Myricetin Potentiates Antibiotics Against Resistant Pseudomonas aeruginosa by Disrupting Biofilm Formation and Inhibiting Motility Through FimX-Mediated c-di-GMP Signaling Interference
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Bacterial Strains
2.2. In Vitro Antibacterial Assay
2.2.1. Minimum Inhibitory Concentration (MIC)
2.2.2. Collaborative Research
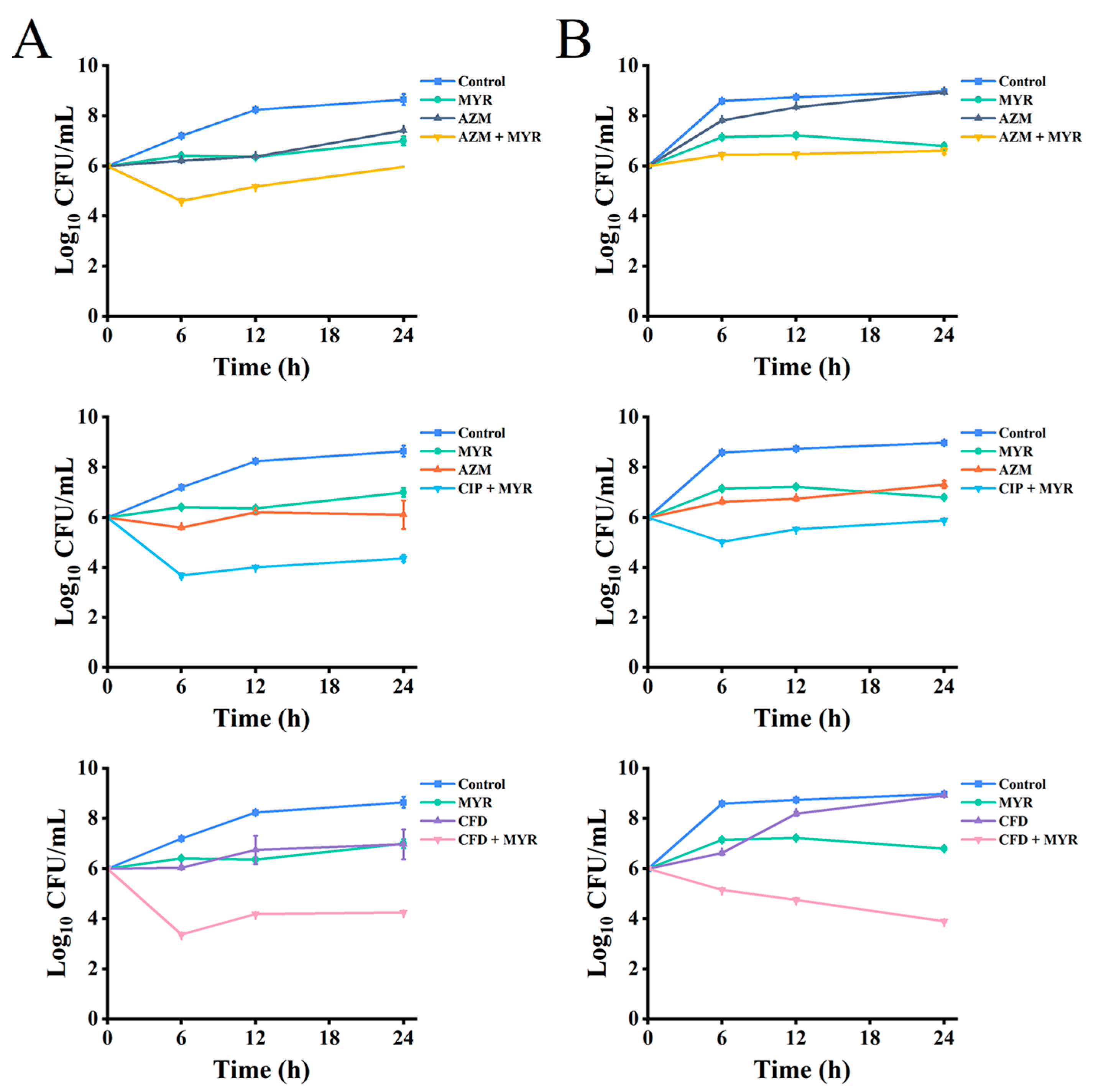
2.2.3. Time-Kill Curves
2.3. Determination of Anti-Biofilm Activity of Myricetin Combined with Antibiotics Against P. aeruginosa
2.3.1. Quantitative Detection of Resistance to Biofilm Formation
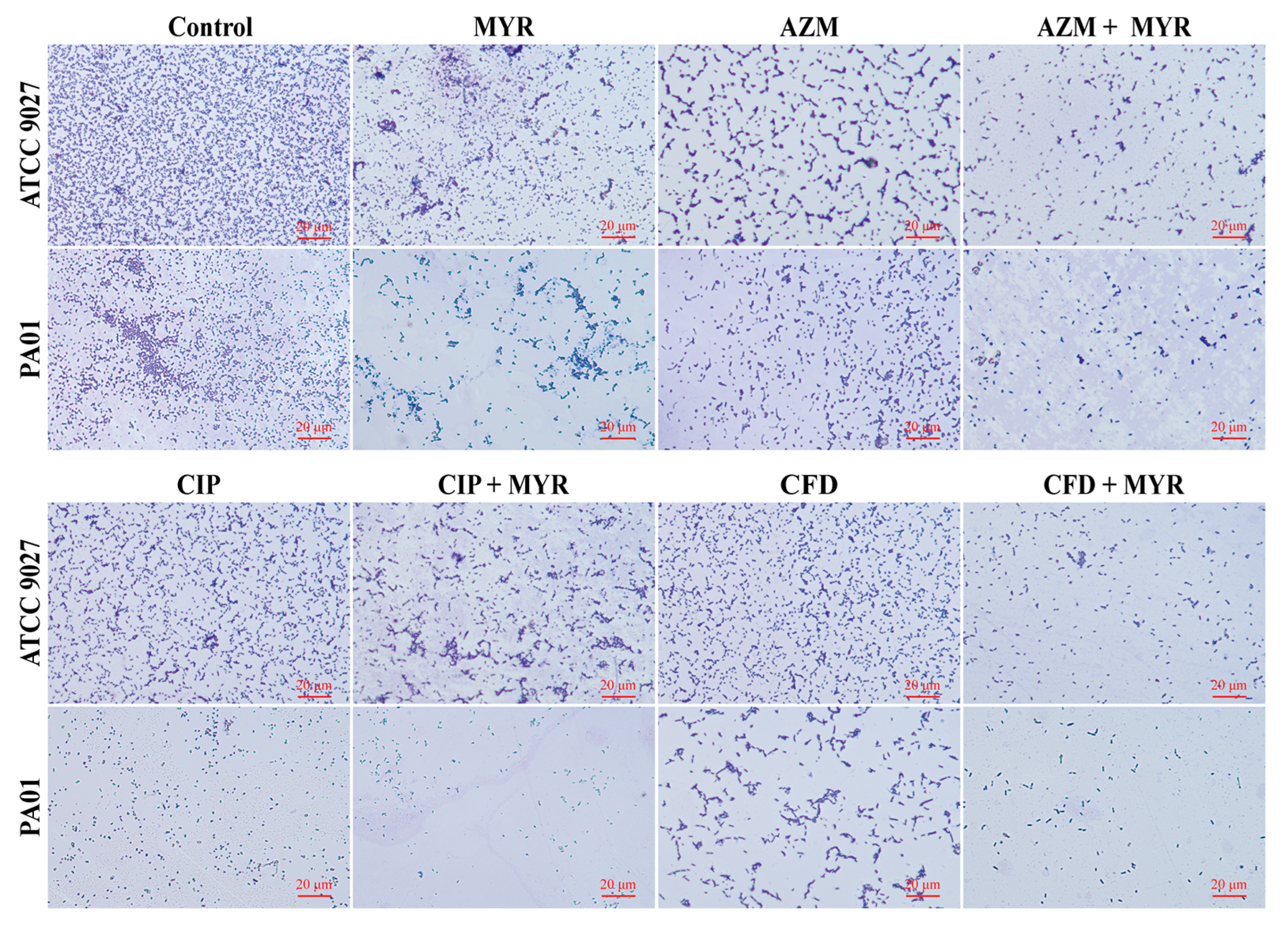
2.3.2. Morphological Analysis of Biofilm Development via Light Microscopy
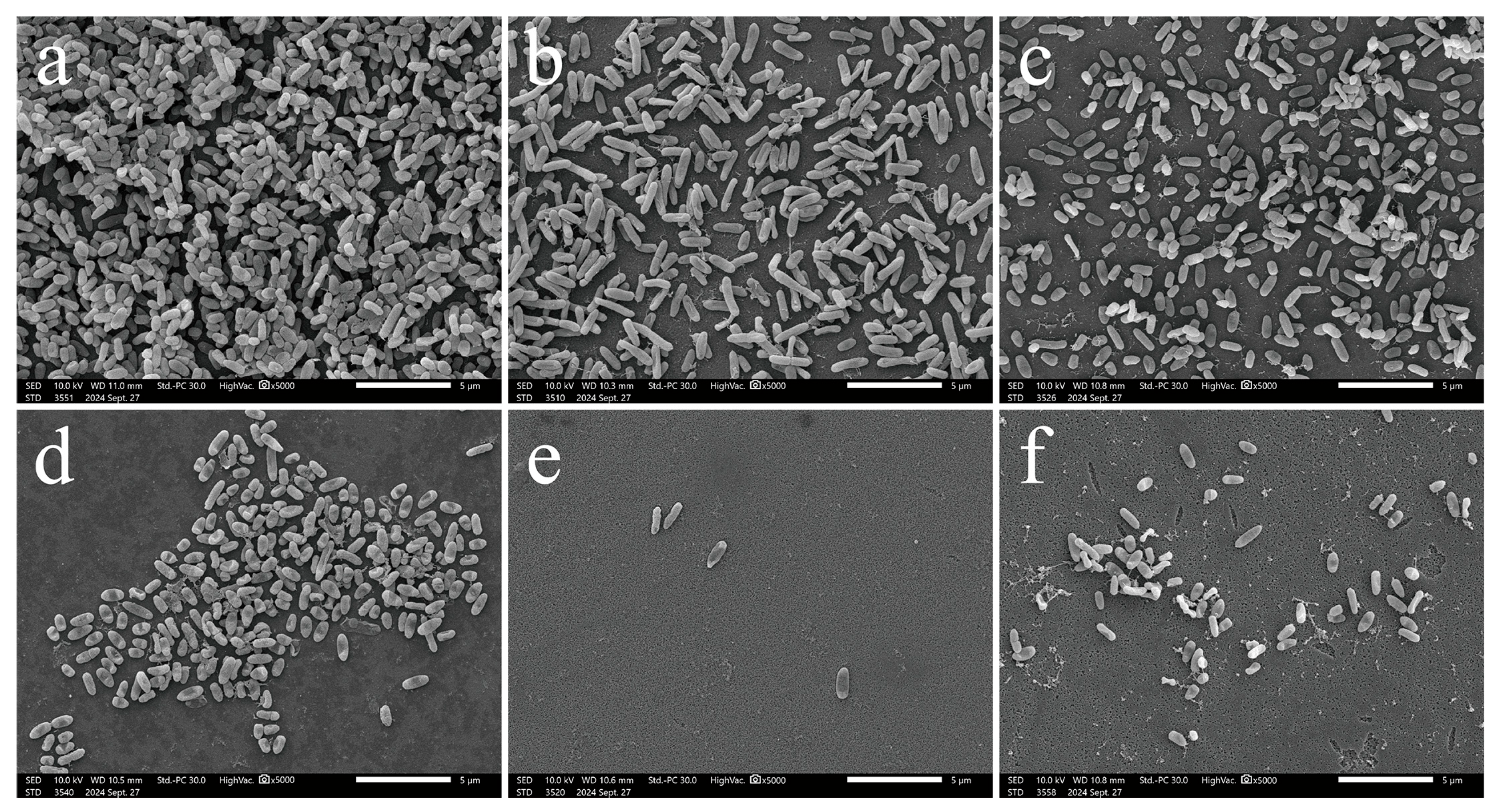
2.3.3. Biofilm Adherence Evaluated by Scanning Electron Microscopy (SEM)
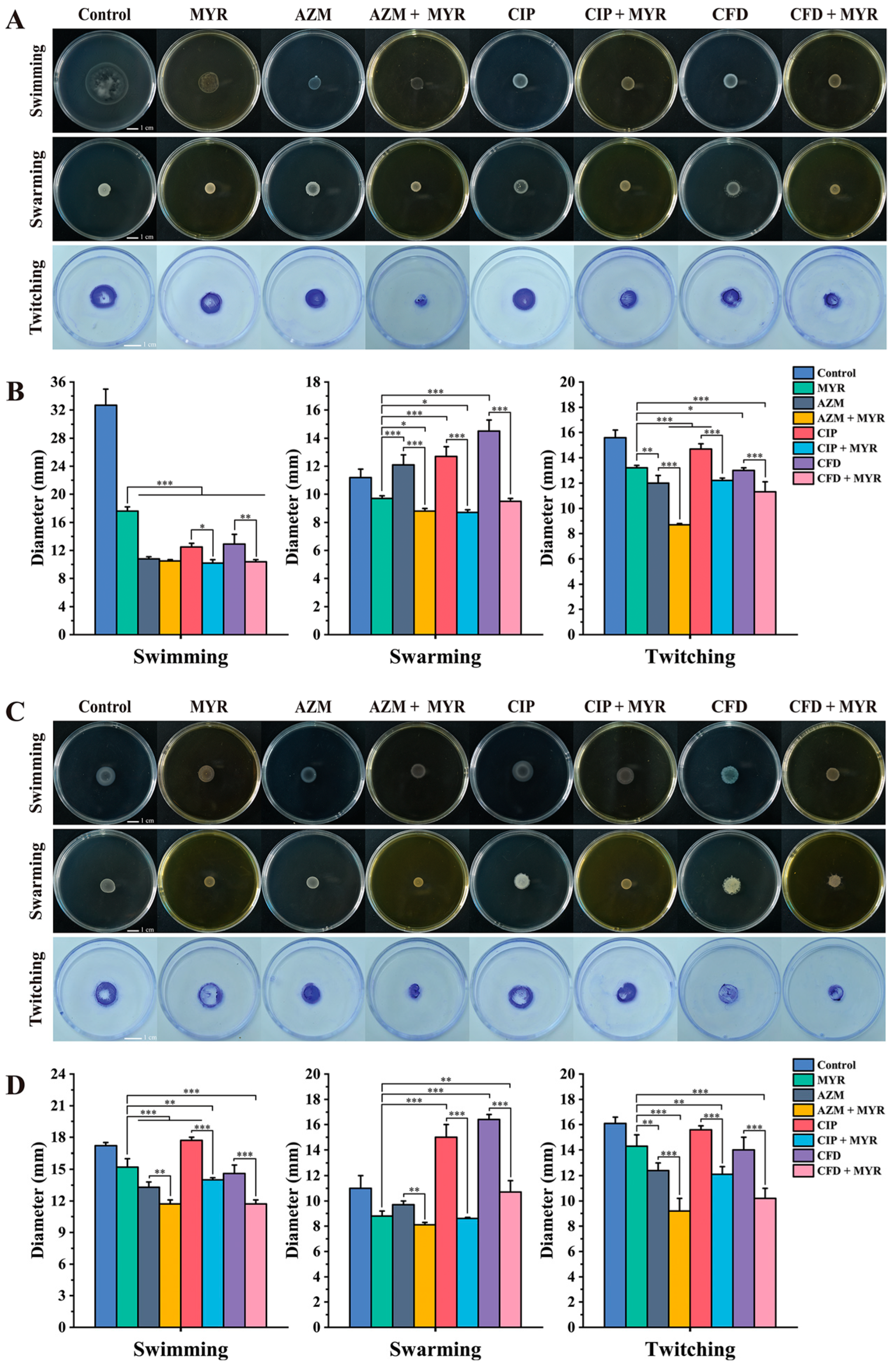
2.4. Assessing the Impact of Myricetin Combined with Antibiotics on the Motility of P. aeruginosa
2.5. Myricetin Combined with Antibiotics for the Extraction and Quantification of Intracellular c-di-GMP in P. aeruginosa
2.6. Statistical Analysis
3. Results
3.1. Antibacterial Activity of Myricetin and Antibiotics
3.2. Synergistic Activity of Myricetin Combined with Antibiotics Against P. aeruginosa
3.3. Synergistic Anti-Biofilm Activity of Myricetin Combined with Antibiotics Against P. aeruginosa
3.4. Microscopic in Situ Visualization of Biofilm Formation by P. aeruginosa
3.5. The Combination of Myricetin and Antibiotics Effectively Restricts the Motility of P. aeruginosa
3.6. Myricetin Synergistically Inhibits the Synthesis of Intracellular c-di-GMP in Combination with Antibiotics
3.7. The Potential Mechanism of Action of Myricetin Targeting FimXEAL via c-di-GMP Signaling Disruption
3.7.1. Significant Reduction in FimXEAL Flexibility due to Ligand Binding
3.7.2. The Binding Mechanism of FimXEAL and Two Molecules (c-di-GMP and Myricetin)
3.7.3. The Ligand Binding Enhances the Internal Coupling of the FimXEAL Protein
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gajdács, M.; Baráth, Z.; Kárpáti, K.; Szabó, D.; Usai, D.; Zanetti, S.; Donadu, M.G. No Correlation between Biofilm Formation, Virulence Factors, and Antibiotic Resistance in Pseudomonas aeruginosa: Results from a Laboratory-Based In Vitro Study. Antibiotics 2021, 10, 1134. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, T.S.; Behrouz, B.; Mousavi Gargari, S.L. Polyclonal Anti-Whole Cell IgY Passive Immunotherapy Shields against P. aeruginosa-Induced Acute Pneumonia and Burn Wound Infections in Murine Models. Sci. Rep. 2024, 14, 405. [Google Scholar] [CrossRef] [PubMed]
- Das, M.C.; Sandhu, P.; Gupta, P.; Rudrapaul, P.; De, U.C.; Tribedi, P.; Akhter, Y.; Bhattacharjee, S. Attenuation of Pseudomonas aeruginosa Biofilm Formation by Vitexin: A Combinatorial Study with Azithromycin and Gentamicin. Sci. Rep. 2016, 6, 23347. [Google Scholar] [CrossRef] [PubMed]
- Elfadadny, A.; Ragab, R.F.; AlHarbi, M.; Badshah, F.; Ibáñez-Arancibia, E.; Farag, A.; Hendawy, A.O.; De Los Ríos-Escalante, P.R.; Aboubakr, M.; Zakai, S.A.; et al. Antimicrobial Resistance of Pseudomonas aeruginosa: Navigating Clinical Impacts, Current Resistance Trends, and Innovations in Breaking Therapies. Front. Microbiol. 2024, 15, 1374466. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Xiao, W.; Zhou, C.; Pu, Q.; Deng, X.; Lan, L.; Liang, H.; Song, X.; Wu, M. Pseudomonas aeruginosa: Pathogenesis, Virulence Factors, Antibiotic Resistance, Interaction with Host, Technology Advances and Emerging Therapeutics. Signal Transduct. Target. Ther. 2022, 7, 199. [Google Scholar] [CrossRef] [PubMed]
- Litwin, A.; Rojek, S.; Gozdzik, W.; Duszynska, W. Pseudomonas aeruginosa Device Associated—Healthcare Associated Infections and Its Multidrug Resistance at Intensive Care Unit of University Hospital: Polish, 8.5-Year, Prospective, Single-Centre Study. BMC Infect. Dis. 2021, 21, 180. [Google Scholar] [CrossRef] [PubMed]
- Olivares, E.; Badel-Berchoux, S.; Provot, C.; Prévost, G.; Bernardi, T.; Jehl, F. Clinical Impact of Antibiotics for the Treatment of Pseudomonas aeruginosa Biofilm Infections. Front. Microbiol. 2019, 10, 2894. [Google Scholar] [CrossRef] [PubMed]
- Haidar, A.; Muazzam, A.; Nadeem, A.; Atique, R.; Saeed, H.A.; Naveed, A.; Sharif, J.; Perveen, A.; Fatima, H.R.; Samad, A. Biofilm Formation and Antibiotic Resistance in Pseudomonas aeruginosa. Microbe 2024, 3, 100078. [Google Scholar] [CrossRef]
- Baker, A.E.; Webster, S.S.; Diepold, A.; Kuchma, S.L.; Bordeleau, E.; Armitage, J.P.; O’Toole, G.A. Flagellar Stators Stimulate C-Di-GMP Production by Pseudomonas aeruginosa. J. Bacteriol. 2019, 201, e00741-18. [Google Scholar] [CrossRef] [PubMed]
- Jenal, U.; Reinders, A.; Lori, C. Cyclic Di-GMP: Second Messenger Extraordinaire. Nat. Rev. Microbiol. 2017, 15, 271–284. [Google Scholar] [CrossRef] [PubMed]
- Conrad, J.C.; Gibiansky, M.L.; Jin, F.; Gordon, V.D.; Motto, D.A.; Mathewson, M.A.; Stopka, W.G.; Zelasko, D.C.; Shrout, J.D.; Wong, G.C.L. Flagella and Pili-Mediated near-Surface Single-Cell Motility Mechanisms in P. aeruginosa. Biophys. J. 2011, 100, 1608–1616. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, G.A.; Kolter, R. Flagellar and Twitching Motility Are Necessary for Pseudomonas aeruginosa Biofilm Development. Mol. Microbiol. 1998, 30, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Schniederberend, M.; Williams, J.F.; Shine, E.; Shen, C.; Jain, R.; Emonet, T.; Kazmierczak, B.I. Modulation of Flagellar Rotation in Surface-Attached Bacteria: A Pathway for Rapid Surface-Sensing after Flagellar Attachment. PLoS Pathog. 2019, 15, e1008149. [Google Scholar] [CrossRef] [PubMed]
- Lichtenberg, M.; Kragh, K.N.; Fritz, B.; Kirkegaard, J.B.; Tolker-Nielsen, T.; Bjarnsholt, T. Cyclic-Di-GMP Signaling Controls Metabolic Activity in Pseudomonas aeruginosa. Cell Rep. 2022, 41, 111515. [Google Scholar] [CrossRef] [PubMed]
- Ha, D.-G.; O’Toole, G.A. C-Di-GMP and Its Effects on Biofilm Formation and Dispersion: A Pseudomonas aeruginosa Review. Microbiol. Spectr. 2015, 3, MB-0003-2014. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Cao, B.; Yang, L.; Gu, J.-D. Biofilm Control by Interfering with C-Di-GMP Metabolism and Signaling. Biotechnol. Adv. 2022, 56, 107915. [Google Scholar] [CrossRef] [PubMed]
- Navarro, M.V.A.S.; De, N.; Bae, N.; Wang, Q.; Sondermann, H. Structural Analysis of the GGDEF-EAL Domain-Containing c-Di-GMP Receptor FimX. Structure 2009, 17, 1104–1116. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Stock, A.M. Catalytically Incompetent by Design. Structure 2009, 17, 1038–1040. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.; Sliusarenko, O.; Kazmierczak, B.I. Interaction of the Cyclic-Di-GMP Binding Protein FimX and the Type 4 Pilus Assembly ATPase Promotes Pilus Assembly. PLoS Pathog. 2017, 13, e1006594. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.; Sahoo, P.K.; Sheenu; Adhikary, A.; Ruhal, R.; Jain, D. Molecular and Structural Facets of C-Di-GMP Signalling Associated with Biofilm Formation in Pseudomonas aeruginosa. Mol. Asp. Med. 2021, 81, 101001. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Chuah, M.L.C.; Dong, X.; Xie, K.; Luo, Z.; Tang, K.; Liang, Z.-X. Binding of Cyclic Diguanylate in the Non-Catalytic EAL Domain of FimX Induces a Long-Range Conformational Change. J. Biol. Chem. 2011, 286, 2910–2917. [Google Scholar] [CrossRef] [PubMed]
- Shariati, A.; Noei, M.; Askarinia, M.; Khoshbayan, A.; Farahani, A.; Chegini, Z. Inhibitory Effect of Natural Compounds on Quorum Sensing System in Pseudomonas aeruginosa: A Helpful Promise for Managing Biofilm Community. Front. Pharmacol. 2024, 15, 1350391. [Google Scholar] [CrossRef] [PubMed]
- Rasamiravaka, T.; Labtani, Q.; Duez, P.; El Jaziri, M. The Formation of Biofilms by Pseudomonas aeruginosa: A Review of the Natural and Synthetic Compounds Interfering with Control Mechanisms. BioMed Res. Int. 2015, 2015, 759348. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jiao, F.; Zeng, D.; Yu, X.; Zhou, Y.; Xue, J.; Yang, W.; Guo, J. Synergistic Effects of Pyrrosia Lingua Caffeoylquinic Acid Compounds with Levofloxacin Against Uropathogenic Escherichia Coli: Insights from Molecular Dynamics Simulations, Antibiofilm, and Antimicrobial Assessments. Molecules 2024, 29, 5679. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Han, W.; Gu, J.; Qiu, C.; Jiang, Q.; Dong, J.; Lei, L.; Li, F. Recent Advances on the Regulation of Bacterial Biofilm Formation by Herbal Medicines. Front. Microbiol. 2022, 13, 1039297. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Huang, Y.; Zuo, X.; Li, Z.; Zhang, L.; Tang, J.; Lu, L.; Huang, Y. Exploring the Molecular Mechanism of Action of Polygonum Capitatum Buch-Ham. Ex D. Don for the Treatment of Bacterial Prostatitis Based on Network Pharmacology and Experimental Verification. J. Ethnopharmacol. 2022, 291, 115007. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; He, L.; Chen, X.-J.; Zhang, X.; Yan, X.-L.; Tu, B.; Zeng, Z.; He, M.-H. Polygonum Capitatum, the Hmong Medicinal Flora: A Comprehensive Review of Its Phytochemical, Pharmacological and Pharmacokinetic Characteristics. Molecules 2022, 27, 6407. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Tan, S.; Su, H.; Wei, L.; Chen, W.; Yuan, D.; Yan, B.; Sun, F.; Li, C.; Jiang, K.; et al. The Extract of Polygonum capitatum Alleviates Hyperoxaluria-Induced Oxidative Stress by Targeting the PI3K-ERK-NRF2 Signaling Pathway. Food Sci. Hum. Wellness 2025, 1–28. [Google Scholar] [CrossRef]
- Gupta, G.; Siddiqui, M.A.; Khan, M.M.; Ajmal, M.; Ahsan, R.; Rahaman, M.A.; Ahmad, M.A.; Arshad, M.; Khushtar, M. Current Pharmacological Trends on Myricetin. Drug Res. 2020, 70, 448–454. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Tan, L.; Wang, M.; Ren, C.; Guo, C.; Yang, B.; Ren, Y.; Cao, Z.; Li, Y.; Pei, J. Myricetin: A Review of the Most Recent Research. Biomed. Pharmacother. 2021, 134, 111017. [Google Scholar] [CrossRef] [PubMed]
- Agraharam, G.; Girigoswami, A.; Girigoswami, K. Myricetin: A Multifunctional Flavonol in Biomedicine. Curr. Pharmacol. Rep. 2022, 8, 48–61. [Google Scholar] [CrossRef] [PubMed]
- Krzyżek, P.; Migdał, P.; Paluch, E.; Karwańska, M.; Wieliczko, A.; Gościniak, G. Myricetin as an Antivirulence Compound Interfering with a Morphological Transformation into Coccoid Forms and Potentiating Activity of Antibiotics against Helicobacter Pylori. Int. J. Mol. Sci. 2021, 22, 2695. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, M.P.; Lewis, J.S. The Clinical and Laboratory Standards Institute Subcommittee on Antimicrobial Susceptibility Testing: Background, Organization, Functions, and Processes. J. Clin. Microbiol. 2020, 58, e01864-19. [Google Scholar] [CrossRef] [PubMed]
- Yi, K.; Liu, S.; Liu, P.; Luo, X.; Zhao, J.; Yan, F.; Pan, Y.; Liu, J.; Zhai, Y.; Hu, G. Synergistic Antibacterial Activity of Tetrandrine Combined with Colistin against MCR-Mediated Colistin-Resistant Salmonella. Biomed. Pharmacother. 2022, 149, 112873. [Google Scholar] [CrossRef] [PubMed]
- López-Gavín, A.; Tudó, G.; Rey-Jurado, E.; Vergara, A.; Hurtado, J.C.; Gonzalez-Martín, J. In Vitro Time-Kill Curves Study of Three Antituberculous Combinations against Mycobacterium Tuberculosis Clinical Isolates. Int. J. Antimicrob. Agents 2016, 47, 97–100. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Hao, S.; Zhao, L.; Shi, F.; Ye, G.; Zou, Y.; Song, X.; Li, L.; Yin, Z.; He, X.; et al. Paeonol Attenuates Quorum-Sensing Regulated Virulence and Biofilm Formation in Pseudomonas aeruginosa. Front. Microbiol. 2021, 12, 692474. [Google Scholar] [CrossRef] [PubMed]
- Sathiya Deepika, M.; Thangam, R.; Sakthidhasan, P.; Arun, S.; Sivasubramanian, S.; Thirumurugan, R. Combined Effect of a Natural Flavonoid Rutin from Citrus Sinensis and Conventional Antibiotic Gentamicin on Pseudomonas aeruginosa Biofilm Formation. Food Control 2018, 90, 282–294. [Google Scholar] [CrossRef]
- Wang, S.; Feng, Y.; Han, X.; Cai, X.; Yang, L.; Liu, C.; Shen, L. Inhibition of Virulence Factors and Biofilm Formation by Wogonin Attenuates Pathogenicity of Pseudomonas aeruginosa PAO1 via Targeting Pqs Quorum-Sensing System. Int. J. Mol. Sci. 2021, 22, 12699. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Ye, L.; Li, D.; Lin, S.; Deng, W.; Zhang, L.; Liang, J.; Li, J.; Wei, Q.; Wang, K. Effects of Daphnetin on Biofilm Formation and Motility of Pseudomonas aeruginosa. Front. Cell Infect. Microbiol. 2022, 12, 1033540. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, W.; Cui, H.; Qin, S. Phylogenetic Analysis and Characterization of Diguanylate Cyclase and Phosphodiesterase in Planktonic Filamentous Cyanobacterium Arthrospira Sp. Int. J. Mol. Sci. 2023, 24, 15210. [Google Scholar] [CrossRef] [PubMed]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA Methods to Estimate Ligand-Binding Affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Tuon, F.F.; Dantas, L.R.; Suss, P.H.; Tasca Ribeiro, V.S. Pathogenesis of the Pseudomonas aeruginosa Biofilm: A Review. Pathogens 2022, 11, 300. [Google Scholar] [CrossRef] [PubMed]
- Barman, S.; Kurnaz, L.B.; Leighton, R.; Hossain, M.W.; Decho, A.W.; Tang, C. Intrinsic Antimicrobial Resistance: Molecular Biomaterials to Combat Microbial Biofilms and Bacterial Persisters. Biomaterials 2024, 311, 122690. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.; Pham, D.T.N.; Oloketuyi, S.F.; Kim, Y.-M. Regulation and Controlling the Motility Properties of Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 2020, 104, 33–49. [Google Scholar] [CrossRef] [PubMed]
- Jayaraman, P.; Sakharkar, M.K.; Lim, C.S.; Tang, T.H.; Sakharkar, K.R. Activity and Interactions of Antibiotic and Phytochemical Combinations against Pseudomonas aeruginosa in Vitro. Int. J. Biol. Sci. 2010, 6, 556–568. [Google Scholar] [CrossRef] [PubMed]
- Hanci, H.; Igan, H. Antimicrobial Synergistic Effects of Apigenin, (-)-Epigallocatechin-3-Gallate, Myricetin and Luteolin in Combination with Some Antibiotics. Ann. Agric. Environ. Med. 2023, 30, 61–64. [Google Scholar] [CrossRef] [PubMed]
- Pruteanu, M.; Hernández Lobato, J.I.; Stach, T.; Hengge, R. Common Plant Flavonoids Prevent the Assembly of Amyloid Curli Fibres and Can Interfere with Bacterial Biofilm Formation. Environ. Microbiol. 2020, 22, 5280–5299. [Google Scholar] [CrossRef] [PubMed]
- Pinto, H.B.; Brust, F.R.; Macedo, A.J.; Trentin, D.S. The Antivirulence Compound Myricetin Possesses Remarkable Synergistic Effect with Antibacterials upon Multidrug Resistant Staphylococcus Aureus. Microb. Pathog. 2020, 149, 104571. [Google Scholar] [CrossRef] [PubMed]
- Topa, S.H.; Subramoni, S.; Palombo, E.A.; Kingshott, P.; Rice, S.A.; Blackall, L.L. Cinnamaldehyde Disrupts Biofilm Formation and Swarming Motility of Pseudomonas aeruginosa. Microbiol. 2018, 164, 1087–1097. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Zhang, K.; Wang, C.; Fan, Q.; Tang, X.; Zhang, X.; Wang, K.; Fu, Y.; Liang, H. A C-Di-GMP Signaling Module Controls Responses to Iron in Pseudomonas aeruginosa. Nat. Commun. 2024, 15, 1860. [Google Scholar] [CrossRef] [PubMed]
- Merighi, M.; Lee, V.T.; Hyodo, M.; Hayakawa, Y.; Lory, S. The Second Messenger Bis-(3′-5′)-Cyclic-GMP and Its PilZ Domain-Containing Receptor Alg44 Are Required for Alginate Biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 2007, 65, 876–895. [Google Scholar] [CrossRef] [PubMed]
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Cheatham, T.E.; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Ryckaert, J.-P.; Ciccotti, G.; Berendsen, H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Matsuyama, B.Y.; Krasteva, P.V.; Baraquet, C.; Harwood, C.S.; Sondermann, H.; Navarro, M.V.A.S. Mechanistic insights into c-di-GMP–dependent control of the biofilm regulator FleQ from Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2015, 113, E209–E218. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; He, Q.; Yin, J.; Xu, S.; Hu, W.; Gu, L. BrlR from Pseudomonas aeruginosa is a receptor for both cyclic di-GMP and pyocyanin. Nat. Commun. 2018, 9, 2563. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein−Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [PubMed]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Luo, L.-S. Theory of the lattice Boltzmann method: From the Boltzmann equation to the lattice Boltzmann equation. Phys. Rev. E 1997, 56, 6811–6817. [Google Scholar] [CrossRef]
- Eargle, J.; Luthey-Schulten, Z. NetworkView: 3D display and analysis of protein·RNA interaction networks. Bioinformatics 2012, 28, 3000–3001. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]


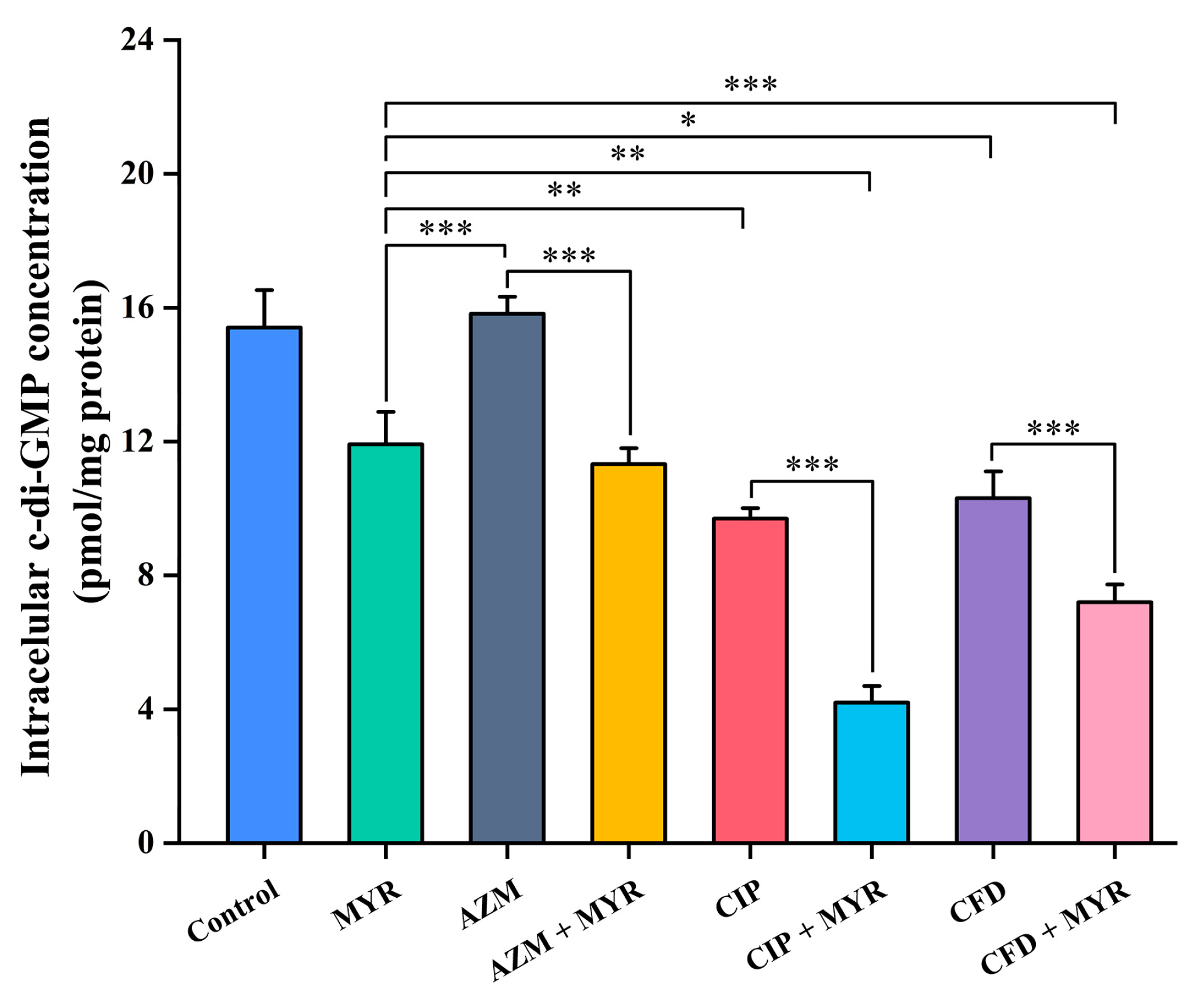

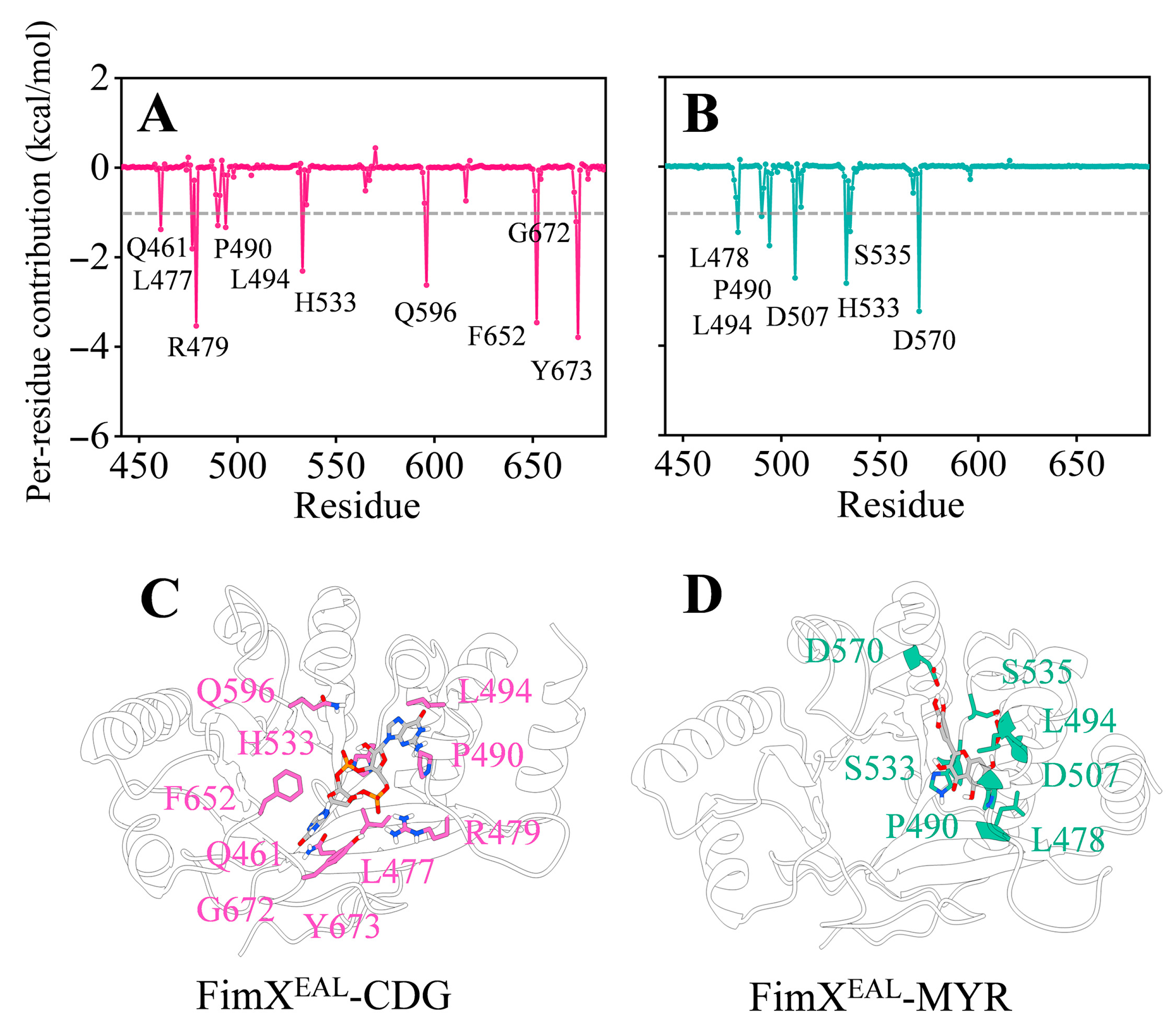
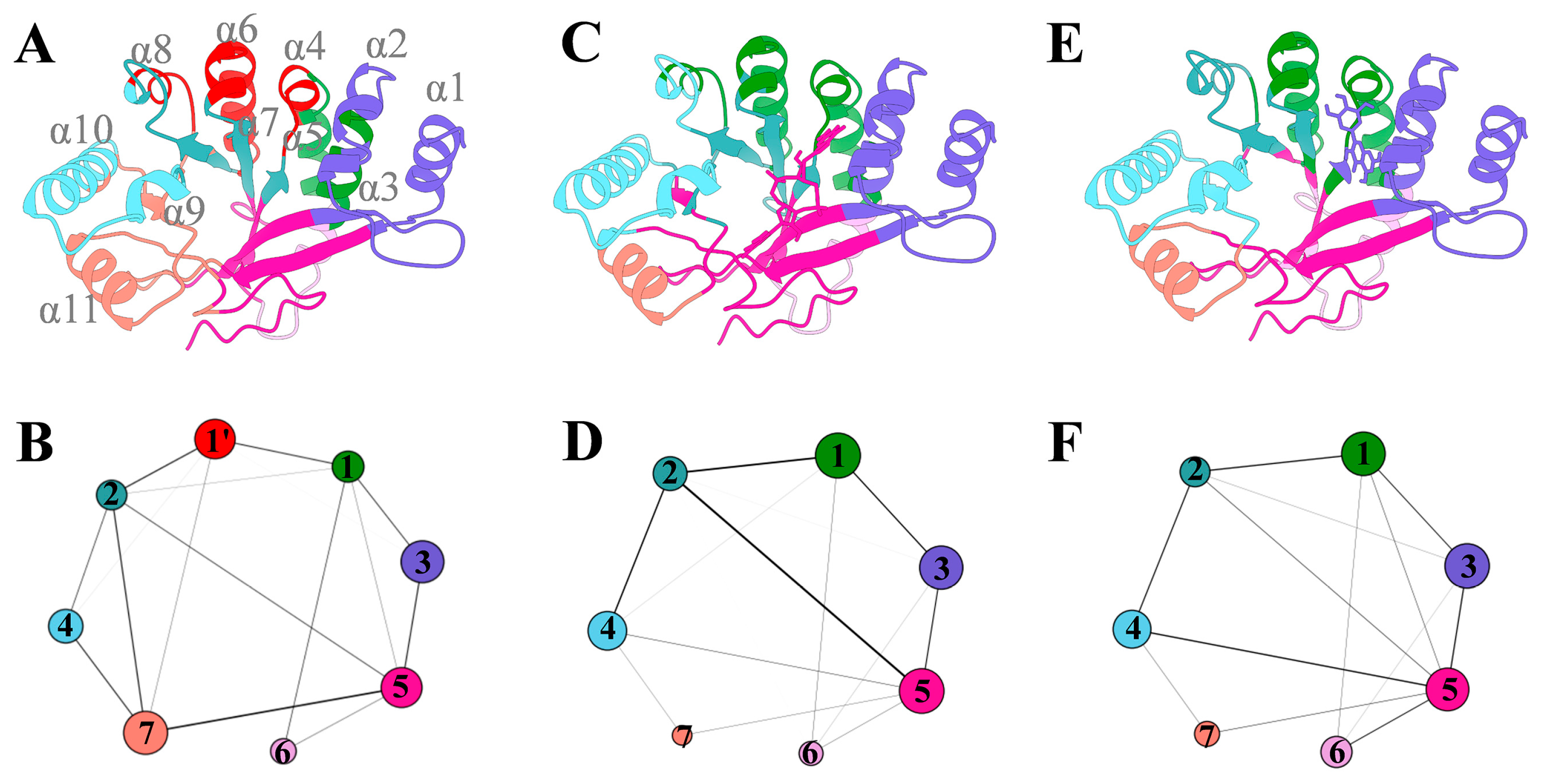
| Strains | Myricetin | Azithromycin | Ciprofloxacin | Cefdinir | |
|---|---|---|---|---|---|
| P. aeruginosa | ATCC 9027 | 512 | 32 | 32 | 256 |
| PA01 | 512 | 128 | 128 | 1024 | |
| PA02 | 512 | 256 | 256 | >1024 | |
| PA03 | 512 | 256 | 256 | 256 | |
| PA04 | 1024 | 512 | 256 | >1024 | |
| E. coli | CICC 10389 | 256 | 128 | 0.5 | 128 |
| EC01 | 1024 | 32 | 32 | 1024 | |
| EC02 | 512 | 128 | 32 | 256 | |
| EC03 | 1024 | 256 | 128 | 0.5 | |
| EC04 | 1024 | 8 | 32 | 16 | |
| EC05 | 1024 | 4 | 0.5 | 256 | |
| EC06 | 1024 | 64 | 512 | 256 | |
| EC07 | 256 | 8 | 0.5 | 256 | |
| E. faecalis | ATCC 19433 | 64 | 128 | 0.5 | 0.5 |
| EF01 | 1024 | >1024 | 64 | >1024 | |
| EF02 | 1024 | 1024 | 256 | 128 | |
| EF03 | 512 | 8 | 2 | 16 | |
| EF04 | 1024 | 64 | 128 | 16 | |
| EF05 | 1024 | 512 | 64 | 0.5 | |
| Compounds | MYR | AZM | CIP | CFD | MYR + AZM | MYR + CIP | MYR + CFD |
|---|---|---|---|---|---|---|---|
| Inhibition Zone (mm) | - | - | 26.0 ± 0.4 | 22.8 ± 0.6 | 13.7 ± 0.7 ** | 30.5 ± 0.9 ** | 24.9 ± 0.4 ** |
| Strains | MIC (μg/L) in Combination | Potentiation a | FICI | Inf | MIC (μg/L) in Combination | Potentiation | FICI | Inf | MIC (μg/L) in Combination | Potentiation | FICI | Inf | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MYR | AZM | MYR | CIP | MYR | CFD | ||||||||||
| ATCC 9027 | 128 | 4 | 8-fold | 0.375 | SN | 256 | 16 | 2-fold | 1 | AD | 256 | 64 | 4-fold | 0.75 | PS |
| PA01 | 32 | 8 | 16-fold | 0.125 | SN | 16 | 32 | 4-fold | 0.28125 | SN | 64 | 32 | 32-fold | 0.15625 | SN |
| PA02 | 256 | 32 | 8-fold | 0.625 | PS | 256 | 256 | 1-fold | 1.5 | AN | 256 | 64 | 16-fold | 0.5625 | PS |
| PA03 | 256 | 16 | 16-fold | 0.5625 | PS | 512 | 128 | 2-fold | 1.5 | AN | 256 | 64 | 4-fold | 0.75 | PS |
| PA04 | 256 | 16 | 32-fold | 0.28125 | SN | 256 | 16 | 16-fold | 0.3125 | SN | 256 | 64 | 4-fold | 0.5 | SN |
| Contribution | FimXEAL-CDG | FimXEAL-MYR |
|---|---|---|
| ΔEvdw | −51.12 ± 3.08 | −28.13 ± 1.44 |
| ΔEele | 53.34 ± 7.38 | −59.69 ± 0.72 |
| ΔGpol,sol | −35.46 ± 5.70 | 64.19 ± 0.58 |
| ΔGnpol,sol | −6.17 ± 0.38 | −4.62 ± 0.15 |
| ΔEMM | 2.23 ± 8.91 | −87.81 ± 1.87 |
| ΔGsol | −41.63 ± 5.57 | 59.57 ± 0.46 |
| ΔGMM/GBSA | −39.40 ± 3.36 | −28.24 ± 1.61 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, D.; Jiao, F.; Yang, Y.; Dou, S.; Yu, J.; Yu, X.; Zhou, Y.; Xue, J.; Li, X.; Duan, H.; et al. Myricetin Potentiates Antibiotics Against Resistant Pseudomonas aeruginosa by Disrupting Biofilm Formation and Inhibiting Motility Through FimX-Mediated c-di-GMP Signaling Interference. Biology 2025, 14, 859. https://doi.org/10.3390/biology14070859
Zeng D, Jiao F, Yang Y, Dou S, Yu J, Yu X, Zhou Y, Xue J, Li X, Duan H, et al. Myricetin Potentiates Antibiotics Against Resistant Pseudomonas aeruginosa by Disrupting Biofilm Formation and Inhibiting Motility Through FimX-Mediated c-di-GMP Signaling Interference. Biology. 2025; 14(7):859. https://doi.org/10.3390/biology14070859
Chicago/Turabian StyleZeng, Derong, Fangfang Jiao, Yuqi Yang, Shuai Dou, Jiahua Yu, Xiang Yu, Yongqiang Zhou, Juan Xue, Xue Li, Hongliang Duan, and et al. 2025. "Myricetin Potentiates Antibiotics Against Resistant Pseudomonas aeruginosa by Disrupting Biofilm Formation and Inhibiting Motility Through FimX-Mediated c-di-GMP Signaling Interference" Biology 14, no. 7: 859. https://doi.org/10.3390/biology14070859
APA StyleZeng, D., Jiao, F., Yang, Y., Dou, S., Yu, J., Yu, X., Zhou, Y., Xue, J., Li, X., Duan, H., Zhang, Y., Guo, J., & Yang, W. (2025). Myricetin Potentiates Antibiotics Against Resistant Pseudomonas aeruginosa by Disrupting Biofilm Formation and Inhibiting Motility Through FimX-Mediated c-di-GMP Signaling Interference. Biology, 14(7), 859. https://doi.org/10.3390/biology14070859






