Genome-Wide Identification, Evolutionary Expansion, and Expression Analyses of Aux/IAA Gene Family in Castanea mollissima During Seed Kernel Development
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification, Molecular Characteristics, and Phylogenetic Analysis
2.2. Chromosomal Distribution and Collinear Analysis
2.3. GO/KEGG Enrichment, Transcription Factors (TFs) Regulatory and Protein-Protein Interaction (PPI) Network Analysis
2.4. Plant Materials and Phenotypic Determination
2.5. Expression Analysis of CmAux/IAA Genes
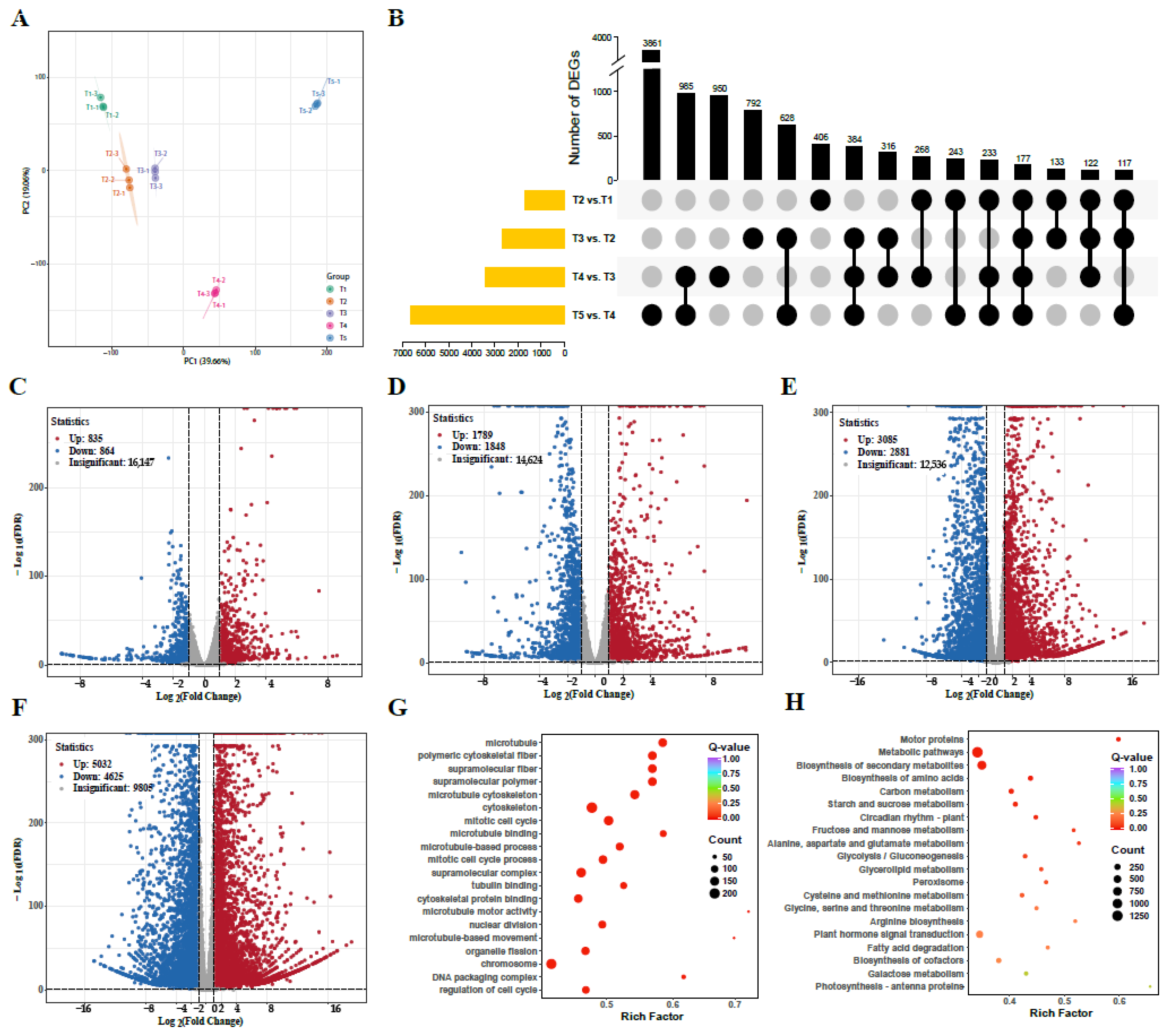
2.6. RNA Sequencing and DEG Analysis
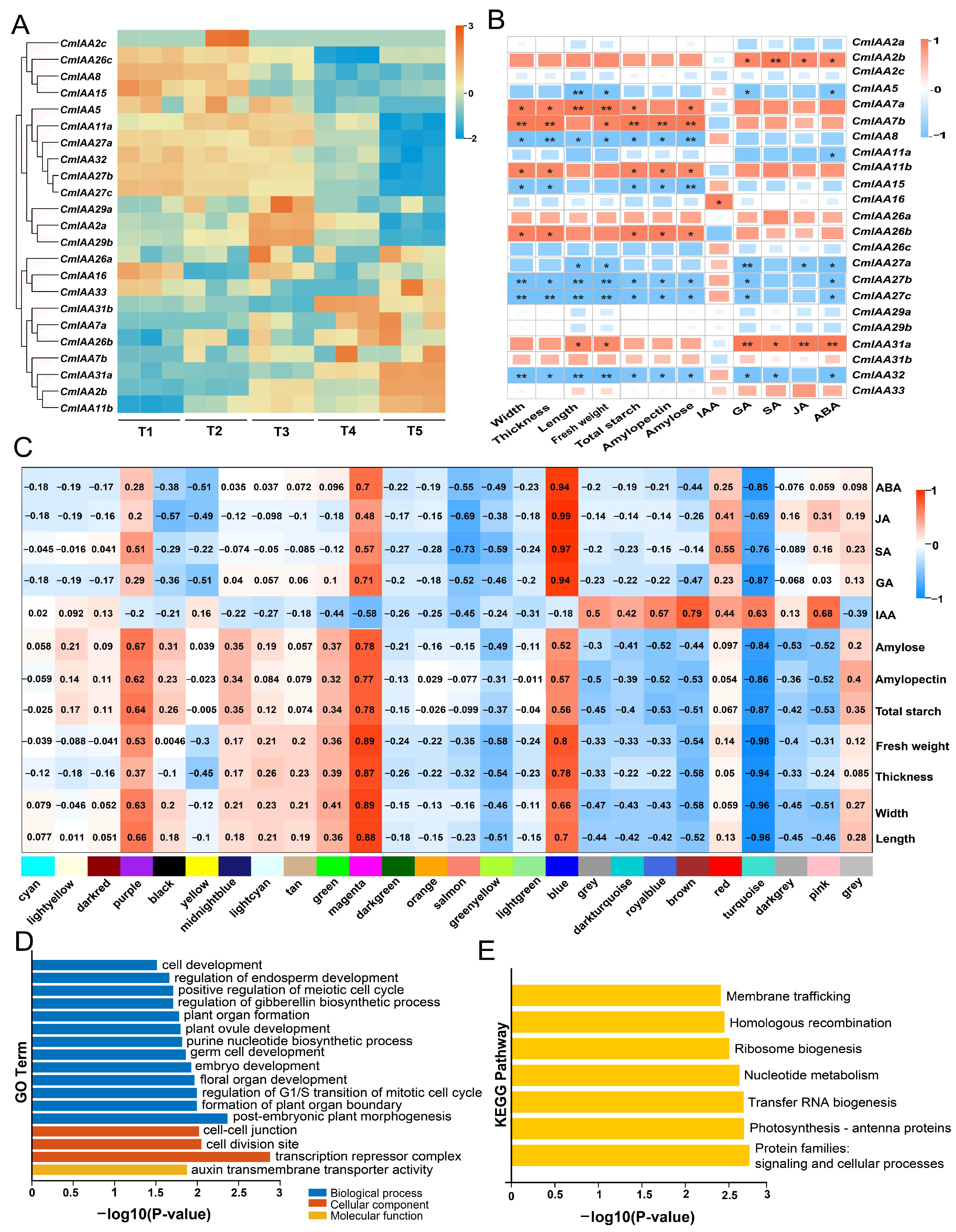
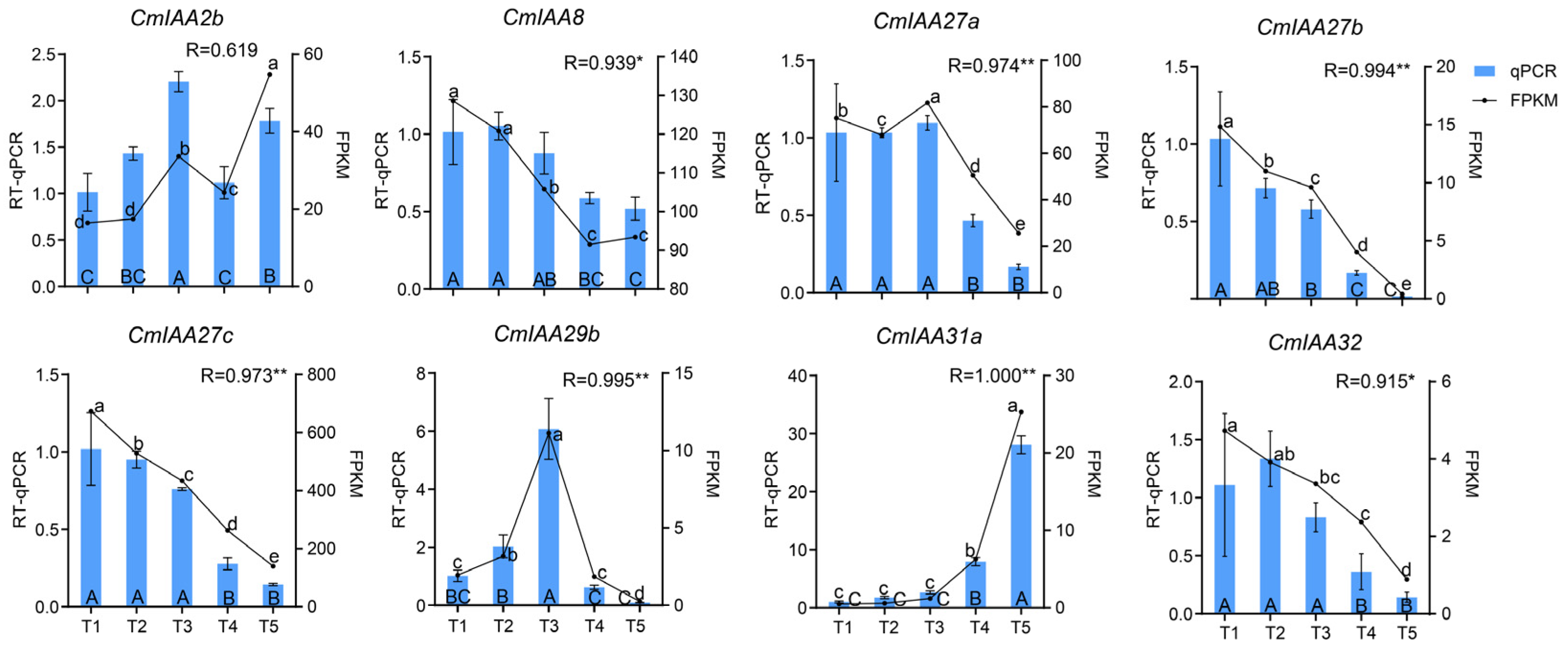
2.7. WGCNA Analysis and RT-qPCR Validation
2.8. Statistical Analysis
3. Results
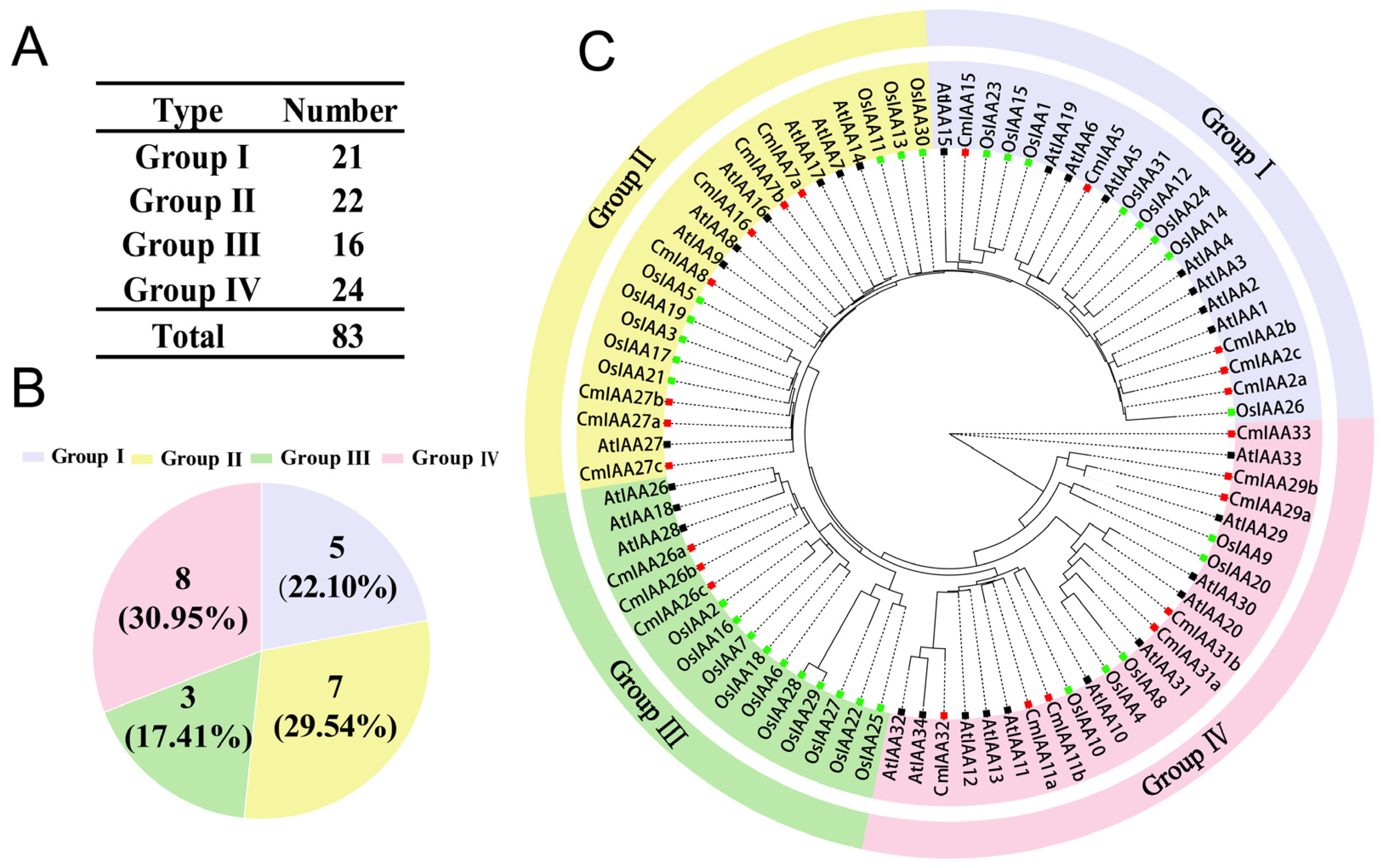
3.1. Identification and Phylogenetic Analysis
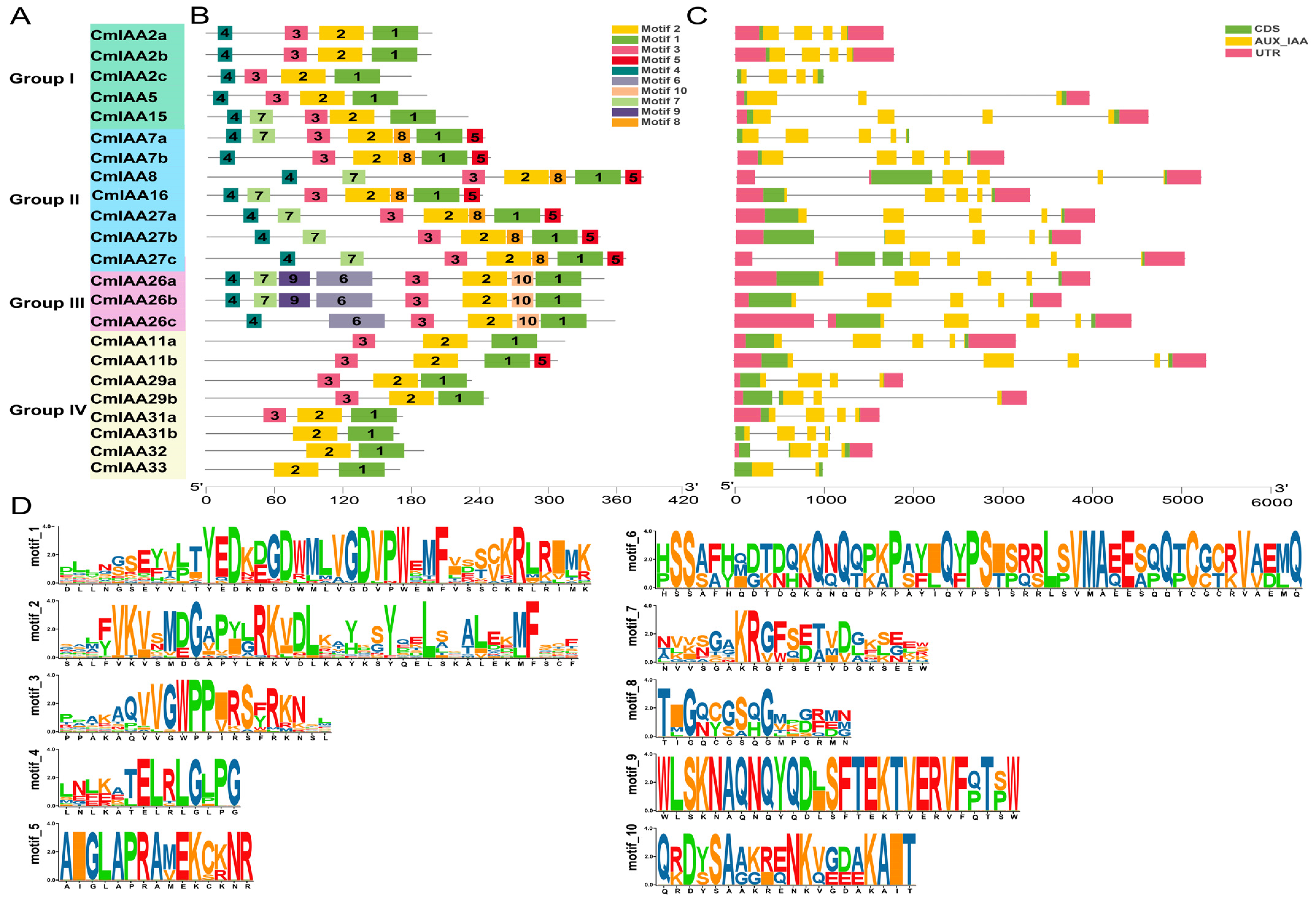
3.2. Gene Structure and Conserved Motif Analysis
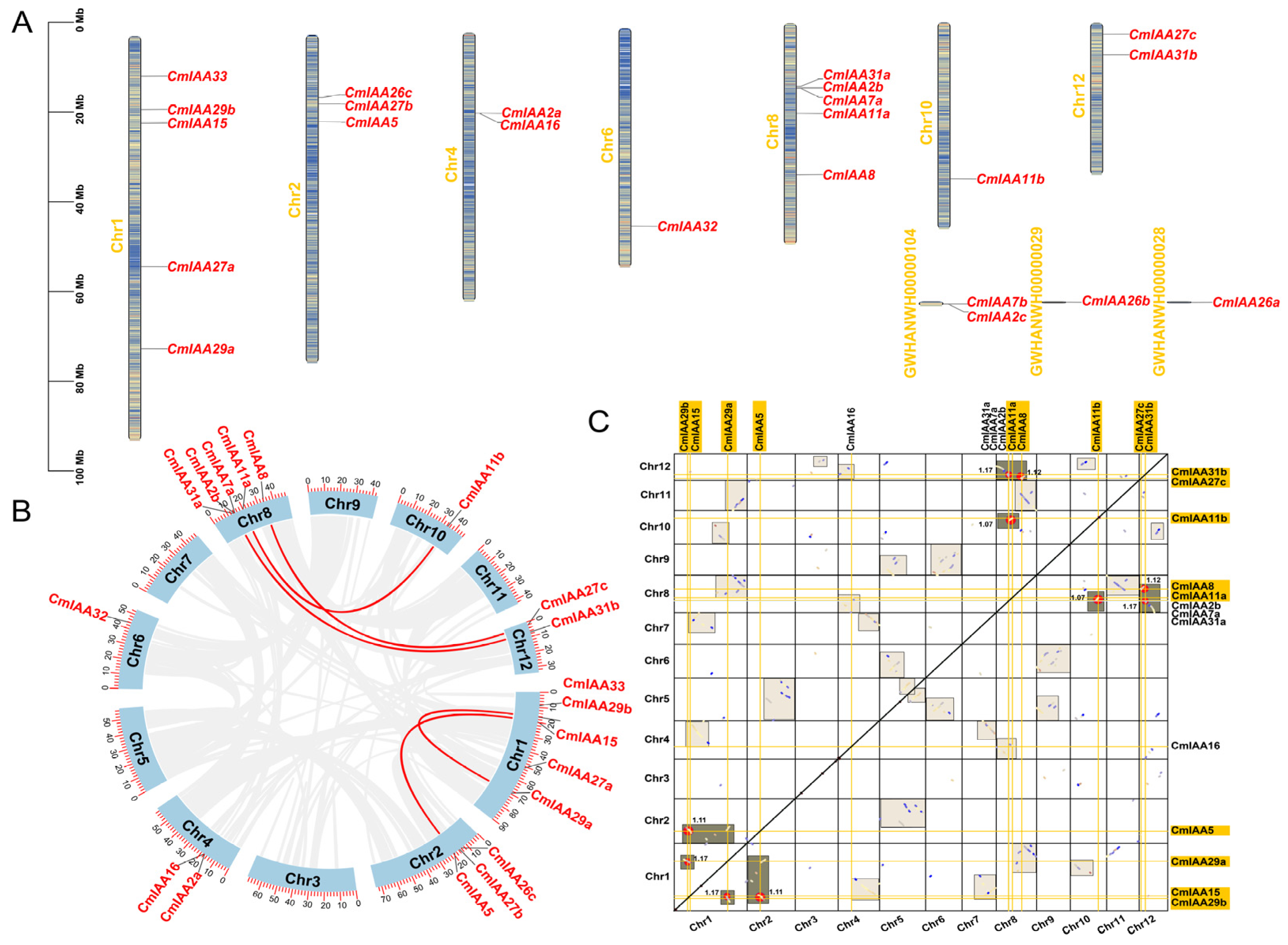
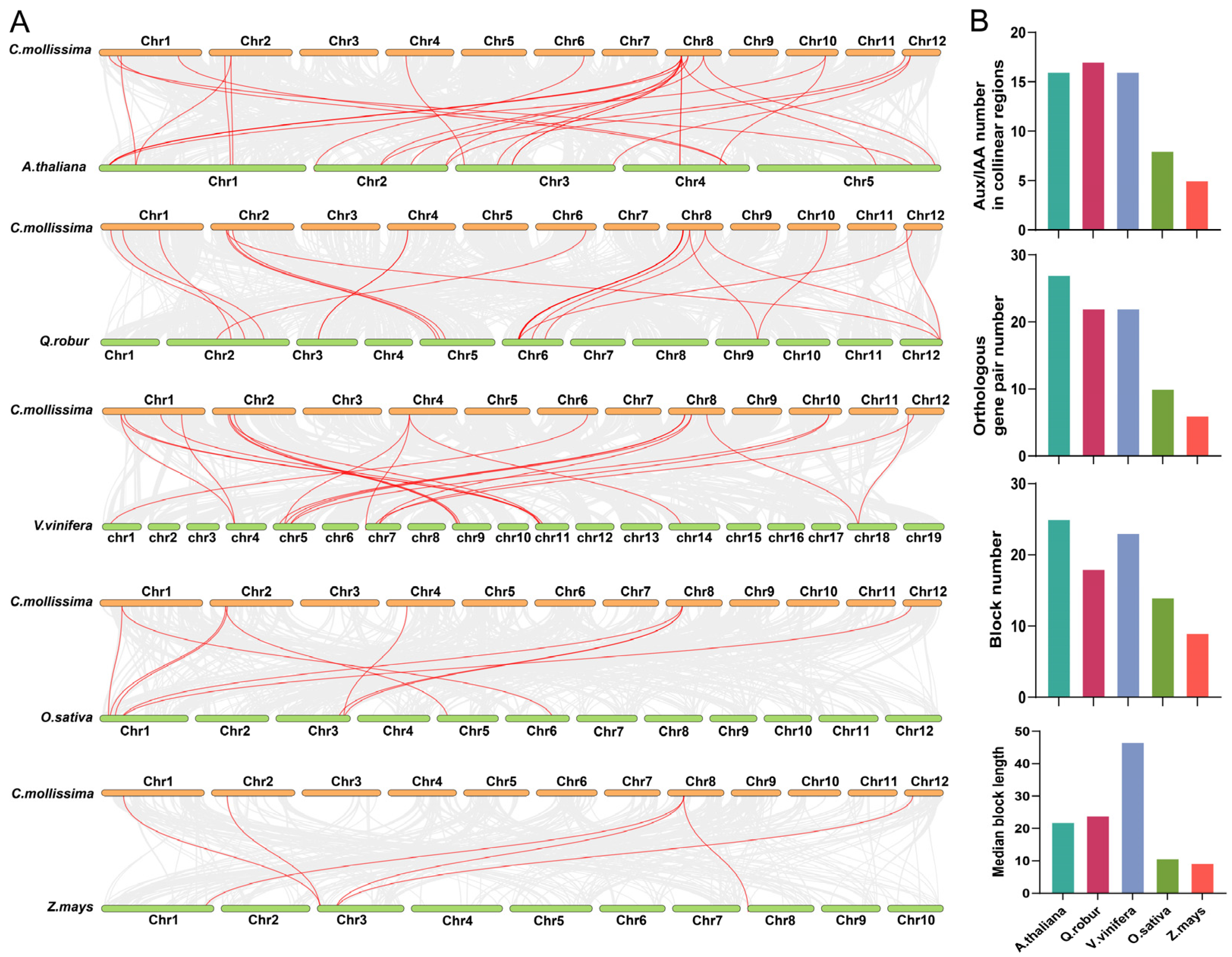
3.3. Chromosome Location and Collinear Analyses
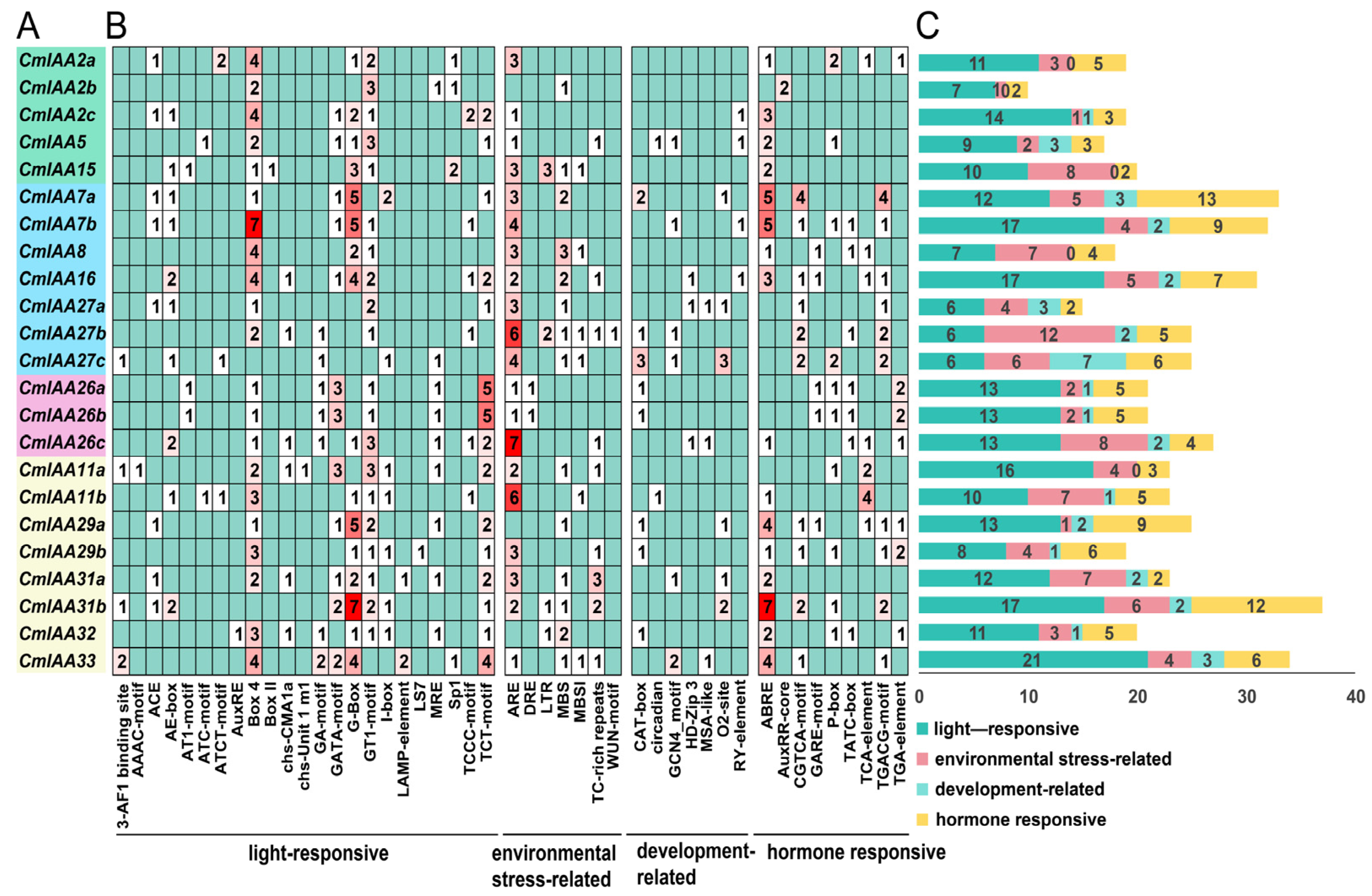
3.4. Cis-Acting Elements Analysis
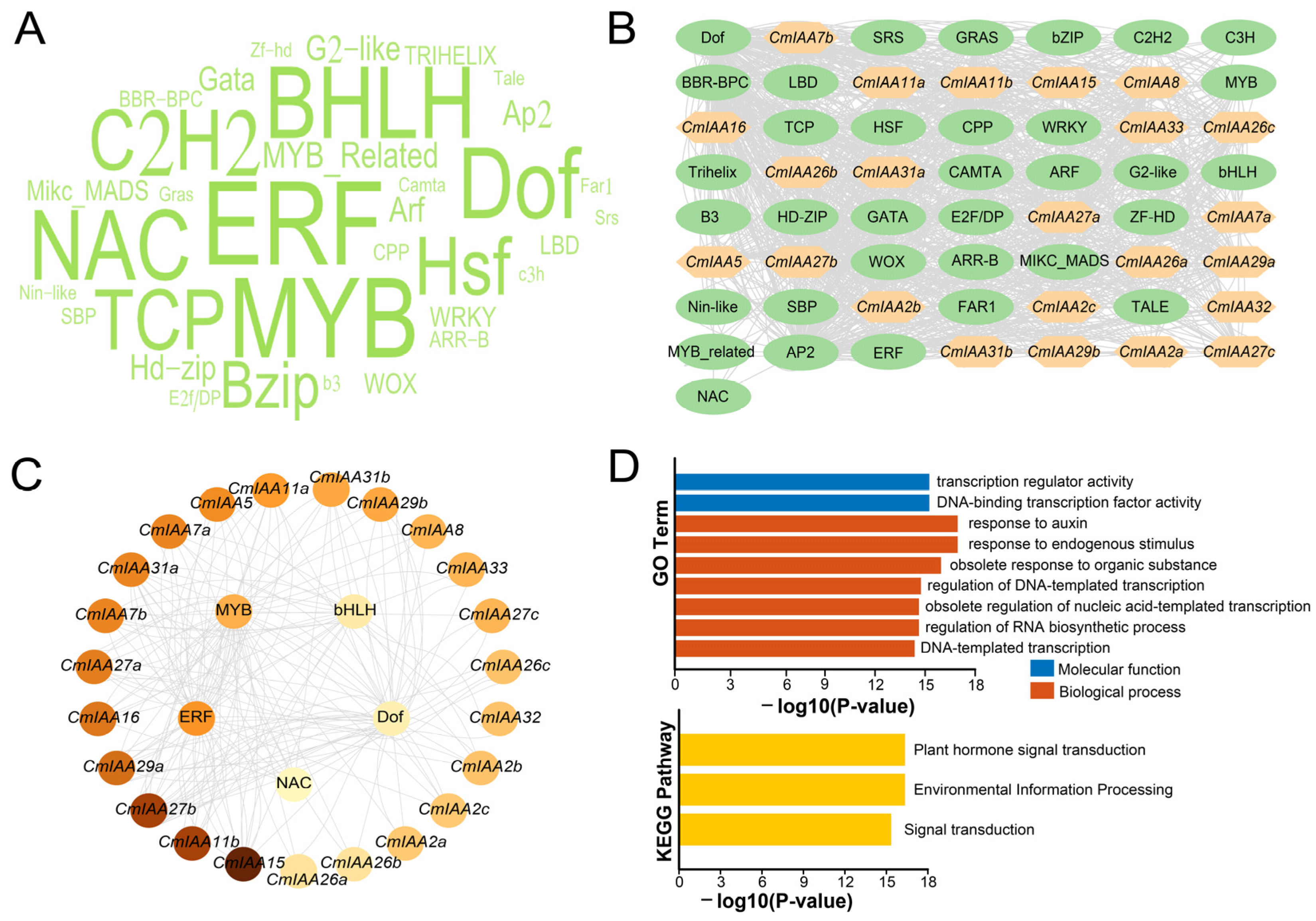
3.5. TFs Regulatory Network Analysis, GO/KEGG Enrichment, and PPI Network Analysis
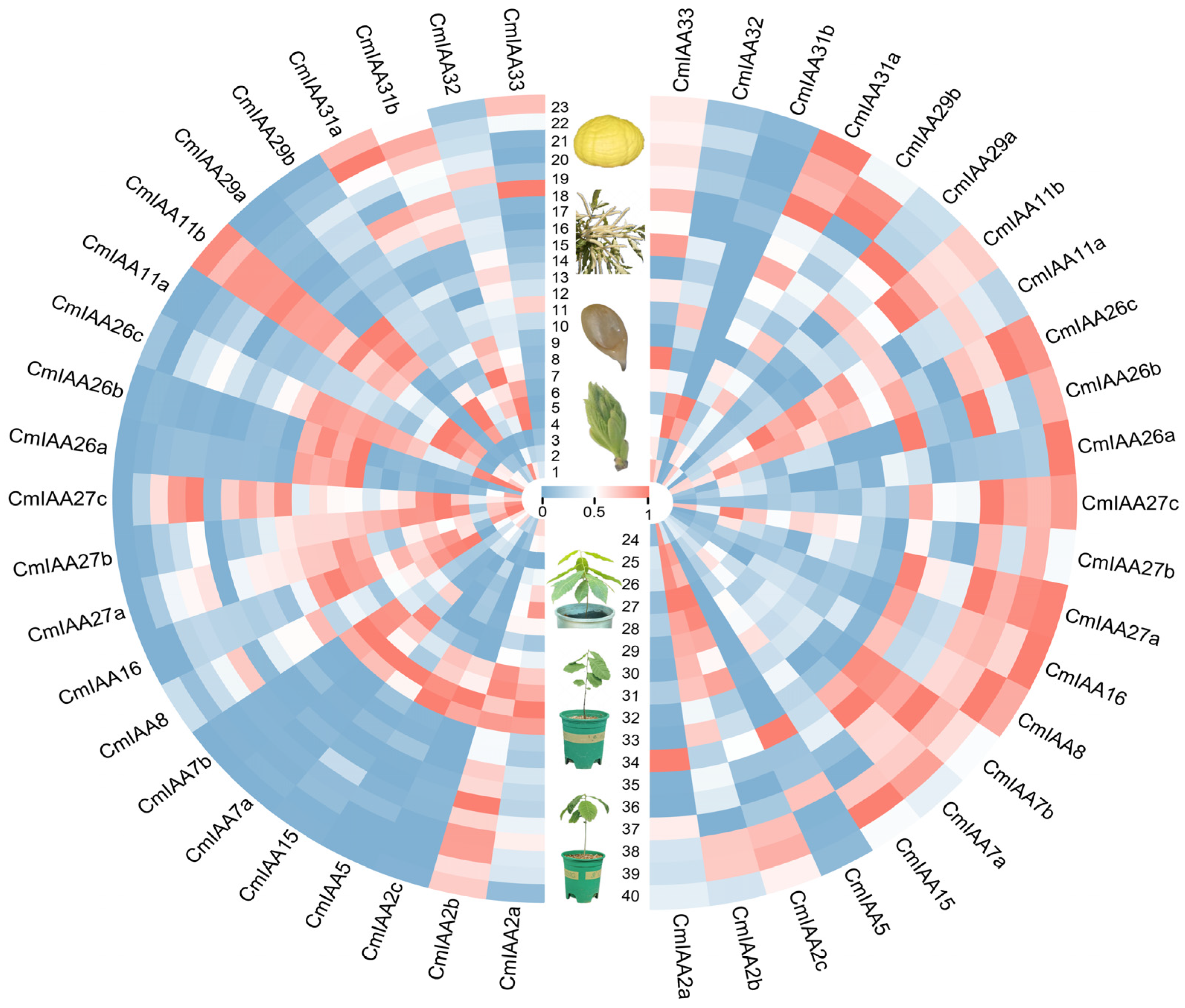
3.6. Expression Patterns of CmAux/IAA Genes in Different Tissues and Under Abiotic Stress
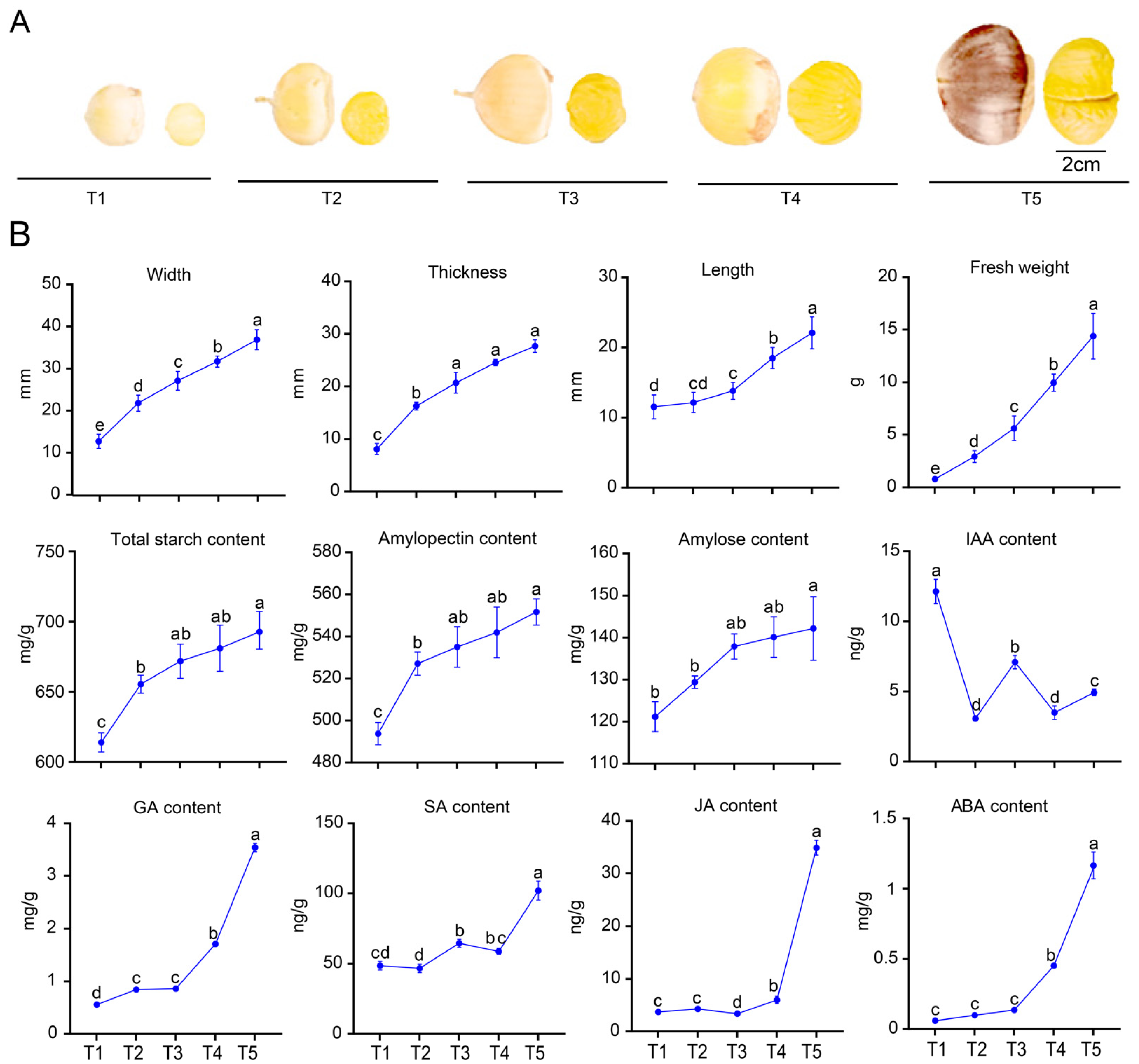
3.7. Phenotypic Changes During the Development
3.8. RNA Sequencing and DEGs Identification
3.9. Expression Patterns of CmAux/IAAs and Their Connections with Phenotypic Indicators
3.10. Expression Analysis of CmAux/IAA Genes During Seed Kernel Development by RT-qPCR
4. Discussion
4.1. The Molecular Characteristics of Aux/IAA Genes
4.2. Analysis of the Evolution and Expansion of the CmAux/IAA Gene Family
4.3. Expression Pattern of CmAux/IAA Genes During Seed Kernel Development
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Luo, J.; Zhou, J.J.; Zhang, J.Z. Aux/IAA Gene Family in Plants: Molecular Structure, Regulation, and Function. Int. J. Mol. Sci. 2018, 19, 259. [Google Scholar] [CrossRef]
- Kalluri, U.C.; Difazio, S.P.; Brunner, A.M.; Tuskan, G.A. Genome-wide analysis of Aux/IAA and ARF gene families in Populus trichocarpa. BMC Plant Biol. 2007, 7, 59. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liu, Y.; Yan, H.; Tian, T.; You, Q.; Zhang, L.; Xu, W.; Su, Z. PlantEAR: Functional Analysis Platform for Plant EAR Motif-Containing Proteins. Front. Genet. 2018, 9, 590. [Google Scholar] [CrossRef] [PubMed]
- Mockaitis, K.; Estelle, M. Auxin receptors and plant development: A new signaling paradigm. Annu. Rev. Cell Dev. Biol. 2008, 24, 55–80. [Google Scholar] [CrossRef] [PubMed]
- Woodward, A.W.; Bartel, B. Auxin: Regulation, action, and interaction. Ann. Bot. 2005, 95, 707–735. [Google Scholar] [CrossRef]
- Morgan, K.E.; Zarembinski, T.I.; Theologis, A.; Abel, S. Biochemical characterization of recombinant polypeptides corresponding to the predicted betaalphaalpha fold in Aux/IAA proteins. FEBS Lett. 1999, 454, 283–287. [Google Scholar] [CrossRef]
- Guilfoyle, T.J.; Hagen, G. Auxin response factors. Curr. Opin. Plant Biol. 2007, 10, 453–460. [Google Scholar] [CrossRef]
- Smalle, J.; Vierstra, R.D. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 2004, 55, 555–590. [Google Scholar] [CrossRef]
- Cao, M.; Chen, R.; Li, P.; Yu, Y.; Zheng, R.; Ge, D.; Zheng, W.; Wang, X.; Gu, Y.; Gelová, Z.; et al. TMK1-mediated auxin signalling regulates differential growth of the apical hook. Nature 2019, 568, 240–243. [Google Scholar] [CrossRef]
- Xu, C.; Shen, Y.; He, F.; Fu, X.; Yu, H.; Lu, W.; Li, Y.; Li, C.; Fan, D.; Wang, H.C.; et al. Auxin-mediated Aux/IAA-ARF-HB signaling cascade regulates secondary xylem development in Populus. New Phytol. 2019, 222, 752–767. [Google Scholar] [CrossRef]
- Wang, C.K.; Han, P.L.; Zhao, Y.W.; Ji, X.L.; Yu, J.Q.; You, C.X.; Hu, D.G.; Hao, Y.J. Auxin regulates anthocyanin biosynthesis through the auxin repressor protein MdIAA26. Biochem. Biophys. Res. Commun. 2020, 533, 717–722. [Google Scholar] [CrossRef] [PubMed]
- Molesini, B.; Pennisi, F.; Vitulo, N.; Pandolfini, T. MicroRNAs associated with AGL6 and IAA9 function in tomato fruit set. BMC Res. Notes 2023, 16, 242. [Google Scholar] [CrossRef] [PubMed]
- Ku, S.J.; Park, J.Y.; Ha, S.B.; Kim, J. Overexpression of IAA1 with domain II mutation impairs cell elongation and cell division in inflorescences and leaves of Arabidopsis. J. Plant Physiol. 2009, 166, 548–553. [Google Scholar] [CrossRef]
- Chen, H.; Song, Z.; Wang, L.; Lai, X.; Chen, W.; Li, X.; Zhu, X. Auxin-responsive protein MaIAA17-like modulates fruit ripening and ripening disorders induced by cold stress in ‘Fenjiao’ banana. Int. J. Biol. Macromol. 2023, 247, 125750. [Google Scholar] [CrossRef]
- Liu, D.J.; Chen, J.Y.; Lu, W.J. Expression and regulation of the early auxin-responsive Aux/IAA genes during strawberry fruit development. Mol. Biol. Rep. 2011, 38, 1187–1193. [Google Scholar] [CrossRef]
- Overvoorde, P.J.; Okushima, Y.; Alonso, J.M.; Chan, A.; Chang, C.; Ecker, J.R.; Hughes, B.; Liu, A.; Onodera, C.; Quach, H.; et al. Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell 2005, 17, 3282–3300. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Kaur, N.; Garg, R.; Thakur, J.K.; Tyagi, A.K.; Khurana, J.P. Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct. Integr. Genom. 2006, 6, 47–59. [Google Scholar] [CrossRef]
- Audran-Delalande, C.; Bassa, C.; Mila, I.; Regad, F.; Zouine, M.; Bouzayen, M. Genome-wide identification, functional analysis and expression profiling of the Aux/IAA gene family in tomato. Plant Cell Physiol. 2012, 53, 659–672. [Google Scholar] [CrossRef]
- Wang, L.; Xu, K.; Li, Y.; Cai, W.; Zhao, Y.; Yu, B.; Zhu, Y. Genome-Wide Identification of the Aux/IAA Family Genes (MdIAA) and Functional Analysis of MdIAA18 for Apple Tree Ideotype. Biochem. Genet. 2019, 57, 709–733. [Google Scholar] [CrossRef]
- Wang, Y.; Deng, D.; Bian, Y.; Lv, Y.; Xie, Q. Genome-wide analysis of primary auxin-responsive Aux/IAA gene family in maize (Zea mays L.). Mol. Biol. Rep. 2010, 37, 3991–4001. [Google Scholar] [CrossRef]
- Wu, J.; Peng, Z.; Liu, S.; He, Y.; Cheng, L.; Kong, F.; Wang, J.; Lu, G. Genome-wide analysis of Aux/IAA gene family in Solanaceae species using tomato as a model. Mol. Genet. Genom. 2012, 287, 295–311. [Google Scholar] [CrossRef] [PubMed]
- Jia, S.S.; Ren, X.Y.; Tong, M.N.; Jiang, S.Y.; Zhang, C.Q.; Liu, Q.Q.; Li, Q.F. OsIAA19, an Aux/IAA Family Gene, Involved in the Regulation of Seed-Specific Traits in Rice. Plants 2024, 13, 3538. [Google Scholar] [CrossRef]
- Huang, R.; Peng, F.; Wang, D.; Cao, F.; Guo, C.; Yu, L.; Zhang, J.; Yang, Y. Transcriptome analysis of differential sugar accumulation in the developing embryo of contrasting two Castanea mollissima cultivars. Front. Plant Sci. 2023, 14, 1206585. [Google Scholar] [CrossRef]
- Santos, M.J.; Pinto, T.; Vilela, A. Sweet Chestnut (Castanea sativa Mill.) Nutritional and Phenolic Composition Interactions with Chestnut Flavor Physiology. Foods 2022, 11, 4052. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Huang, X.; Zhang, C.; Zhang, M.; Huang, C.; Yang, H. Amino acid composition and nutritional value evaluation of Chinese chestnut (Castanea mollissima Blume) and its protein subunit. RSC Adv. 2018, 8, 2653–2659. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Newberg, L.A. Error statistics of hidden Markov model and hidden Boltzmann model results. BMC Bioinform. 2009, 10, 212. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Yu, L.; Tian, Y.; Wang, X.; Cao, F.; Wang, H.; Huang, R.; Guo, C.; Zhang, H.; Zhang, J. Genome-wide identification, phylogeny, evolutionary expansion, and expression analyses of ABC gene family in Castanea mollissima under temperature stress. Plant Physiol. Biochem. 2025, 219, 109450. [Google Scholar] [CrossRef] [PubMed]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Gustavsson, E.K.; Zhang, D.; Reynolds, R.H.; Garcia-Ruiz, S.; Ryten, M. ggtranscript: An R package for the visualization and interpretation of transcript isoforms using ggplot2. Bioinformatics 2022, 38, 3844–3846. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Hu, M.; Xie, M.; Cui, X.; Huang, J.; Cheng, X.; Liu, L.; Yan, S.; Liu, S.; Tong, C. Characterization and Potential Function Analysis of the SRS Gene Family in Brassica napus. Genes 2023, 14, 1421. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Hadar, N.; Weintraub, G.; Gudes, E.; Dolev, S.; Birk, O.S. GeniePool: Genomic database with corresponding annotated samples based on a cloud data lake architecture. Database 2023, 2023, baad043. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Liu, S.; Wang, Z.; Zhu, R.; Wang, F.; Cheng, Y.; Liu, Y. Three Differential Expression Analysis Methods for RNA Sequencing: Limma, EdgeR, DESeq2. J. Vis. Exp. 2021, 18, 175. [Google Scholar] [CrossRef]
- Singh, V.K.; Mangalam, A.K.; Dwivedi, S.; Naik, S. Primer premier: Program for design of degenerate primers from a protein sequence. BioTechniques 1998, 24, 318–319. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Mitteer, D.R.; Greer, B.D.; Randall, K.R.; Briggs, A.M. Further Evaluation of Teaching Behavior Technicians to Input Data and Graph Using GraphPad Prism. Behav. Anal. 2020, 20, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Weaver, B.; Wuensch, K.L. SPSS and SAS programs for comparing Pearson correlations and OLS regression coefficients. Behav. Res. Methods 2013, 45, 880–895. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S. Evolution by Gene Duplication; Springer: Berlin/Heidelberg, Germany, 1970. [Google Scholar]
- Yu, L.; Diao, S.; Zhang, G.; Yu, J.; Zhang, T.; Luo, H.; Duan, A.; Wang, J.; He, C.; Zhang, J. Genome sequence and population genomics provide insights into chromosomal evolution and phytochemical innovation of Hippophae rhamnoides. Plant Biotechnol. J. 2022, 20, 1257–1273. [Google Scholar] [CrossRef]
- Birchler, J.A.; Veitia, R.A. The gene balance hypothesis: From classical genetics to modern genomics. Plant Cell 2007, 19, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Wickett, N.J.; Ayyampalayam, S.; Chanderbali, A.S.; Landherr, L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.; Soltis, P.S.; et al. Ancestral polyploidy in seed plants and angiosperms. Nature 2011, 473, 97–100. [Google Scholar] [CrossRef]
- Van de Peer, Y.; Mizrachi, E.; Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 2017, 18, 411–424. [Google Scholar] [CrossRef]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217–218, 109–119. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, Y.H.; Wang, S.; Zhang, Y.; Huang, T.; Cai, Y.D. Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways. PLoS ONE 2017, 12, e0184129. [Google Scholar] [CrossRef]
- Wang, T.; Long, C.; Chang, M.; Wu, Y.; Su, S.; Wei, J.; Jiang, S.; Wang, X.; He, J.; Xing, D.; et al. Genome-wide identification of the B3 transcription factor family in pepper (Capsicum annuum) and expression patterns during fruit ripening. Sci. Rep. 2024, 14, 2226. [Google Scholar] [CrossRef] [PubMed]
- Qi, T.; Yang, W.; Hassan, M.J.; Liu, J.; Yang, Y.; Zhou, Q.; Li, H.; Peng, Y. Genome-wide identification of Aux/IAA gene family in white clover (Trifolium repens L.) and functional verification of TrIAA18 under different abiotic stress. BMC Plant Biol. 2024, 24, 346. [Google Scholar] [CrossRef] [PubMed]
- Bu, H.; Sun, X.; Yue, P.; Qiao, J.; Sun, J.; Wang, A.; Yuan, H.; Yu, W. The MdAux/IAA2 Transcription Repressor Regulates Cell and Fruit Size in Apple Fruit. Int. J. Mol. Sci. 2022, 23, 9454. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, S.; Hayat, F.; Mushtaq, N.; Khalil-Ur-Rehman, M.; Khan, U.; Yasoob, T.B.; Khan, M.N.; Ni, Z.; Ting, S.; Gao, Z. Bioinformatics Study of Aux/IAA Family Genes and Their Expression in Response to Different Hormones Treatments during Japanese Apricot Fruit Development and Ripening. Plants 2022, 11, 1898. [Google Scholar] [CrossRef]
- Molesini, B.; Dusi, V.; Pennisi, F.; Pandolfini, T. How Hormones and MADS-Box Transcription Factors Are Involved in Controlling Fruit Set and Parthenocarpy in Tomato. Genes 2020, 11, 1441. [Google Scholar] [CrossRef]
- Li, R.; Huang, X.; Yang, L.; Liao, J.; Wei, X.; Li, J.; Zeng, G.; Liu, D.; Shi, Z.; Zhao, Z. Whole genome sequencing of Castanea mollissima and molecular mechanisms of sugar and starch synthesis. Front. Plant Sci. 2024, 15, 1455885. [Google Scholar] [CrossRef]
- Liu, K.; Yuan, C.; Feng, S.; Zhong, S.; Li, H.; Zhong, J.; Shen, C.; Liu, J. Genome-wide analysis and characterization of Aux/IAA family genes related to fruit ripening in papaya (Carica papaya L.). BMC Genom. 2017, 18, 351. [Google Scholar] [CrossRef]
- Nozawa, R.S.; Gilbert, N. RNA: Nuclear Glue for Folding the Genome. Trends Cell Biol. 2019, 29, 201–211. [Google Scholar] [CrossRef]
- Sato, A.; Yamamoto, K.T. Overexpression of the non-canonical Aux/IAA genes causes auxin-related aberrant phenotypes in Arabidopsis. Physiol. Plant 2008, 133, 397–405. [Google Scholar] [CrossRef]
- Feng, K.; Hou, X.L.; Xing, G.M.; Liu, J.X.; Duan, A.Q.; Xu, Z.S.; Li, M.Y.; Zhuang, J.; Xiong, A.S. Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef]
- Yan, H.; Pei, X.; Zhang, H.; Li, X.; Zhang, X.; Zhao, M.; Chiang, V.L.; Sederoff, R.R.; Zhao, X. MYB-Mediated Regulation of Anthocyanin Biosynthesis. Int. J. Mol. Sci. 2021, 22, 3103. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Dubos, C. The arabidopsis bHLH transcription factor family. Trends Plant Sci. 2024, 29, 668–680. [Google Scholar] [CrossRef] [PubMed]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Christophe Liseron-Monfils, D.W. Revealing gene regulation and associations through biological networks. Curr. Plant Biol. 2015, 3–4, 30–39. [Google Scholar] [CrossRef]
- Wen, S.; Ying, J.; Ye, Y.; Cai, Y.; Li, L.; Qian, R. Genome-wide identification and salt stress-responsive expression profiling of Aux/IAA gene family in Asparagus officinalis. BMC Plant Biol. 2025, 25, 759. [Google Scholar] [CrossRef]
- Alves, S.; Braga, Â.; Parreira, D.; Alhinho, A.T.; Silva, H.; Ramos, M.J.N.; Costa, M.M.R.; Morais-Cecílio, L. Genome-wide identification, phylogeny, and gene duplication of the epigenetic regulators in Fagaceae. Physiol. Plant 2022, 174, e13788. [Google Scholar] [CrossRef]
- Street, N.R. Structural Genomics of Angiosperm Trees: Genome Duplications, Ploidy, and Repeat Sequences. In Comparative and Evolutionary Genomics of Angiosperm Trees; Groover, A., Cronk, Q., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 101–120. [Google Scholar]
- Nystedt, B.; Street, N.R.; Wetterbom, A.; Zuccolo, A.; Lin, Y.C.; Scofield, D.G.; Vezzi, F.; Delhomme, N.; Giacomello, S.; Alexeyenko, A.; et al. The Norway spruce genome sequence and conifer genome evolution. Nature 2013, 497, 579–584. [Google Scholar] [CrossRef]
- Tuskan, G.A.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [CrossRef]
- Hollister, J.D.; Smith, L.M.; Guo, Y.L.; Ott, F.; Weigel, D.; Gaut, B.S. Transposable elements and small RNAs contribute to gene expression divergence between Arabidopsis thaliana and Arabidopsis lyrata. Proc. Natl. Acad. Sci. USA 2011, 108, 2322–2327. [Google Scholar] [CrossRef]
- Huang, Z.; Duan, W.; Song, X.; Tang, J.; Wu, P.; Zhang, B.; Hou, X. Retention, Molecular Evolution, and Expression Divergence of the Auxin/Indole Acetic Acid and Auxin Response Factor Gene Families in Brassica Rapa Shed Light on Their Evolution Patterns in Plants. Genome Biol. Evol. 2015, 8, 302–316. [Google Scholar] [CrossRef]
- Hou, Y.; Li, H.; Zhai, L.; Xie, X.; Li, X.; Bian, S. Identification and functional characterization of the Aux/IAA gene VcIAA27 in blueberry. Plant Signal Behav. 2020, 15, 1700327. [Google Scholar] [CrossRef] [PubMed]
- Rusak, G.; Cerni, S.; Stupin Polancec, D.; Ludwig-Müller, J. The responsiveness of the IAA2 promoter to IAA and IBA is differentially affected in Arabidopsis roots and shoots by flavonoids. Biol. Plant. 2010, 54, 403–414. [Google Scholar] [CrossRef]
- Wang, J.; Yan, D.W.; Yuan, T.T.; Gao, X.; Lu, Y.T. A gain-of-function mutation in IAA8 alters Arabidopsis floral organ development by change of jasmonic acid level. Plant Mol. Biol. 2013, 82, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, Z.; Eaglesfield, R.; Carr, C.; Amtmann, A. Cryptic variation in RNA-directed DNA-methylation controls lateral root development when auxin signalling is perturbed. Nat. Commun. 2020, 11, 218. [Google Scholar] [CrossRef]
- Padmanabhan, M.S.; Goregaoker, S.P.; Golem, S.; Shiferaw, H.; Culver, J.N. Interaction of the tobacco mosaic virus replicase protein with the Aux/IAA protein PAP1/IAA26 is associated with disease development. J. Virol. 2005, 79, 2549–2558. [Google Scholar] [CrossRef]
- Takahashi, N.; Ogita, N.; Koike, T.; Nishimura, K.; Inagaki, S.; Umeda, M. Local induction of IAA5 and IAA29 promotes DNA damage-triggered stem cell death in Arabidopsis roots. bioRxiv 2022. [Google Scholar] [CrossRef]
- Zimmermann, P.; Hirsch-Hoffmann, M.; Hennig, L.; Gruissem, W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004, 136, 2621–2632. [Google Scholar] [CrossRef]
- De Grassi, A.; Lanave, C.; Saccone, C. Genome duplication and gene-family evolution: The case of three OXPHOS gene families. Gene 2008, 421, 1–6. [Google Scholar] [CrossRef]
- Lallemand, T.; Leduc, M.; Desmazières, A.; Aubourg, S.; Rizzon, C.; Landès, C.; Celton, J.M. Insights into the Evolution of Ohnologous Sequences and Their Epigenetic Marks Post-WGD in Malus Domestica. Genome Biol. Evol. 2023, 15, evad178. [Google Scholar] [CrossRef]
- Schranz, M.E.; Mohammadin, S.; Edger, P.P. Ancient whole genome duplications, novelty and diversification: The WGD Radiation Lag-Time Model. Curr. Opin. Plant Biol. 2012, 15, 147–153. [Google Scholar] [CrossRef]
- Abdullaev, E.T.; Haridoss, D.A.; Arndt, P.F. Reconstruction of Segmental Duplication Rates and Associated Genomic Features by Network Analysis. Genome Biol. Evol. 2025, 17, evaf011. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ficklin, S.P.; Wang, X.; Feltus, F.A.; Paterson, A.H. Large-Scale Gene Relocations following an Ancient Genome Triplication Associated with the Diversification of Core Eudicots. PLoS ONE 2016, 11, e0155637. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Bassa, C.; Audran, C.; Mila, I.; Cheniclet, C.; Chevalier, C.; Bouzayen, M.; Roustan, J.P.; Chervin, C. The auxin Sl-IAA17 transcriptional repressor controls fruit size via the regulation of endoreduplication-related cell expansion. Plant Cell Physiol. 2014, 55, 1969–1976. [Google Scholar] [CrossRef]
- Zhao, X.; Muhammad, N.; Zhao, Z.; Yin, K.; Liu, Z.; Wang, L.; Luo, Z.; Wang, L.; Liu, M. Molecular regulation of fruit size in horticultural plants: A review. Sci. Hortic. 2021, 288. [Google Scholar] [CrossRef]
- Zhang, L.; Lin, Q.; Feng, Y.; Fan, X.; Zou, F.; Yuan, D.Y.; Zeng, X.; Cao, H. Transcriptomic identification and expression of starch and sucrose metabolism genes in the seeds of Chinese chestnut (Castanea mollissima). J. Agric. Food Chem. 2015, 63, 929–942. [Google Scholar] [CrossRef]
- Bassa, C.; Mila, I.; Bouzayen, M.; Audran-Delalande, C. Phenotypes associated with down-regulation of Sl-IAA27 support functional diversity among Aux/IAA family members in tomato. Plant Cell Physiol. 2012, 53, 1583–1595. [Google Scholar] [CrossRef]
- Venturini, L.; Ferrarini, A.; Zenoni, S.; Tornielli, G.B.; Fasoli, M.; Dal Santo, S.; Minio, A.; Buson, G.; Tononi, P.; Zago, E.D.; et al. De novo transcriptome characterization of Vitis vinifera cv. Corvina unveils varietal diversity. BMC Genom. 2013, 14, 41. [Google Scholar] [CrossRef]
- Hu, J.; Israeli, A.; Ori, N.; Sun, T.P. The Interaction between DELLA and ARF/IAA Mediates Crosstalk between Gibberellin and Auxin Signaling to Control Fruit Initiation in Tomato. Plant Cell 2018, 30, 1710–1728. [Google Scholar] [CrossRef]
- Pattison, R.J.; Csukasi, F.; Catalá, C. Mechanisms regulating auxin action during fruit development. Physiol. Plant 2014, 151, 62–72. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, Y.; Huang, J.; Wang, J.; Wang, D.; Huang, R.; Liu, X.; Zhang, H.; Zhang, J.; Wang, X.; Yu, L. Genome-Wide Identification, Evolutionary Expansion, and Expression Analyses of Aux/IAA Gene Family in Castanea mollissima During Seed Kernel Development. Biology 2025, 14, 806. https://doi.org/10.3390/biology14070806
Tian Y, Huang J, Wang J, Wang D, Huang R, Liu X, Zhang H, Zhang J, Wang X, Yu L. Genome-Wide Identification, Evolutionary Expansion, and Expression Analyses of Aux/IAA Gene Family in Castanea mollissima During Seed Kernel Development. Biology. 2025; 14(7):806. https://doi.org/10.3390/biology14070806
Chicago/Turabian StyleTian, Yujuan, Jingmiao Huang, Jinxin Wang, Dongsheng Wang, Ruimin Huang, Xia Liu, Haie Zhang, Jingzheng Zhang, Xiangyu Wang, and Liyang Yu. 2025. "Genome-Wide Identification, Evolutionary Expansion, and Expression Analyses of Aux/IAA Gene Family in Castanea mollissima During Seed Kernel Development" Biology 14, no. 7: 806. https://doi.org/10.3390/biology14070806
APA StyleTian, Y., Huang, J., Wang, J., Wang, D., Huang, R., Liu, X., Zhang, H., Zhang, J., Wang, X., & Yu, L. (2025). Genome-Wide Identification, Evolutionary Expansion, and Expression Analyses of Aux/IAA Gene Family in Castanea mollissima During Seed Kernel Development. Biology, 14(7), 806. https://doi.org/10.3390/biology14070806





