Genetic Signatures of Competitive Performance in Burmese Gamecocks: A Transcriptomic Analysis
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data and Sample Collection
2.2. RNA Extraction and Quality Control
2.3. RNA Sequencing
2.4. Bioinformatic Analyses
2.5. Differentially Expressed Gene Analysis
3. Results
3.1. RNA Sequencing and Data Preprocessing
3.2. Bioinformatic Analysis: Read Alignment to Reference Genome
3.3. Differential Gene Expression Analysis Between High- and Low-Performing Cocks
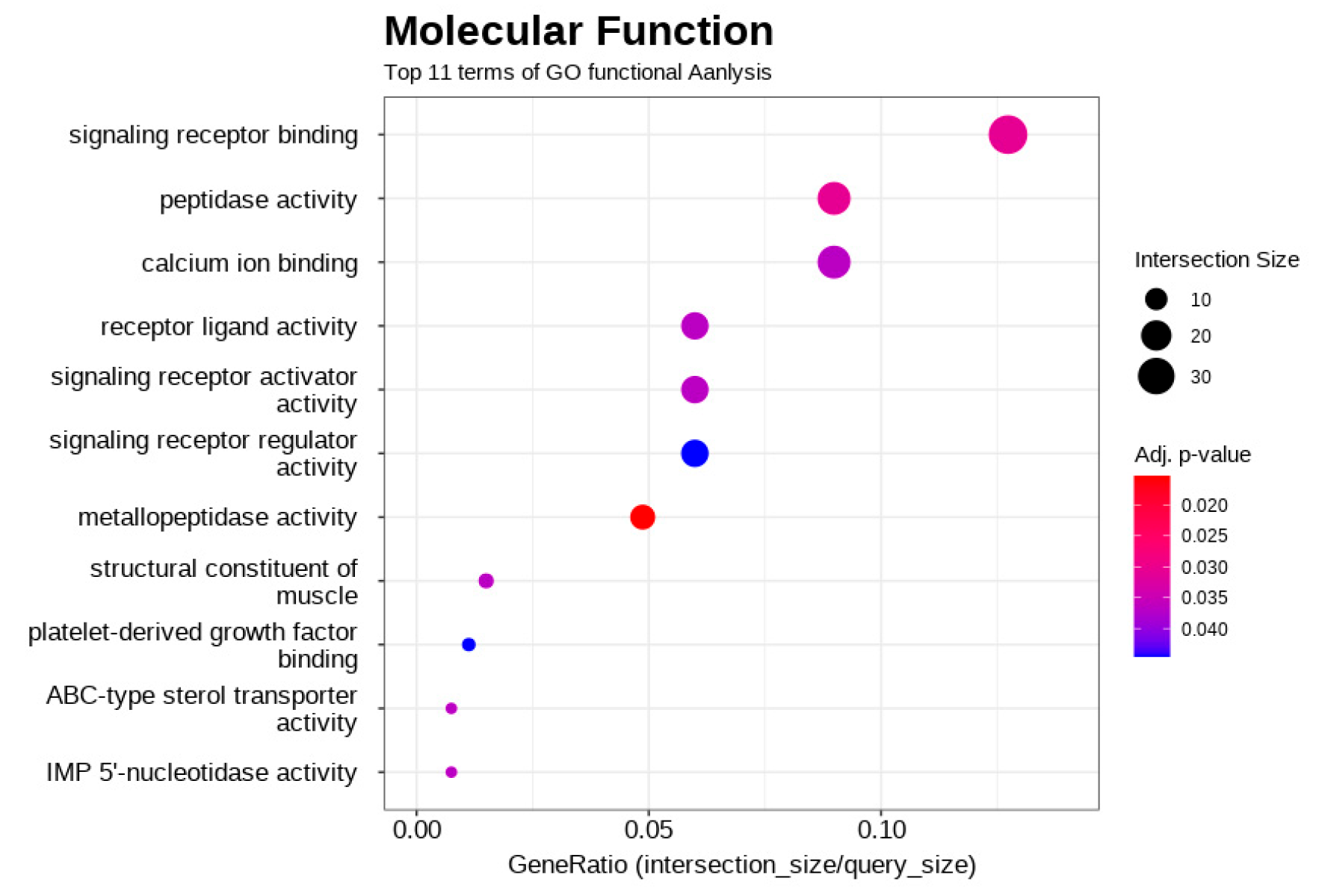
3.4. Functional Enrichment Analysis of DEGs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ibiapina Neto, V.; Santos, N.P.D.S.; Campelo, J.E.G.; Barbosa, F.J.V.; Sarmento, J.L.R.; Carvalho, M.D.F.D. Phenotypic Diversity between Brazilian Fighting Cocks and Naturalized Roosters. Rev. Bras. De Zootec. 2019, 48, e20180271. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, X.; Xu, H.; Nie, Q.; Zhang, Z.; Xu, Z.; Ye, Q.; Zheng, M.; Abdalla, B.A.; Luo, W. Genome-Wide Association Study of Aggressive Behaviour in Chicken. Sci. Rep. 2016, 6, 30981. [Google Scholar] [CrossRef] [PubMed]
- Crawford, R.D.; Smyth, J.R. Social and Sexual Behavior as Related to Plumage Pattern in the Fayoumi Fowl. Poult. Sci. 1964, 43, 1193–1199. [Google Scholar] [CrossRef]
- Komiyama, T.; Kobayashi, H.; Yokoyama, K.; Yoshikawa, M. Analysis of the Source of Aggressiveness in Gamecocks. Sci. Rep. 2020, 10, 7005. [Google Scholar] [CrossRef]
- Favati, A.; Løvlie, H.; Leimar, O. Effects of Social Experience, Aggressiveness and Comb Size on Contest Success in Male Domestic Fowl. R. Soc. Open Sci. 2021, 8, 201213. [Google Scholar] [CrossRef]
- Smagin, D.A.; Galyamina, A.G.; Kovalenko, I.L.; Kudryavtseva, N.N. Altered Expression of Genes Associated with Major Neurotransmitter Systems in the Reward-Related Brain Regions of Mice with Positive Fighting Experience. Int. J. Mol. Sci. 2022, 23, 13644. [Google Scholar] [CrossRef]
- Kim, G.; Shin, S.P.; Song, S.; Park, T.S.; Kim, S.W.; Park, B.; Han, J.S.; Lee, J.H. Generation of Myostatin-Knockout Chickens Mediated by D10A-Cas9 Nickase. FASEB J. 2020, 34, 5688–5696. [Google Scholar] [CrossRef]
- Ren, X.; Zhao, X.; Ning, Z.; Liu, Y.; Zhang, X.; Guan, Z.; Cheng, H.; Zhang, Y.; Cheng, X.; Wen, J.; et al. Systematic Selection Signature Analysis of Chinese Gamecocks Based on Genomic and Transcriptomic Data. Int. J. Mol. Sci. 2023, 24, 5868. [Google Scholar] [CrossRef] [PubMed]
- Sahraeian, S.; Mohiyuddin, M.; Sebra, R.; Tilgner, H.; Afshar, P.; Au, K.; Asadi, N.B.; Gerstein, M.; Wong, W.; Snyder, M.; et al. Gaining Comprehensive Biological Insight into the Transcriptome by Performing a Broad-Spectrum RNA-Seq Analysis. Nat. Commun. 2017, 8, 59. [Google Scholar] [CrossRef]
- Schurch, N.J.; Schofield, P.; Gierliński, M.; Cole, C.; Sherstnev, A.; Singh, V.; Wrobel, N.; Gharbi, K.; Simpson, G.G.; Owen-Hughes, T. How Many Biological Replicates Are Needed in an RNA-Seq Experiment and Which Differential Expression Tool Should You Use? RNA 2016, 22, 839–851. [Google Scholar] [CrossRef]
- Ng, C.S.; Wu, P.; Fan, W.-L.; Yan, J.; Chen, C.-K.; Lai, Y.-T.; Wu, S.-M.; Mao, C.-T.; Chen, J.-J.; Lu, M.-Y.J. Genomic Organization, Transcriptomic Analysis, and Functional Characterization of Avian α-and β-Keratins in Diverse Feather Forms. Genome Biol. Evol. 2014, 6, 2258–2273. [Google Scholar] [CrossRef]
- Dyomin, A.G.; Koshel, E.I.; Kiselev, A.M.; Saifitdinova, A.F.; Galkina, S.A.; Fukagawa, T.; Kostareva, A.A.; Gaginskaya, E.R. Chicken rRNA Gene Cluster Structure. PLoS ONE 2016, 11, e0157464. [Google Scholar] [CrossRef]
- Schroeder, A.; Mueller, O.; Stocker, S.; Salowsky, R.; Leiber, M.; Gassmann, M.; Lightfoot, S.; Menzel, W.; Granzow, M.; Ragg, T. The RIN: An RNA Integrity Number for Assigning Integrity Values to RNA Measurements. BMC Mol. Biol. 2006, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Liu, J.; Zhao, M.; Su, Y.; Cong, B.; Wang, Z. RNA Quality Score Evaluation: A Preliminary Study of RNA Integrity Number (RIN) and RNA Integrity and Quality Number (RNA IQ). Forensic Sci. Int. 2024, 357, 111976. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of mRNA Abundance Using RNA-Seq Data: RPKM Measure Is Inconsistent among Samples. Theory Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Adler, P.; Vilo, J.; Peterson, H. G: Profiler—Interoperable Web Service for Functional Enrichment Analysis and Gene Identifier Mapping (2023 Update). Nucleic Acids Res. 2023, 51, W207–W212. [Google Scholar] [CrossRef]
- Breher, S.S.; Mavridou, E.; Brenneis, C.; Froese, A.; Arnold, H.; Brand, T. Popeye Domain Containing Gene 2 (Popdc2) Is a Myocyte-specific Differentiation Marker during Chick Heart Development. Dev. Dyn. An. Off. Publ. Am. Assoc. Anat. 2004, 229, 695–702. [Google Scholar] [CrossRef]
- Andrée, B.; Hillemann, T.; Kessler-Icekson, G.; Schmitt-John, T.; Jockusch, H.; Arnold, H.-H.; Brand, T. Isolation and Characterization of the Novel Popeye Gene Family Expressed in Skeletal Muscle and Heart. Dev. Biol. 2000, 223, 371–382. [Google Scholar] [CrossRef]
- Jin, J.; Du, M.; Wang, J.; Guo, Y.; Zhang, J.; Zuo, H.; Hou, Y.; Wang, S.; Lv, W.; Bai, W. Conservative Analysis of Synaptopodin-2 Intron Sense-overlapping lncRNA Reveals Its Novel Function in Promoting Muscle Atrophy. J. Cachexia Sarcopenia Muscle 2022, 13, 2017–2030. [Google Scholar] [CrossRef]
- Lohanadan, K.; Assent, M.; Linnemann, A.; Schuld, J.; Heukamp, L.C.; Krause, K.; Vorgerd, M.; Reimann, J.; Schänzer, A.; Kirfel, G. Synaptopodin-2 Isoforms Have Specific Binding Partners and Display Distinct, Muscle Cell Type-Specific Expression Patterns. Cells 2023, 13, 85. [Google Scholar] [CrossRef]
- Zheng, Z.; Song, Y. Synaptopodin-2: A Potential Tumor Suppressor. Cancer Cell Int. 2023, 23, 158. [Google Scholar] [CrossRef]
- Jahejo, A.R.; Zhang, D.; Niu, S.; Mangi, R.A.; Khan, A.; Qadir, M.F.; Khan, A.; Chen, H.; Tian, W. Transcriptome-Based Screening of Intracellular Pathways and Angiogenesis Related Genes at Different Stages of Thiram Induced Tibial Lesions in Broiler Chickens. BMC Genom. 2020, 21, 50. [Google Scholar] [CrossRef]
- Lu, J.; Li, Z.; Zhao, Q.; Liu, D.; Mei, Y. Neuritin Improves the Neurological Functional Recovery after Experimental Intracerebral Hemorrhage in Mice. Neurobiol. Dis. 2021, 156, 105407. [Google Scholar] [CrossRef]
- Yao, J.; Zhao, Q.-R.; Liu, D.-D.; Chow, C.-W.; Mei, Y.-A. Neuritin Up-Regulates Kv4. 2 α-Subunit of Potassium Channel Expression and Affects Neuronal Excitability by Regulating the Calcium-Calcineurin-NFATc4 Signaling Pathway. J. Biol. Chem. 2016, 291, 17369–17381. [Google Scholar] [CrossRef]
- Sharma, T.; Liu, Y.; Wordinger, R.; Pang, I.; Clark, A. Neuritin 1 Promotes Retinal Ganglion Cell Survival and Axonal Regeneration Following Optic Nerve Crush. Cell Death Dis. 2015, 6, e1661. [Google Scholar] [CrossRef]
- Christians, J.K.; King, A.Y.; Rogowska, M.D.; Hessels, S.M. Pappa2 Deletion in Mice Affects Male but Not Female Fertility. Reprod. Biol. Endocrinol. 2015, 13, 109. [Google Scholar] [CrossRef]
- Forbes, B.E.; Blyth, A.J.; Wit, J.M. Disorders of IGFs and IGF-1R Signaling Pathways. Mol. Cell. Endocrinol. 2020, 518, 111035. [Google Scholar] [CrossRef]
- Conover, C.A.; Boldt, H.B.; Bale, L.K.; Clifton, K.B.; Grell, J.A.; Mader, J.R.; Mason, E.J.; Powell, D.R. Pregnancy-Associated Plasma Protein-A2 (PAPP-A2): Tissue Expression and Biological Consequences of Gene Knockout in Mice. Endocrinology 2011, 152, 2837–2844. [Google Scholar] [CrossRef]
- Canali, G.; Garcia, M.; Hivert, B.; Pinatel, D.; Goullancourt, A.; Oguievetskaia, K.; Saint-Martin, M.; Girault, J.-A.; Faivre-Sarrailh, C.; Goutebroze, L. Genetic Variants in Autism-Related CNTNAP2 Impair Axonal Growth of Cortical Neurons. Human. Mol. Genet. 2018, 27, 1941–1954. [Google Scholar] [CrossRef]
- Vogt, D.; Cho, K.K.A.; Shelton, S.M.; Paul, A.; Huang, Z.J.; Sohal, V.S.; Rubenstein, J.L.R. Mouse Cntnap2 and Human CNTNAP2 ASD Alleles Cell Autonomously Regulate PV+ Cortical Interneurons. Cereb. Cortex 2018, 28, 3868–3879. [Google Scholar] [CrossRef]
- Scott, K.E.; Mann, R.S.; Schormans, A.L.; Schmid, S.; Allman, B.L. Hyperexcitable and Immature-like Neuronal Activity in the Auditory Cortex of Adult Rats Lacking the Language-Linked CNTNAP2 Gene. Cereb. Cortex 2022, 32, 4797–4817. [Google Scholar] [CrossRef]
- Lindahl, M.; Timmusk, T.; Rossi, J.; Saarma, M.; Airaksinen, M.S. Expression and Alternative Splicing of Mouse Gfra4 Suggest Roles in Endocrine Cell Development. Mol. Cell. Neurosci. 2000, 15, 522–533. [Google Scholar] [CrossRef]
- Wang, G.; Wang, H.; Zhang, L.; Guo, F.; Wu, X.; Liu, Y. MiR-195-5p Inhibits Proliferation and Invasion of Nerve Cells in Hirschsprung Disease by Targeting GFRA4. Mol. Cell. Biochem. 2021, 476, 2061–2073. [Google Scholar] [CrossRef]
- Zhou, J.; Chang, Y.; Li, J.; Bao, H.; Wu, C. Integrating Whole-Genome Resequencing and RNA Sequencing Data Reveals Selective Sweeps and Differentially Expressed Genes Related to Nervous System Changes in Luxi Gamecocks. Genes 2023, 14, 584. [Google Scholar] [CrossRef]
- St. George-Hyslop, F.; Kivisild, T.; Livesey, F.J. The Role of Contactin-Associated Protein-like 2 in Neurodevelopmental Disease and Human Cerebral Cortex Evolution. Front. Mol. Neurosci. 2022, 15, 1017144. [Google Scholar] [CrossRef]
- Scott, K.E.; Kazazian, K.; Mann, R.S.; Möhrle, D.; Schormans, A.L.; Schmid, S.; Allman, B.L. Loss of Cntnap2 in the Rat Causes Autism-Related Alterations in Social Interactions, Stereotypic Behavior, and Sensory Processing. Autism Res. 2020, 13, 1698–1717. [Google Scholar] [CrossRef]
- Berchtold, N.C.; Coleman, P.D.; Cribbs, D.H.; Rogers, J.; Gillen, D.L.; Cotman, C.W. Synaptic Genes Are Extensively Downregulated across Multiple Brain Regions in Normal Human Aging and Alzheimer’s Disease. Neurobiol. Aging 2013, 34, 1653–1661. [Google Scholar] [CrossRef]
- Zheng, Q.; Zhang, Y.; Chen, Y.; Yang, N.; Wang, X.-J.; Zhu, D. Systematic Identification of Genes Involved in Divergent Skeletal Muscle Growth Rates of Broiler and Layer Chickens. BMC Genom. 2009, 10, 87. [Google Scholar] [CrossRef]
- Meyer, E.; Manahan, D. Gene Expression Profiling of Genetically Determined Growth Variation in Bivalve Larvae (Crassostrea Gigas). J. Exp. Biol. 2010, 213, 749–758. [Google Scholar] [CrossRef]
- Dalton, C.M.; Schlegel, C.; Hunter, C.J. Caveolin-1: A Review of Intracellular Functions, Tissue-Specific Roles, and Epithelial Tight Junction Regulation. Biology 2023, 12, 1402. [Google Scholar] [CrossRef]
- Sun, M.; Luo, E.Y.; Adams, S.M.; Adams, T.; Ye, Y.; Shetye, S.S.; Soslowsky, L.J.; Birk, D.E. Collagen XI Regulates the Acquisition of Collagen Fibril Structure, Organization and Functional Properties in Tendon. Matrix Biol. 2020, 94, 77–94. [Google Scholar] [CrossRef]
- Slabaugh, E.; Desai, J.; Sartor, R.; Lawas, L.; Jagadish, S.; Doherty, C. Analysis of Differential Gene Expression and Alternative Splicing Is Significantly Influenced by Choice of Reference Genome. RNA 2019, 25, 669–684. [Google Scholar] [CrossRef]
- Guo, W.-J.; Coulter, M.; Waugh, R.; Zhang, R. The Value of Genotype-Specific Reference for Transcriptome Analyses in Barley. Life Sci. Alliance 2021, 5, 8. [Google Scholar] [CrossRef]
- Chen, N.-C.; Solomon, B.; Mun, T.; Iyer, S.; Langmead, B. Reference Flow: Reducing Reference Bias Using Multiple Population Genomes. Genome Biol. 2021, 22, 8. [Google Scholar] [CrossRef]
- Min, J.L.; Barrett, A.; Watts, T.; Pettersson, F.H.; Lockstone, H.E.; Lindgren, C.M.; Taylor, J.M.; Allen, M.; Zondervan, K.T.; McCarthy, M.I. Variability of Gene Expression Profiles in Human Blood and Lymphoblastoid Cell Lines. BMC Genom. 2010, 11, 96. [Google Scholar] [CrossRef]
- Danaher, P.; Warren, S.; Dennis, L.; D’Amico, L.; White, A.; Disis, M.L.; Geller, M.A.; Odunsi, K.; Beechem, J.; Fling, S.P. Gene Expression Markers of Tumor Infiltrating Leukocytes. J. Immunother. Cancer 2017, 5, 18. [Google Scholar] [CrossRef]
- Lopes-Ramos, C.M.; Paulson, J.N.; Chen, C.-Y.; Kuijjer, M.L.; Fagny, M.; Platig, J.; Sonawane, A.R.; DeMeo, D.L.; Quackenbush, J.; Glass, K. Regulatory Network Changes between Cell Lines and Their Tissues of Origin. BMC Genom. 2017, 18, 723. [Google Scholar] [CrossRef]
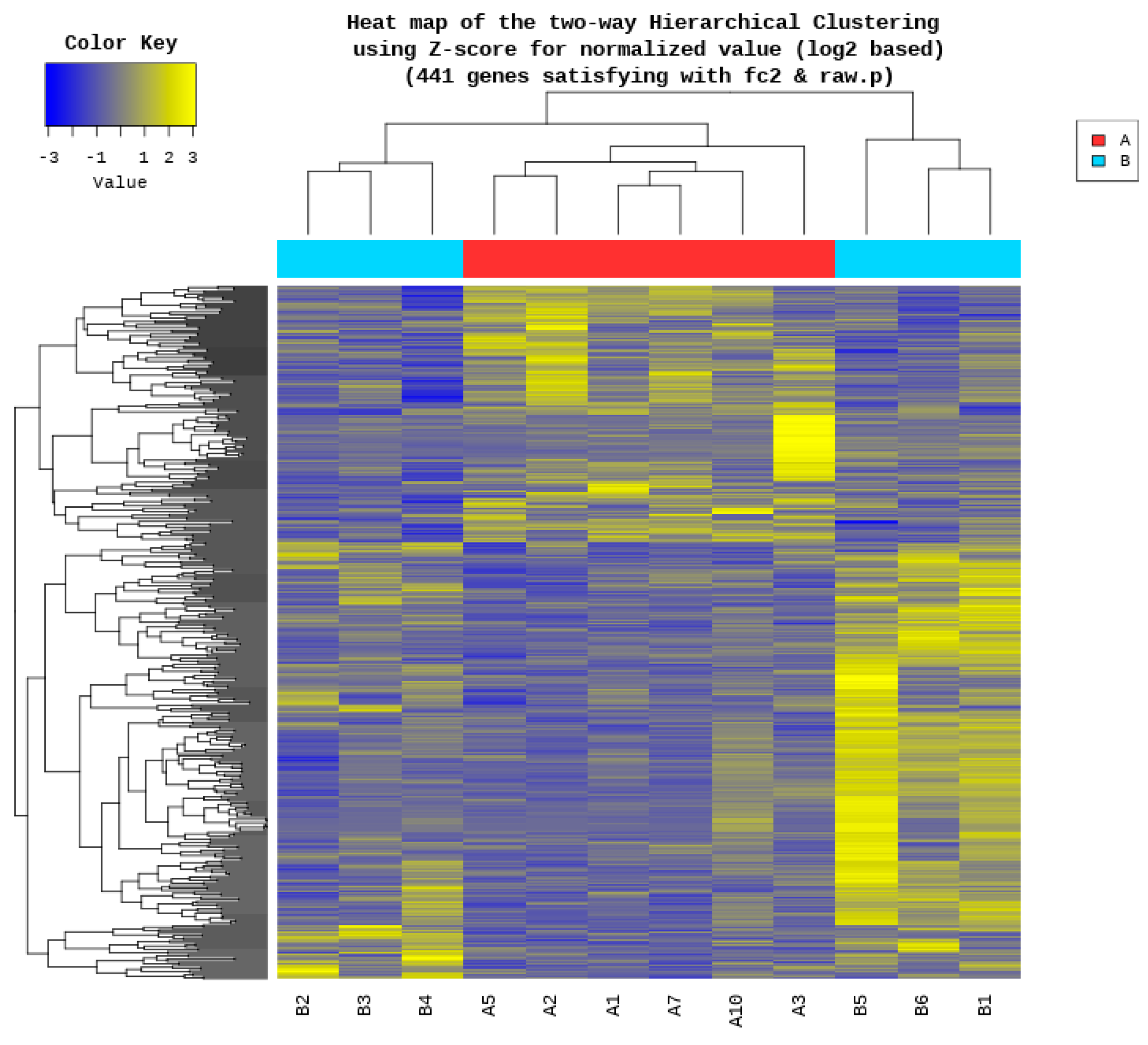
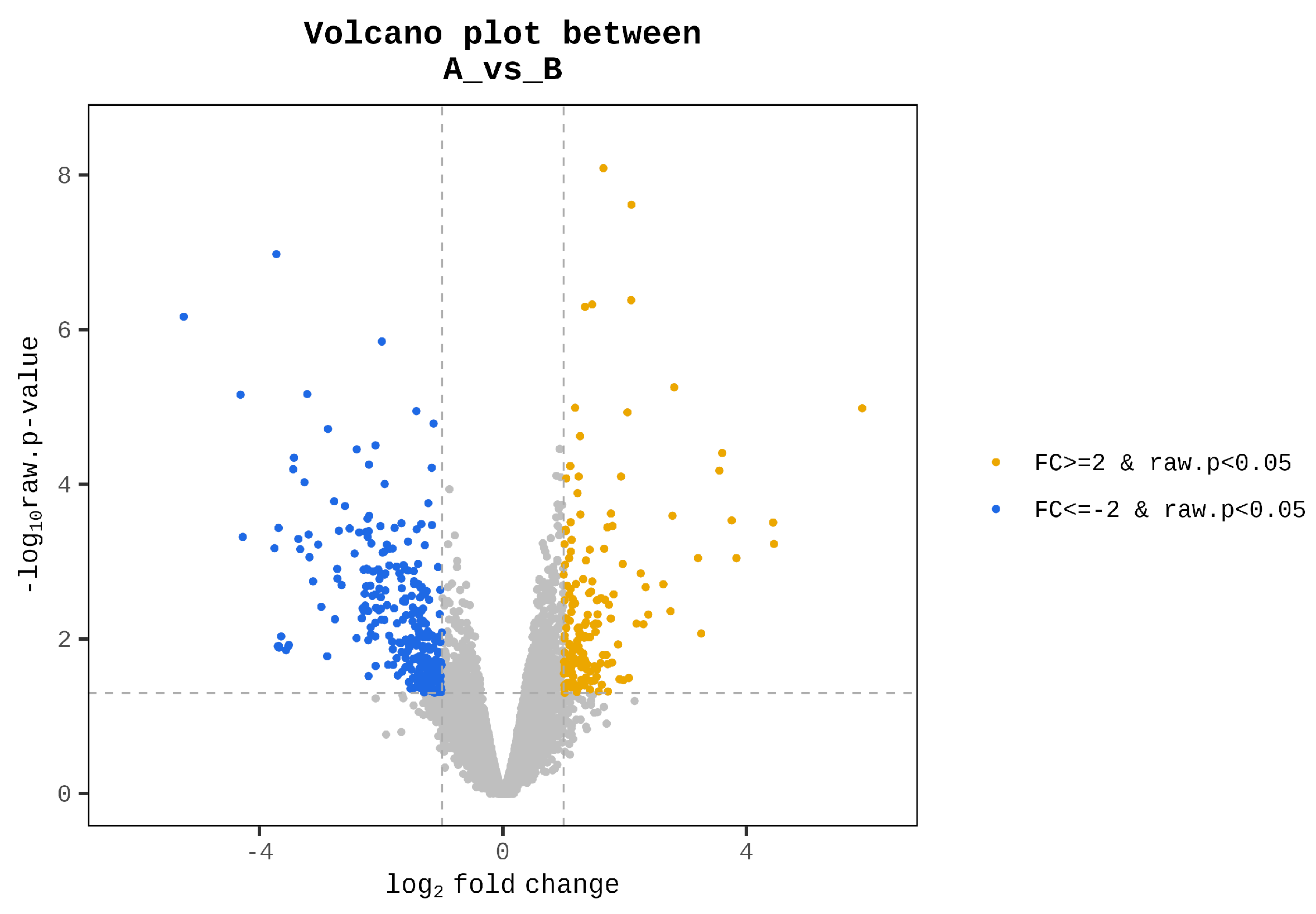
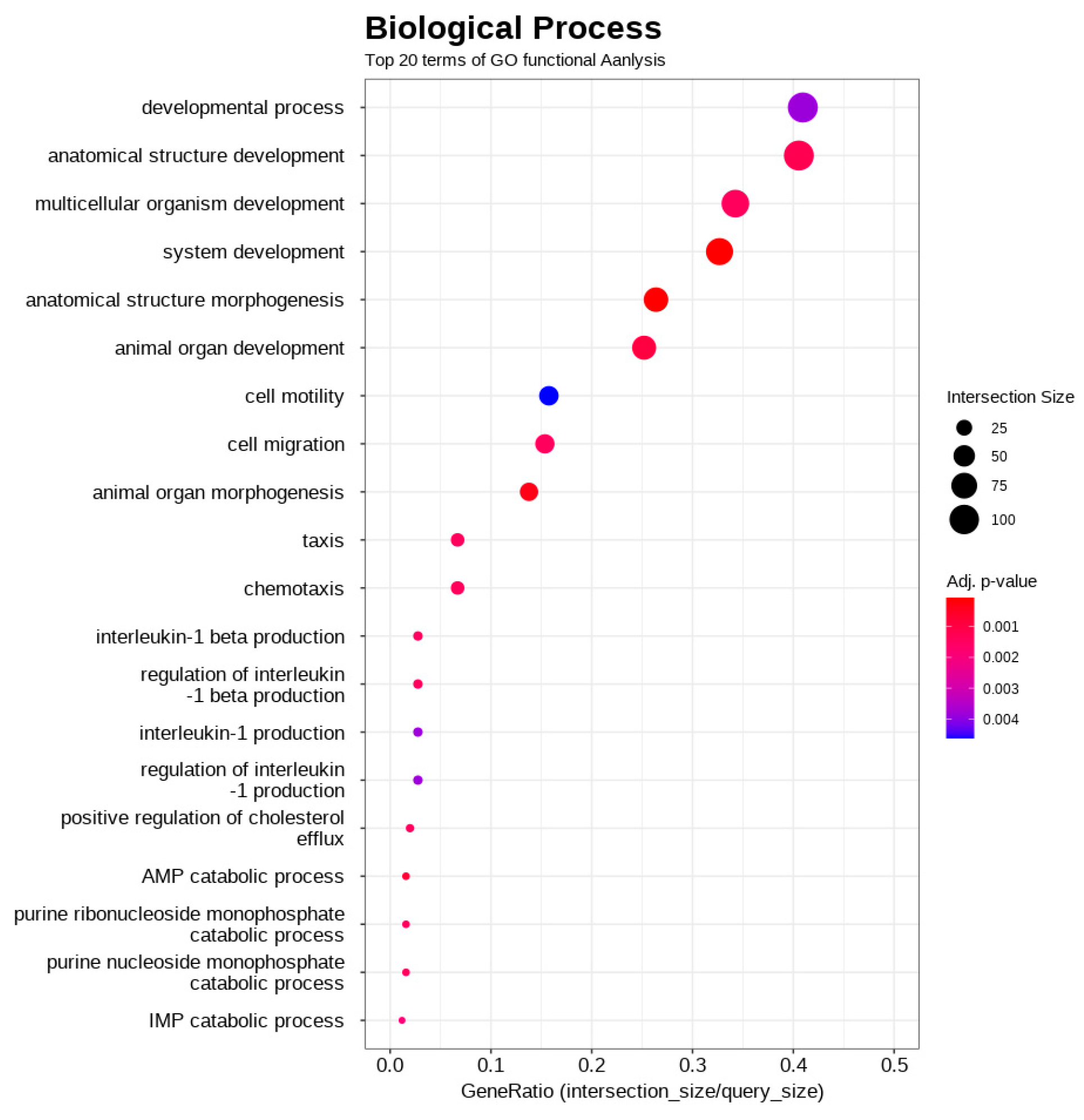
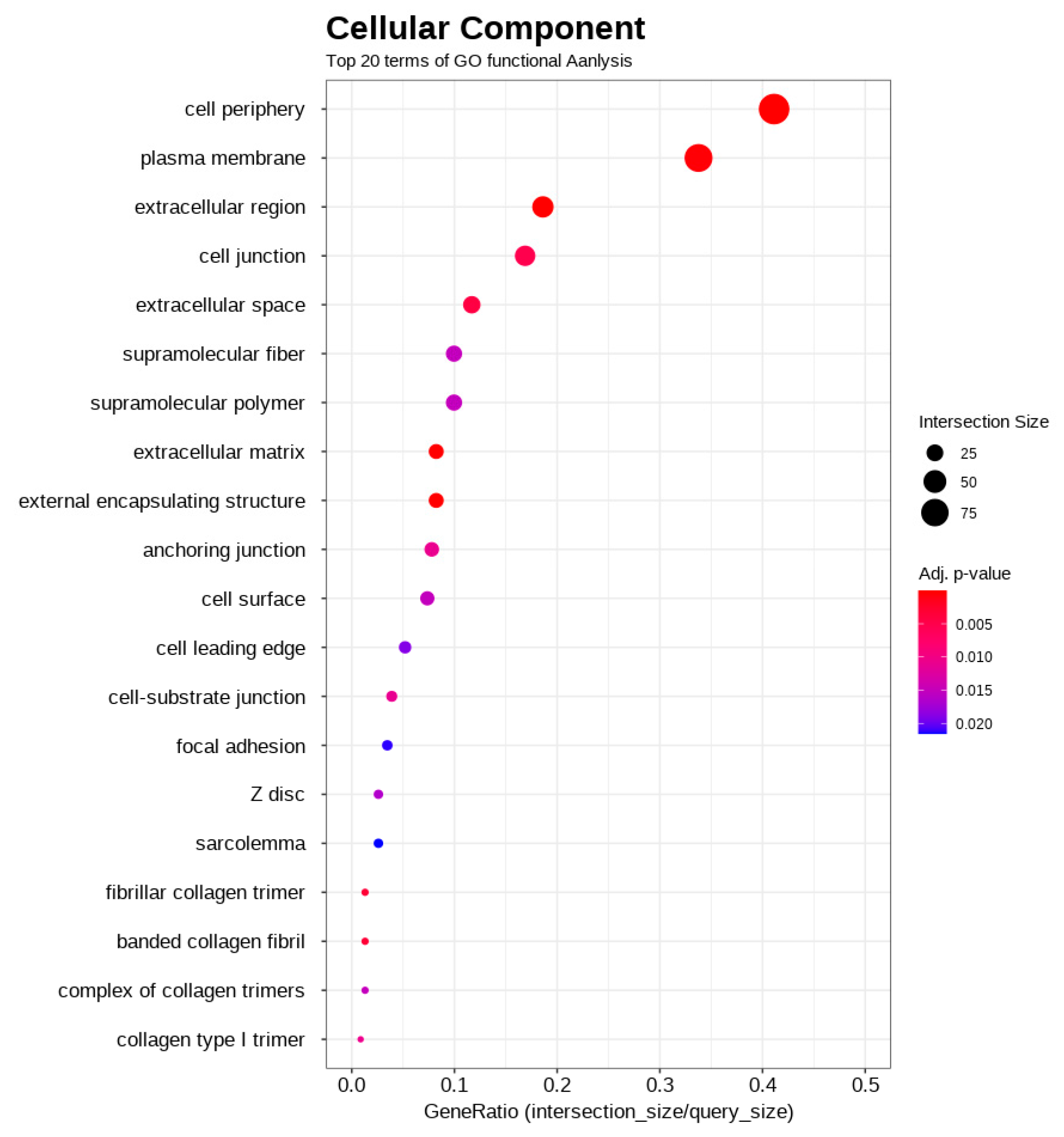
| Rank | Gene Symbol | Description | A/B Fold Change | Adjusted p-Value |
|---|---|---|---|---|
| 1 | SYNPO2 | synaptopodin 2, transcript variant X1 | |59.76| | <0.001 |
| 2 | CNTNAP2 | contactin associated protein-like 2, transcript variant X4 | |−37.80| | <0.001 |
| 3 | GFRA4 | GDNF family receptor alpha 4 | |−19.81| | <0.001 |
| 4 | LOC107055098 | uncharacterized LOC107055098 | |−13.17| | <0.001 |
| 5 | POPDC2 | popeye domain containing 2 | |12.14| | <0.05 |
| 6 | LOC107055485 | 60S ribosomal protein L17-like | |11.76| | <0.001 |
| 7 | FBN2 | fibrillin 2, transcript variant X2 | |−10.88| | <0.05 |
| 8 | ALDH1A2 | aldehyde dehydrogenase 1 family member A2 | |−10.78| | <0.05 |
| 9 | FABP5 | fatty acid binding protein 5 | |−9.57| | <0.05 |
| 10 | KCNS2 | potassium voltage-gated channel modifier subfamily S member 2, transcript variant X1 | |−9.27| | <0.05 |
| 11 | AGBL1 | ATP/GTP binding protein like 1, transcript variant X5 | |−7.32| | <0.05 |
| 12 | NRN1 | neuritin 1 | |7.03| | <0.05 |
| 13 | LOC121106479 | arylacetamide deacetylase-like 3 | |−5.28| | <0.05 |
| 14 | RIMS2 | regulating synaptic membrane exocytosis 2, transcript variant X29 | |−4.59| | <0.05 |
| 15 | PAPPA2 | pappalysin 2, transcript variant X1 | |4.32| | <0.001 |
| 16 | FAM19A4 | family with sequence similarity 19 member A4, C-C motif chemokine like, transcript variant X2 | |4.30| | <0.05 |
| 17 | SLC24A4 | solute carrier family 24 member 4, transcript variant X2 | |−4.23| | <0.05 |
| 18 | SLC6A4 | solute carrier family 6 member 4 | |4.13| | <0.05 |
| 19 | SLC6A15 | solute carrier family 6 member 15, transcript variant X8 | |−3.97| | <0.05 |
| 20 | CRISPLD2 | cysteine rich secretory protein LCCL domain containing 2, transcript variant X2 | |−3.84| | <0.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piratae, S.; Yamtubtim, C.; Nonsri, T.; Poomprasert, P.; Purisotayo, T. Genetic Signatures of Competitive Performance in Burmese Gamecocks: A Transcriptomic Analysis. Biology 2025, 14, 1066. https://doi.org/10.3390/biology14081066
Piratae S, Yamtubtim C, Nonsri T, Poomprasert P, Purisotayo T. Genetic Signatures of Competitive Performance in Burmese Gamecocks: A Transcriptomic Analysis. Biology. 2025; 14(8):1066. https://doi.org/10.3390/biology14081066
Chicago/Turabian StylePiratae, Supawadee, Chanistha Yamtubtim, Thanitaporn Nonsri, Panpanit Poomprasert, and Tarid Purisotayo. 2025. "Genetic Signatures of Competitive Performance in Burmese Gamecocks: A Transcriptomic Analysis" Biology 14, no. 8: 1066. https://doi.org/10.3390/biology14081066
APA StylePiratae, S., Yamtubtim, C., Nonsri, T., Poomprasert, P., & Purisotayo, T. (2025). Genetic Signatures of Competitive Performance in Burmese Gamecocks: A Transcriptomic Analysis. Biology, 14(8), 1066. https://doi.org/10.3390/biology14081066







