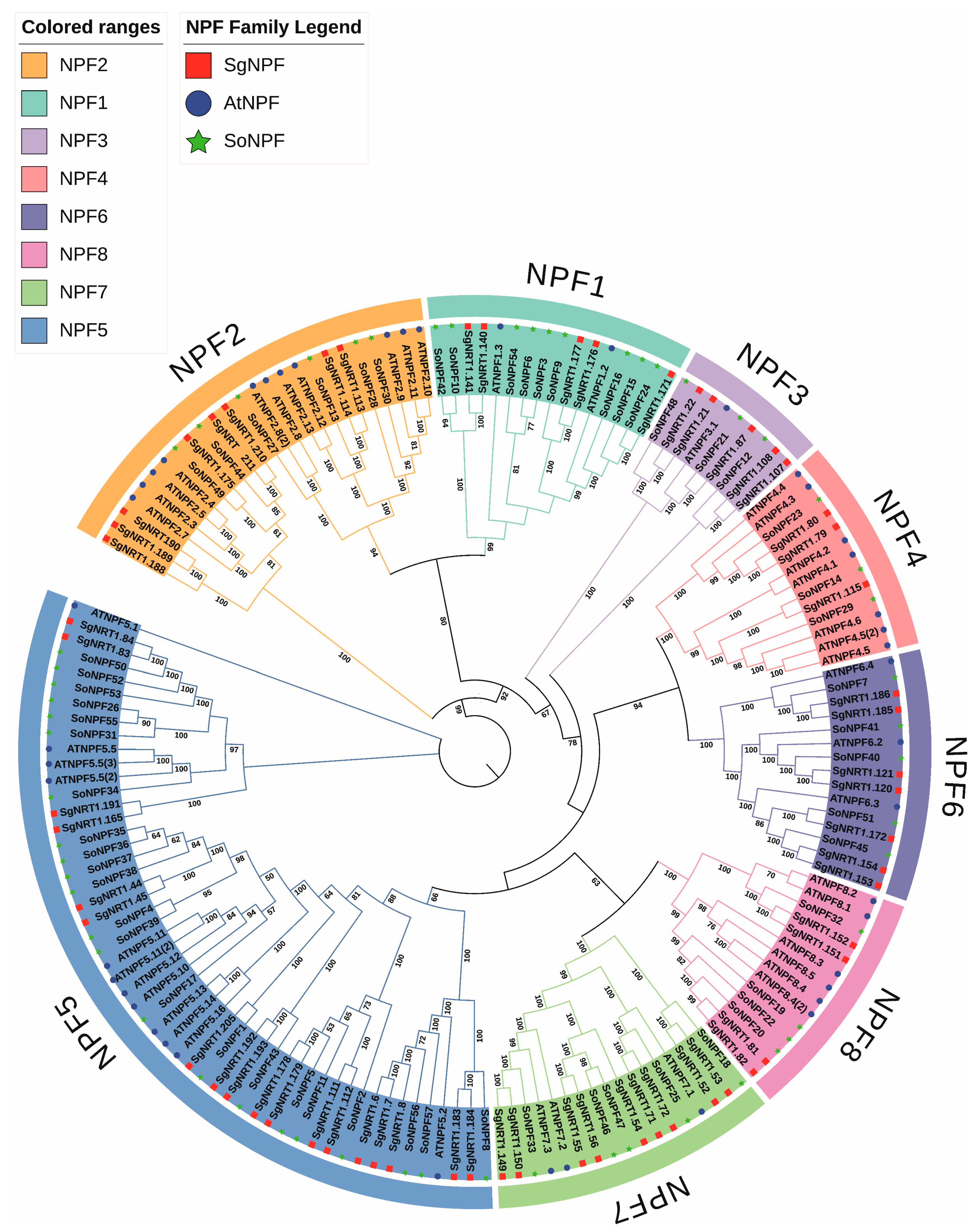




| SgNPF Family | ID | SgNPF Family | ID |
|---|---|---|---|
| SgNPF1 | SgNRT1.140/SgNPF1.1 | SgNPF5 | SgNRT1.165/SgNPF5.10 |
| SgNPF1 | SgNRT1.141/SgNPF1.2 | SgNPF5 | SgNRT1.178/SgNPF5.11 |
| SgNPF1 | SgNRT1.171/SgNPF1.3 | SgNPF5 | SgNRT1.179/SgNPF5.12 |
| SgNPF1 | SgNRT1.176/SgNPF1.4 | SgNPF5 | SgNRT1.183/SgNPF5.13 |
| SgNPF1 | SgNRT1.177/SgNPF1.5 | SgNPF5 | SgNRT1.184/SgNPF5.14 |
| SgNPF2 | SgNRT1.113/SgNPF2.1 | SgNPF5 | SgNRT1.191/SgNPF5.15 |
| SgNPF2 | SgNRT1.114/SgNPF2.2 | SgNPF5 | SgNRT1.192/SgNPF5.16 |
| SgNPF2 | SgNRT1.175/SgNPF2.3 | SgNPF5 | SgNRT1.193/SgNPF5.17 |
| SgNPF2 | SgNRT1.188/SgNPF2.4 | SgNPF5 | SgNRT1.205/SgNPF5.18 |
| SgNPF2 | SgNRT1.189/SgNPF2.5 | SgNPF6 | SgNRT1.120/SgNPF6.1 |
| SgNPF2 | SgNRT1.190/SgNPF2.6 | SgNPF6 | SgNRT1.121/SgNPF6.2 |
| SgNPF2 | SgNRT1.210/SgNPF2.7 | SgNPF6 | SgNRT1.153/SgNPF6.3 |
| SgNPF2 | SgNRT1.211/SgNPF2.8 | SgNPF6 | SgNRT1.154/SgNPF6.4 |
| SgNPF3 | SgNRT1.21/SgNPF3.1 | SgNPF6 | SgNRT1.172/SgNPF6.5 |
| SgNPF3 | SgNRT1.22/SgNPF3.2 | SgNPF6 | SgNRT1.185/SgNPF6.6 |
| SgNPF3 | SgNRT1.87/SgNPF3.3 | SgNPF6 | SgNRT1.186/SgNPF6.7 |
| SgNPF3 | SgNRT1.107/SgNPF3.4 | SgNPF7 | SgNRT1.52/SgNPF7.1 |
| SgNPF3 | SgNRT1.108/SgNPF3.5 | SgNPF7 | SgNRT1.53/SgNPF7.2 |
| SgNPF4 | SgNRT1.79/SgNPF4.1 | SgNPF7 | SgNRT1.54/SgNPF7.3 |
| SgNPF4 | SgNRT1.80/SgNPF4.2 | SgNPF7 | SgNRT1.55/SgNPF7.4 |
| SgNPF4 | SgNRT1.115/SgNPF4.3 | SgNPF7 | SgNRT1.56/SgNPF7.5 |
| SgNPF5 | SgNRT1.6/SgNPF5.1 | SgNPF7 | SgNRT1.71/SgNPF7.6 |
| SgNPF5 | SgNRT1.7/SgNPF5.2 | SgNPF7 | SgNRT1.72/SgNPF7.7 |
| SgNPF5 | SgNRT1.8/SgNPF5.3 | SgNPF7 | SgNRT1.149/SgNPF7.8 |
| SgNPF5 | SgNRT1.44/SgNPF5.4 | SgNPF7 | SgNRT1.150/SgNPF7.9 |
| SgNPF5 | SgNRT1.45/SgNPF5.5 | SgNPF8 | SgNRT1.81/SgNPF8.1 |
| SgNPF5 | SgNRT1.83/SgNPF5.6 | SgNPF8 | SgNRT1.82/SgNPF8.2 |
| SgNPF5 | SgNRT1.84/SgNPF5.7 | SgNPF8 | SgNRT1.141/SgNPF8.3 |
| SgNPF5 | SgNRT1.111/SgNPF5.8 | SgNPF8 | SgNRT1.152/SgNPF8.4 |
| SgNPF5 | SgNRT1.112/SgNPF5.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ou, Z.; Sun, J.; Li, X.; Feng, H.; Chen, X.; Liang, S.; Guo, Z.; Wang, L.; Niu, X.; Ma, J.; et al. Genome-Wide Identification, Characterization, and Expression Analysis of NRT Gene Family in Suaeda glauca. Biology 2025, 14, 1097. https://doi.org/10.3390/biology14081097
Ou Z, Sun J, Li X, Feng H, Chen X, Liang S, Guo Z, Wang L, Niu X, Ma J, et al. Genome-Wide Identification, Characterization, and Expression Analysis of NRT Gene Family in Suaeda glauca. Biology. 2025; 14(8):1097. https://doi.org/10.3390/biology14081097
Chicago/Turabian StyleOu, Zitong, Jin Sun, Xueli Li, Haoran Feng, Xingguang Chen, Sisi Liang, Zhonghua Guo, Lulu Wang, Xiaoping Niu, Jinbiao Ma, and et al. 2025. "Genome-Wide Identification, Characterization, and Expression Analysis of NRT Gene Family in Suaeda glauca" Biology 14, no. 8: 1097. https://doi.org/10.3390/biology14081097
APA StyleOu, Z., Sun, J., Li, X., Feng, H., Chen, X., Liang, S., Guo, Z., Wang, L., Niu, X., Ma, J., Wang, S., Qin, Y., & Cheng, Y. (2025). Genome-Wide Identification, Characterization, and Expression Analysis of NRT Gene Family in Suaeda glauca. Biology, 14(8), 1097. https://doi.org/10.3390/biology14081097







