The Development of Horns in Bovidae and the Genetic Mechanisms Underpinning This Process
Simple Summary
Abstract
1. Introduction
2. Evolution and Functional Diversification of Horns in Bovidae
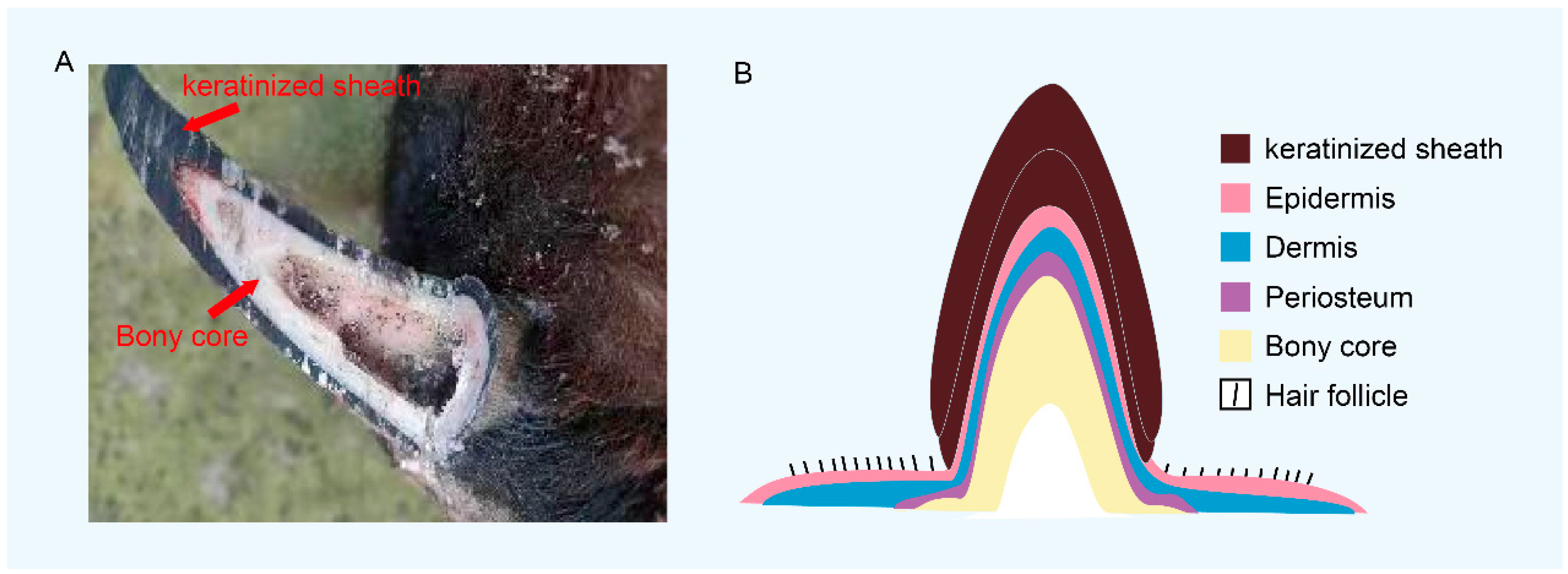
3. Structure of Horns in Bovidae
4. Horn Morphological Development in Bovidae
5. Genetic Regulation of Horns in Bovidae
5.1. Genetic Regulation of Horns in Bovines
5.1.1. Genes Governing Bovine Horn Development
5.1.2. Genetic Regions and Genes Related to Bovine Polledness
5.1.3. Genetic Regions and Genes Linked to Bovine Scurs
5.2. Genetic Regulation of Horns in Sheep
5.2.1. Genes Related to Horn Development in Sheep
5.2.2. Genetic Regions and Genes Controlling Sheep Polledness
5.2.3. Genetic Regions and Genes Controlling Polycerate Sheep
5.3. Genetic Regulation of Horns in Goats
| Species | Phenotypes | Positions | Candidate Genes | Ref. |
|---|---|---|---|---|
| Bovine | Polled | BTA 1: 0.8~2.8 Mb | OLIG1, URB1, OLIG2, IFNAR1, C1H21orf62, GART | [41] |
| BTA 1: 147 kb | C1H21orf62, GCFC1, SYNJ1 | [50] | ||
| BTA 2: 3.7 Mb | ZEB2 | [15] | ||
| Scurs | BTA 19: BMS2142 and IDVGA46 regions | ALOX12, MFAP4 | [57,80] | |
| BTA 4: 1.7 Mb | TWIST1 | [13] | ||
| Sheep | Polled | OAR 10: 1833 bp | RXFP2 | [9] |
| Polycerate | OAR 2: 132.9~133.1 Mb | MTX2, HOXD, EVX2, MIR10B, KIAA1715 | [68] | |
| Goat | Polled | Chr 1: 11.7 kb | FOXL2, PISRT1 | [24] |
| Chr 1: 10.159 kb deletion and 480 kb duplication | KCNJ15, ERG | [71,72,73] |
6. Environmental Influences on Horn Development
7. Cellular Heterogeneity Underlying Horn Development
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, X.; Guan, T.P.; Jiang, W.L.; Li, D.D.; Yang, K.; Li, S. Distribution and population status of bovine species in China based on bibliometric analysis. Biodivers. Sci. 2021, 29, 668–679. [Google Scholar] [CrossRef]
- Davis, E.B.; Brakora, K.A.; Lee, A.H. Evolution of ruminant headgear: A review. Proc. Biol. Sci. 2011, 278, 2857–2865. [Google Scholar] [CrossRef]
- Preston, B.T.; Stevenson, I.R.; Pemberton, J.M.; Coltman, D.W.; Wilson, K. Overt and covert competition in a promiscuous mammal: The importance of weaponry and testes size to male reproductive success. Proc. Biol. Sci. 2003, 270, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.R.; Kruuk, L.E. Function of weaponry in females: The use of horns in intrasexual competition for resources in female Soay sheep. Biol. Lett. 2007, 3, 651–654. [Google Scholar] [CrossRef]
- Simon, R.; Drögemüller, C.; Lühken, G. The Complex and Diverse Genetic Architecture of the Absence of Horns (Polledness) in Domestic Ruminants, including Goats and Sheep. Genes 2022, 13, 832. [Google Scholar] [CrossRef] [PubMed]
- Thompson, K.G.; Bateman, R.S.; Morris, P.J. Cerebral infarction and meningoencephalitis following hot-iron disbudding of goat kids. N. Z. Vet. J. 2005, 53, 368–370. [Google Scholar] [CrossRef]
- Ren, Q. Morphogenesis of Yak Horns and the Expression of SHH and β-Catenin. Master’s Thesis, Northwest A&F University, Yangling, China, 2008. [Google Scholar]
- He, T.; Yu, F. Comparative study on the epidermal changes of horn primordia during the embryonic stage in yak, cattle, and Tibetan goats. Heilongjiang Anim. Sci. Vet. Med. 2010, 61–62. Available online: https://link.cnki.net/doi/10.13881/j.cnki.hljxmsy.2010.19.082 (accessed on 1 August 2025).
- Wiedemar, N.; Drögemüller, C. A 1.8-kb insertion in the 3′-UTR of RXFP2 is associated with polledness in sheep. Anim. Genet. 2015, 46, 457–461. [Google Scholar] [CrossRef]
- Wiedemar, N.; Tetens, J.; Jagannathan, V.; Menoud, A.; Neuenschwander, S.; Bruggmann, R.; Thaller, G.; Drögemüller, C. Independent polled mutations leading to complex gene expression differences in cattle. PLoS ONE 2014, 9, e93435. [Google Scholar] [CrossRef]
- He, X.; Song, S.; Chen, X.; Song, T.; Lobsang, T.; Guan, W.; Pu, Y.; Zhao, Q.; Jiang, L.; Ma, Y. Genome-wide association analysis reveals the common genetic locus for both the typical and atypical polycerate phenotype in Tibetan sheep. Anim. Genet. 2018, 49, 142–143. [Google Scholar] [CrossRef]
- Allais-Bonnet, A.; Hintermann, A.; Deloche, M.C.; Cornette, R.; Bardou, P.; Naval-Sanchez, M.; Pinton, A.; Haruda, A.; Grohs, C.; Zakany, J.; et al. Analysis of Polycerate Mutants Reveals the Evolutionary Co-option of HOXD1 for Horn Patterning in Bovidae. Mol. Biol. Evol. 2021, 38, 2260–2272. [Google Scholar] [CrossRef]
- Capitan, A.; Grohs, C.; Weiss, B.; Rossignol, M.N.; Reversé, P.; Eggen, A. A newly described bovine type 2 scurs syndrome segregates with a frame-shift mutation in TWIST1. PLoS ONE 2011, 6, e22242. [Google Scholar] [CrossRef]
- Gehrke, L.J.; Upadhyay, M.; Heidrich, K.; Kunz, E.; Klaus-Halla, D.; Weber, F.; Zerbe, H.; Seichter, D.; Graf, A.; Krebs, S.; et al. A de novo frameshift mutation in ZEB2 causes polledness, abnormal skull shape, small body stature and subfertility in Fleckvieh cattle. Sci. Rep. 2020, 10, 17032. [Google Scholar] [CrossRef]
- Capitan, A.; Allais-Bonnet, A.; Pinton, A.; Marquant-Le Guienne, B.; Le Bourhis, D.; Grohs, C.; Bouet, S.; Clément, L.; Salas-Cortes, L.; Venot, E.; et al. A 3.7 Mb deletion encompassing ZEB2 causes a novel polled and multisystemic syndrome in the progeny of a somatic mosaic bull. PLoS ONE 2012, 7, e49084. [Google Scholar] [CrossRef]
- Calamari, Z.T.; Flynn, J.J. Gene expression supports a single origin of horns and antlers in hoofed mammals. Commun. Biol. 2024, 7, 509. [Google Scholar] [CrossRef] [PubMed]
- Gentry, A.W. The Miocene differentiation of old world Pecora (Mammalia). Hist. Biol. 1994, 7, 115–158. [Google Scholar] [CrossRef]
- Bibi, F. A multi-calibrated mitochondrial phylogeny of extant Bovidae (Artiodactyla, Ruminantia) and the importance of the fossil record to systematics. BMC Evol. Biol. 2013, 13, 166. [Google Scholar] [CrossRef]
- Mennecart, B.; DeMiguel, D.; Bibi, F.; Rössner, G.E.; Métais, G.; Neenan, J.M.; Wang, S.; Schulz, G.; Müller, B.; Costeur, L. Bony labyrinth morphology clarifies the origin and evolution of deer. Sci. Rep. 2017, 7, 13176. [Google Scholar] [CrossRef]
- Cantalapiedra, J.L.; Fitzjohn, R.G.; Kuhn, T.S.; Fernández, M.H.; DeMiguel, D.; Azanza, B.; Morales, J.; Mooers, A. Dietary innovations spurred the diversification of ruminants during the Caenozoic. Proc. Biol. Sci. 2014, 281, 20132746. [Google Scholar] [CrossRef] [PubMed]
- Vrba, E.S.; Schaller, G.B. Antelopes, Deer, and Relatives: Fossil Record, Behavioral Ecology, Systematics, and Conservation; Yale University Press: New Haven, CT, USA, 2000. [Google Scholar]
- Vander Linden, A.; Dumont, E.R. Intraspecific male combat behaviour predicts morphology of cervical vertebrae in ruminant mammals. Proc. Biol. Sci. 2019, 286, 20192199. [Google Scholar] [CrossRef]
- Picard, K.; Festa-Bianchet, M.; Thomas, D. The cost of horniness: Heat loss may counter sexual selection for large horns in temperate bovids. Écoscience 1996, 3, 280–284. [Google Scholar] [CrossRef]
- Pailhoux, E.; Vigier, B.; Chaffaux, S.; Servel, N.; Taourit, S.; Furet, J.P.; Fellous, M.; Grosclaude, F.; Cribiu, E.P.; Cotinot, C.; et al. A 11.7-kb deletion triggers intersexuality and polledness in goats. Nat. Genet. 2001, 29, 453–458. [Google Scholar] [CrossRef]
- Liu, Z.Z.; Xiang, C.G. Observation and management of intersexual infertility in hornless populations of Guizhou back goats. Heilongjiang Anim. Sci. Vet. Med. 2015, 56–57. Available online: https://link.cnki.net/doi/10.13881/j.cnki.hljxmsy.2015.1257 (accessed on 1 August 2025).
- Zhang, Q.W.; Du, M.M.; Cheng, M.; Dai, Z.H.; Liu, K.D.; Lin, X.K.; Gao, E.S.; Qin, Z.L.; Zhao, J.S.; Li, H.G. Genotypic analysis of polled intersex syndrome reproductive defect gene in Laoshan dairy goats. China Anim. Husb. Vet. Med. 2022, 49, 3054–3061. [Google Scholar]
- Alvarez, L.; Adcock, S.J.J.; Tucker, C.B. Sensitivity and wound healing after hot-iron disbudding in goat kids. J. Dairy Sci. 2019, 102, 10152–10162. [Google Scholar] [CrossRef] [PubMed]
- Dove, W.F. The physiology of horn growth: A study of the morphogenesis, the interaction of tissues, and the evolutionary processes of a Mendelian recessive character by means of transplantation of tissues. J. Exp. Zool. 1935, 69, 347–405. [Google Scholar] [CrossRef]
- Hartwig, H.; Schrudde, J. Experimentelle Untersuchungen zur Bildung der primären Stirnauswüchse beim Reh (Capreolus capreolus L.). Z. Für. Jagdwiss. 1974, 20, 1–13. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, C.; Wang, N.; Li, Z.; Heller, R.; Liu, R.; Zhao, Y.; Han, J.; Pan, X.; Zheng, Z.; et al. Genetic basis of ruminant headgear and rapid antler regeneration. Science 2019, 364, eaav6335. [Google Scholar] [CrossRef]
- Wiener, D.J.; Wiedemar, N.; Welle, M.M.; Drögemüller, C. Novel Features of the Prenatal Horn Bud Development in Cattle (Bos taurus). PLoS ONE 2015, 10, e0127691. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Huang, W.; Hayashi, C.; Gatesy, J.; McKittrick, J. Microstructure and mechanical properties of different keratinous horns. J. R. Soc. Interface 2018, 15, 20180093. [Google Scholar] [CrossRef]
- Lyne, A.; Hollis, D. Development of horns in merino sheep. Aust. J. Zool. 1973, 21, 153–169. [Google Scholar] [CrossRef]
- Evans, H.E.; Sack, W.O. Prenatal development of domestic and laboratory mammals: Growth curves, external features and selected references. Zentralbl. Vet. C 1973, 2, 11–45. [Google Scholar] [CrossRef]
- Aldersey, J.E.; Chen, T.; Petrovski, K.; Williams, J.L.; Bottema, C.D.K. Histological characterisation of the horn bud region in 58 day old bovine fetuses. Int. J. Dev. Biol. 2024, 68, 117–126. [Google Scholar] [CrossRef]
- Li, M.; Wu, X.; Guo, X.; Bao, P.; Ding, X.; Chu, M.; Liang, C.; Yan, P. Comparative iTRAQ proteomics revealed proteins associated with horn development in yak. Proteome Sci. 2018, 16, 14. [Google Scholar] [CrossRef]
- Luan, Y.; Wu, S.; Wang, M.; Pu, Y.; Zhao, Q.; Ma, Y.; Jiang, L.; He, X. Identification of Critical Genes for Ovine Horn Development Based on Transcriptome during the Embryonic Period. Biology 2023, 12, 591. [Google Scholar] [CrossRef]
- Li, H.; Du, M.; Lin, X.; Cao, X.; Leng, L.; Campo, F.M.P.; Xu, D.; Hou, L.; Gao, X.; Zhou, J.; et al. Multiple cell types guided by neurocytes orchestrate horn bud initiation in dairy goats. Genet. Sel. Evol. 2025, 57, 34. [Google Scholar] [CrossRef]
- Côté, S.D.; Festa-Bianchet, M.; Smith, K.G. Horn Growth in Mountain Goats (Oreamnos americanus). J. Mammal. 1998, 79, 406–414. [Google Scholar] [CrossRef]
- Long, C.R.; Gregory, K.E. Inheritance of the horned, scurred, and polled condition in cattle. J. Hered. 1978, 69, 395–400. [Google Scholar] [CrossRef]
- Allais-Bonnet, A.; Grohs, C.; Medugorac, I.; Krebs, S.; Djari, A.; Graf, A.; Fritz, S.; Seichter, D.; Baur, A.; Russ, I.; et al. Novel insights into the bovine polled phenotype and horn ontogenesis in Bovidae. PLoS ONE 2013, 8, e63512. [Google Scholar] [CrossRef] [PubMed]
- Ren, Q.; Yu, F.; Wang, X.Q.; Zhao, Y.H.; Li, J. Temporal-spatial expression of Shh and β-catenin during horn morphogenesis in yak. Northwest J. Agric. Sci. 2009, 18, 24–29. [Google Scholar]
- He, T. The Relationship Between Development of Hair Follicles at the Horn Border and Horn Sheath, and the Expression Pattern of β-Catenin in Yak and Cattle. Master’s Thesis, Northwest A&F University, Yangling, China, 2010. [Google Scholar]
- Georges, M.; Drinkwater, R.; King, T.; Mishra, A.; Moore, S.S.; Nielsen, D.; Sargeant, L.S.; Sorensen, A.; Steele, M.R.; Zhao, X.; et al. Microsatellite mapping of a gene affecting horn development in Bos taurus. Nat. Genet. 1993, 4, 206–210. [Google Scholar] [CrossRef] [PubMed]
- Brenneman, R.A.; Davis, S.K.; Sanders, J.O.; Burns, B.M.; Wheeler, T.C.; Turner, J.W.; Taylor, J.F. The polled locus maps to BTA1 in a Bos indicus x Bos taurus cross. J. Hered. 1996, 87, 156–161. [Google Scholar] [CrossRef]
- Medugorac, I.; Seichter, D.; Graf, A.; Russ, I.; Blum, H.; Göpel, K.H.; Rothammer, S.; Förster, M.; Krebs, S. Bovine polledness--an autosomal dominant trait with allelic heterogeneity. PLoS ONE 2012, 7, e39477. [Google Scholar] [CrossRef]
- Rothammer, S.; Capitan, A.; Mullaart, E.; Seichter, D.; Russ, I.; Medugorac, I. The 80-kb DNA duplication on BTA1 is the only remaining candidate mutation for the polled phenotype of Friesian origin. Genet. Sel. Evol. 2014, 46, 44. [Google Scholar] [CrossRef]
- Medugorac, I.; Graf, A.; Grohs, C.; Rothammer, S.; Zagdsuren, Y.; Gladyr, E.; Zinovieva, N.; Barbieri, J.; Seichter, D.; Russ, I.; et al. Whole-genome analysis of introgressive hybridization and characterization of the bovine legacy of Mongolian yaks. Nat. Genet. 2017, 49, 470–475. [Google Scholar] [CrossRef]
- Utsunomiya, Y.T.; Torrecilha, R.B.P.; Milanesi, M.; Paulan, S.C.; Utsunomiya, A.T.H.; Garcia, J.F. Hornless Nellore cattle (Bos indicus) carrying a novel 110 kbp duplication variant of the polled locus. Anim. Genet. 2019, 50, 187–188. [Google Scholar] [CrossRef]
- Liu, W.B.; Liu, J.; Liang, C.N.; Guo, X.; Bao, P.J.; Chu, M.; Ding, X.Z.; Wang, H.B.; Zhu, X.S.; Yan, P. Associations of single nucleotide polymorphisms in candidate genes with the polled trait in Datong domestic yaks. Anim. Genet. 2014, 45, 138–141. [Google Scholar] [CrossRef]
- Hennig, S.L.; McNabb, B.R.; Trott, J.F.; Van Eenennaam, A.L.; Murray, J.D. LincRNA#1 knockout alone does not affect polled phenotype in cattle heterozygous for the celtic POLLED allele. Sci. Rep. 2022, 12, 7627. [Google Scholar] [CrossRef] [PubMed]
- Hennig, S.L.; Owen, J.R.; Lin, J.C.; McNabb, B.R.; Van Eenennaam, A.L.; Murray, J.D. A deletion at the polled PC locus alone is not sufficient to cause a polled phenotype in cattle. Sci. Rep. 2022, 12, 2067. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Carlson, D.F.; Lancto, C.A.; Garbe, J.R.; Webster, D.A.; Hackett, P.B.; Fahrenkrug, S.C. Efficient nonmeiotic allele introgression in livestock using custom endonucleases. Proc. Natl. Acad. Sci. USA 2013, 110, 16526–16531. [Google Scholar] [CrossRef]
- Carlson, D.F.; Lancto, C.A.; Zang, B.; Kim, E.S.; Walton, M.; Oldeschulte, D.; Seabury, C.; Sonstegard, T.S.; Fahrenkrug, S.C. Production of hornless dairy cattle from genome-edited cell lines. Nat. Biotechnol. 2016, 34, 479–481. [Google Scholar] [CrossRef]
- Schuster, F.; Aldag, P.; Frenzel, A.; Hadeler, K.G.; Lucas-Hahn, A.; Niemann, H.; Petersen, B. CRISPR/Cas12a mediated knock-in of the Polled Celtic variant to produce a polled genotype in dairy cattle. Sci. Rep. 2020, 10, 13570. [Google Scholar] [CrossRef]
- Meng, G.; La, Y.; Bao, Q.; Wu, X.; Ma, X.; Huang, C.; Chu, M.; Liang, C.; Yan, P. Early Growth and Development and Nonlinear Model Fitting Analysis of Ashidan Yak. Animals 2023, 13, 1545. [Google Scholar] [CrossRef]
- Asai, M.; Berryere, T.G.; Schmutz, S.M. The scurs locus in cattle maps to bovine chromosome 19. Anim. Genet. 2004, 35, 34–39. [Google Scholar] [CrossRef]
- Gehrke, L.J.; Capitan, A.; Scheper, C.; König, S.; Upadhyay, M.; Heidrich, K.; Russ, I.; Seichter, D.; Tetens, J.; Medugorac, I.; et al. Are scurs in heterozygous polled (Pp) cattle a complex quantitative trait? Genet. Sel. Evol. 2020, 52, 6. [Google Scholar] [CrossRef] [PubMed]
- Tetens, J.; Wiedemar, N.; Menoud, A.; Thaller, G.; Drögemüller, C. Association mapping of the scurs locus in polled Simmental cattle—evidence for genetic heterogeneity. Anim. Genet. 2015, 46, 224–225. [Google Scholar] [CrossRef] [PubMed]
- Ferlin, A.; Pepe, A.; Gianesello, L.; Garolla, A.; Feng, S.; Giannini, S.; Zaccolo, M.; Facciolli, A.; Morello, R.; Agoulnik, A.I.; et al. Mutations in the insulin-like factor 3 receptor are associated with osteoporosis. J. Bone Miner. Res. 2008, 23, 683–693. [Google Scholar] [CrossRef]
- He, X.H.; Chen, X.F.; Pu, Y.B.; Guan, W.J.; Shen, S.; Zhao, Q.J.; Li, X.C.; Jiang, L.; Ma, Y.H. iTRAQ-based quantitative proteomic analysis reveals key pathways responsible for scurs in sheep (Ovis aries). J. Integr. Agric. 2018, 17, 1843–1851. [Google Scholar] [CrossRef]
- Johnston, S.E.; Beraldi, D.; McRae, A.F.; Pemberton, J.M.; Slate, J. Horn type and horn length genes map to the same chromosomal region in Soay sheep. Heredity 2010, 104, 196–205. [Google Scholar] [CrossRef]
- Montgomery, G.W.; Henry, H.M.; Dodds, K.G.; Beattie, A.E.; Wuliji, T.; Crawford, A.M. Mapping the Horns (Ho) locus in sheep: A further locus controlling horn development in domestic animals. J. Hered. 1996, 87, 358–363. [Google Scholar] [CrossRef] [PubMed]
- Kijas, J.W.; Lenstra, J.A.; Hayes, B.; Boitard, S.; Porto Neto, L.R.; San Cristobal, M.; Servin, B.; McCulloch, R.; Whan, V.; Gietzen, K.; et al. Genome-wide analysis of the world’s sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS Biol. 2012, 10, e1001258. [Google Scholar] [CrossRef]
- Dominik, S.; Henshall, J.M.; Hayes, B.J. A single nucleotide polymorphism on chromosome 10 is highly predictive for the polled phenotype in Australian Merino sheep. Anim. Genet. 2012, 43, 468–470. [Google Scholar] [CrossRef]
- Kijas, J.W.; Hadfield, T.; Naval Sanchez, M.; Cockett, N. Genome-wide association reveals the locus responsible for four-horned ruminant. Anim. Genet. 2016, 47, 258–262. [Google Scholar] [CrossRef]
- He, X.; Zhou, Z.; Pu, Y.; Chen, X.; Ma, Y.; Jiang, L. Mapping the four-horned locus and testing the polled locus in three Chinese sheep breeds. Anim. Genet. 2016, 47, 623–627. [Google Scholar] [CrossRef]
- Ren, X.; Yang, G.L.; Peng, W.F.; Zhao, Y.X.; Zhang, M.; Chen, Z.H.; Wu, F.A.; Kantanen, J.; Shen, M.; Li, M.H. A genome-wide association study identifies a genomic region for the polycerate phenotype in sheep (Ovis aries). Sci. Rep. 2016, 6, 21111. [Google Scholar] [CrossRef] [PubMed]
- Greyvenstein, O.F.; Reich, C.M.; van Marle-Koster, E.; Riley, D.G.; Hayes, B.J. Polyceraty (multi-horns) in Damara sheep maps to ovine chromosome 2. Anim. Genet. 2016, 47, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Eaton, O.N. The Relation between Polled and Hermaphroditic Characters in Dairy Goats. Genetics 1945, 30, 51–61. [Google Scholar] [CrossRef]
- Simon, R.; Lischer, H.E.L.; Pieńkowska-Schelling, A.; Keller, I.; Häfliger, I.M.; Letko, A.; Schelling, C.; Lühken, G.; Drögemüller, C. New genomic features of the polled intersex syndrome variant in goats unraveled by long-read whole-genome sequencing. Anim. Genet. 2020, 51, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Jiang, R.; Mao, A.; Liu, G.E.; Zhan, S.; Li, L.; Zhong, T.; Wang, L.; Cao, J.; Chen, Y.; et al. Genome-wide association study reveals 14 new SNPs and confirms two structural variants highly associated with the horned/polled phenotype in goats. BMC Genom. 2021, 22, 769. [Google Scholar] [CrossRef]
- E, G.-X.; Zhou, D.K.; Zheng, Z.Q.; Yang, B.G.; Li, X.L.; Li, L.H.; Zhou, R.Y.; Nai, W.H.; Jiang, X.P.; Zhang, J.H.; et al. Identification of a Goat Intersexuality-Associated Novel Variant Through Genome-Wide Resequencing and Hi-C. Front. Genet. 2020, 11, 616743. [Google Scholar] [CrossRef]
- Zhang, Y. Study on PISRT1, FOXL2 Genes and PIS Region in Polled Intersex Dairy Goats. Master’s Thesis, Hebei Agricultural University, Baoding, China, 2014. [Google Scholar]
- Zhang, J.J. Study on FOXL2 Gene and PIS Region Variation in Polled Intersex Dairy Goats. Master’s Thesis, Hebei Agricultural University, Baoding, China, 2010. [Google Scholar]
- Pannetier, M.; Renault, L.; Jolivet, G.; Cotinot, C.; Pailhoux, E. Ovarian-specific expression of a new gene regulated by the goat PIS region and transcribed by a FOXL2 bidirectional promoter. Genomics 2005, 85, 715–726. [Google Scholar] [CrossRef]
- Li, X.; Zhang, J.; Zhou, R.; Li, L.; Zheng, G. Special variations within 11.7 kb fragment in goat polled intersex syndrome. Afr. J. Biotechnol. 2011, 10, 6695–6699. [Google Scholar]
- Vaiman, D.; Koutita, O.; Oustry, A.; Elsen, J.-M.; Manfredi, E.; Fellous, M.; Cribiu, E. Genetic mapping of the autosomal region involved in XX sex-reversal and horn development in goats. Mamm. Genome 1996, 7, 133–137. [Google Scholar] [CrossRef]
- Asdell, S.A. The genetic sex of intersexual goats and a probable linkage with the gene for hornlessness. Science 1944, 99, 124. [Google Scholar] [CrossRef] [PubMed]
- Capitan, A.; Grohs, C.; Gautier, M.; Eggen, A. The scurs inheritance: New insights from the French Charolais breed. BMC Genet. 2009, 10, 33. [Google Scholar] [CrossRef] [PubMed]
- Festa-Bianchet, M.; Coltman, D.W.; Turelli, L.; Jorgenson, J.T. Relative allocation to horn and body growth in bighorn rams varies with resource availability. Behav. Ecol. 2004, 15, 305–312. [Google Scholar] [CrossRef]
- Giacometti, M.; Willing, R.; Defila, C. Ambient Temperature in Spring Affects Horn Growth in Male Alpine Ibexes. J. Mammal. 2002, 83, 245–251. [Google Scholar] [CrossRef][Green Version]
- Rughetti, M.; Festa-Bianchet, M. Effects of early horn growth on reproduction and hunting mortality in female chamois. J. Anim. Ecol. 2011, 80, 438–447. [Google Scholar] [CrossRef]
- Miura, S.; Kita, I.; Sugimura, M. Horn Growth and Reproductive History in Female Japanese Serow. J. Mammal. 1987, 68, 826–836. [Google Scholar] [CrossRef]
- Chirichella, R.; Ciuti, S.; Grignolio, S.; Rocca, M.; Apollonio, M. The role of geological substrate for horn growth in ungulates: A case study on Alpine chamois. Evol. Ecol. 2013, 27, 145–163. [Google Scholar] [CrossRef]
- Qin, T.; Zhang, G.; Zheng, Y.; Li, S.; Yuan, Y.; Li, Q.; Hu, M.; Si, H.; Wei, G.; Gao, X.; et al. A population of stem cells with strong regenerative potential discovered in deer antlers. Science 2023, 379, 840–847. [Google Scholar] [CrossRef] [PubMed]
- Ba, H.; Wang, X.; Wang, D.; Ren, J.; Wang, Z.; Sun, H.X.; Hu, P.; Zhang, G.; Wang, S.; Ma, C.; et al. Single-cell transcriptome reveals core cell populations and androgen-RXFP2 axis involved in deer antler full regeneration. Cell Regen. 2022, 11, 43. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Berg, D.; Ba, H.; Sun, H.; Wang, Z.; Li, C. Deer antler stem cells are a novel type of cells that sustain full regeneration of a mammalian organ-deer antler. Cell Death Dis. 2019, 10, 443. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Yan, W.; Guo, J.; Dai, D.; Li, L.; Zhang, H. The Development of Horns in Bovidae and the Genetic Mechanisms Underpinning This Process. Biology 2025, 14, 1027. https://doi.org/10.3390/biology14081027
Xu X, Yan W, Guo J, Dai D, Li L, Zhang H. The Development of Horns in Bovidae and the Genetic Mechanisms Underpinning This Process. Biology. 2025; 14(8):1027. https://doi.org/10.3390/biology14081027
Chicago/Turabian StyleXu, Xiaoli, Wenwen Yan, Jiazhong Guo, Dinghui Dai, Li Li, and Hongping Zhang. 2025. "The Development of Horns in Bovidae and the Genetic Mechanisms Underpinning This Process" Biology 14, no. 8: 1027. https://doi.org/10.3390/biology14081027
APA StyleXu, X., Yan, W., Guo, J., Dai, D., Li, L., & Zhang, H. (2025). The Development of Horns in Bovidae and the Genetic Mechanisms Underpinning This Process. Biology, 14(8), 1027. https://doi.org/10.3390/biology14081027






