Genomic Analysis of Adaptability and Genetic Structure of Jabal Akhdar Goats: Evidence of Positive Selection in an Indigenous Omani Breed
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Goat Populations SNP Genotype Data
2.2. Genetic Structure Analyses
2.3. ROH and Inbreeding Level Estimation
2.4. LD and Ne Analyses
2.5. Signatures of Selection Analyses
2.6. Functional Characterization of the Candidate Selection Regions
3. Results
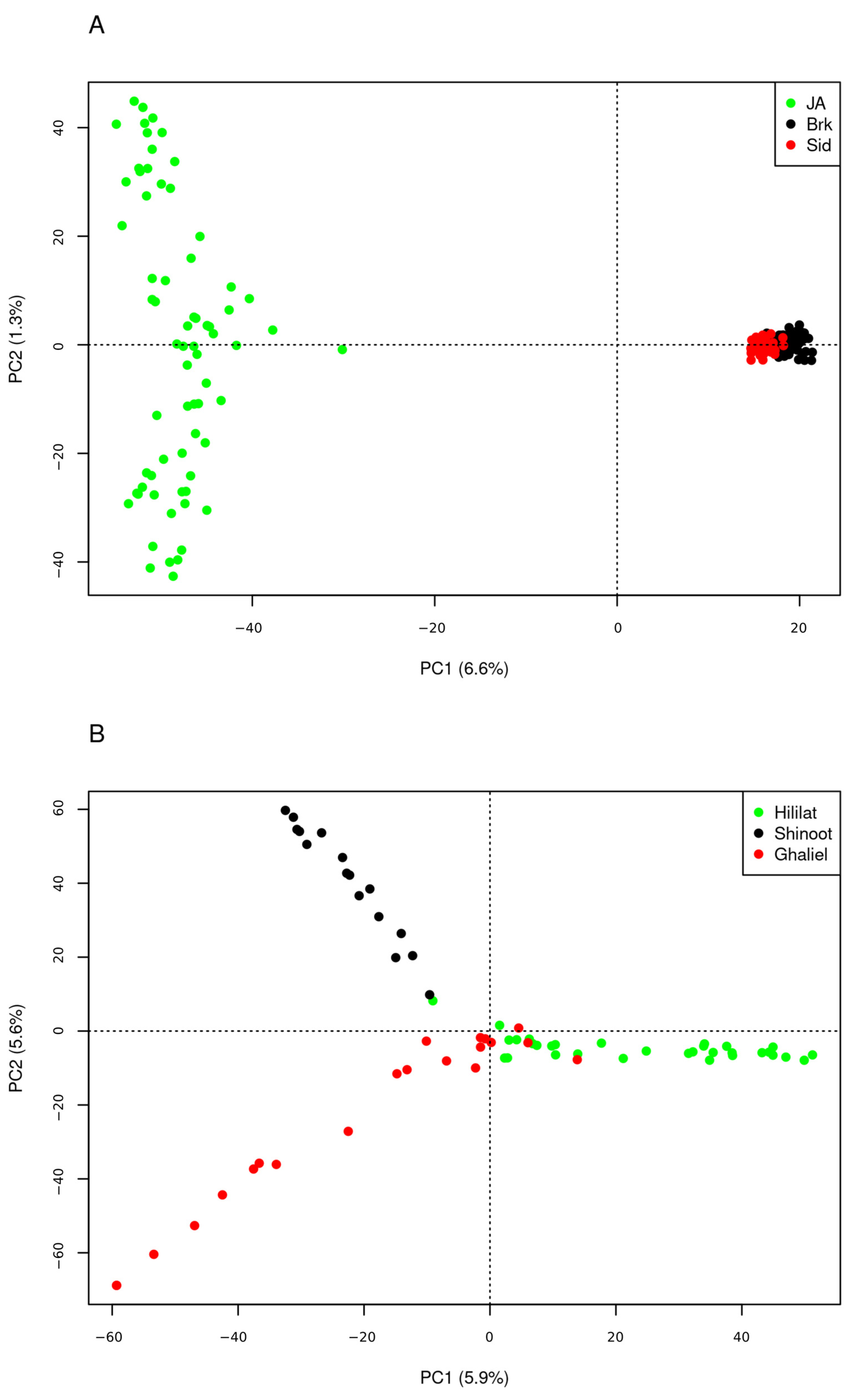
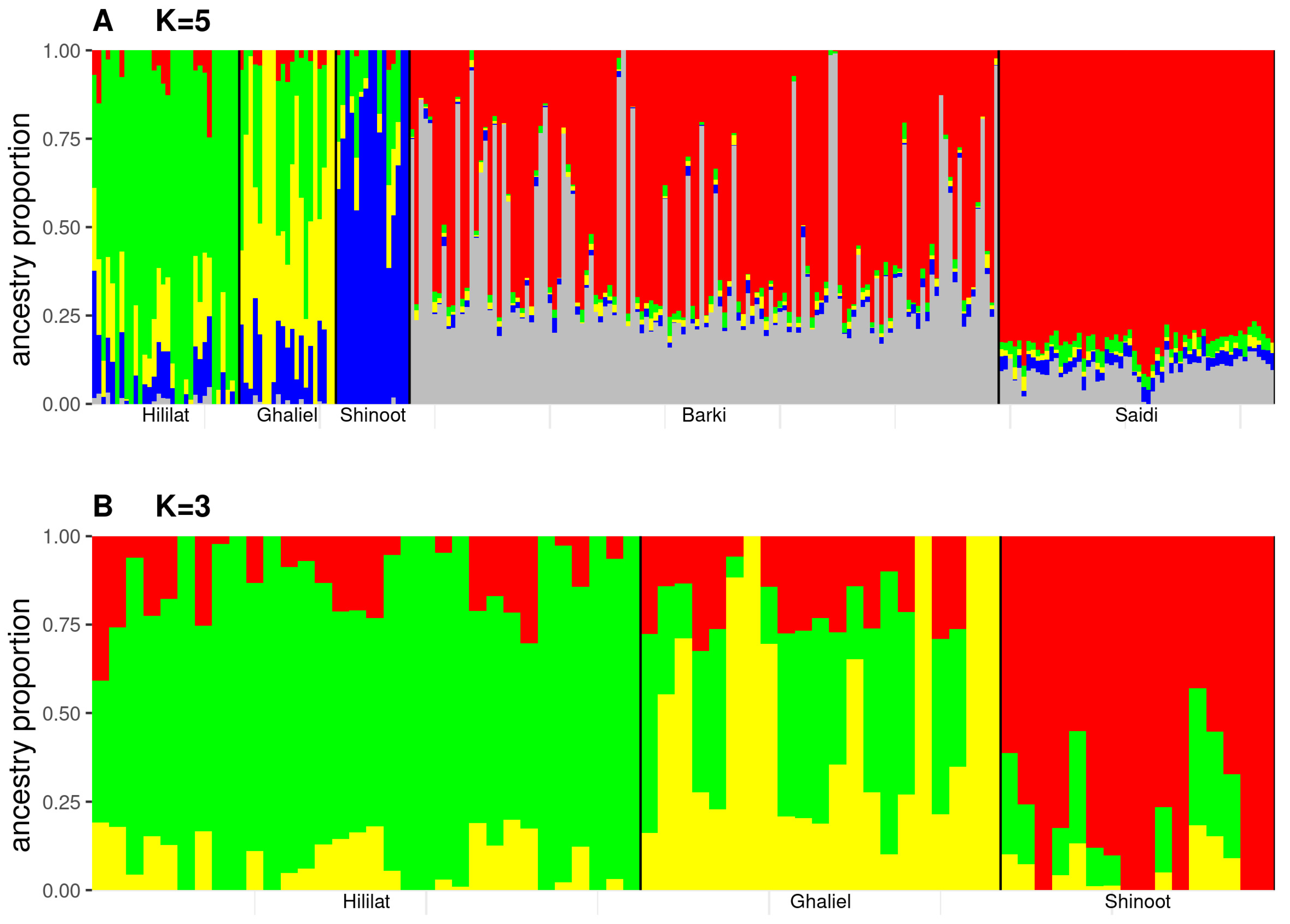
3.1. Genetic Structure Analyses
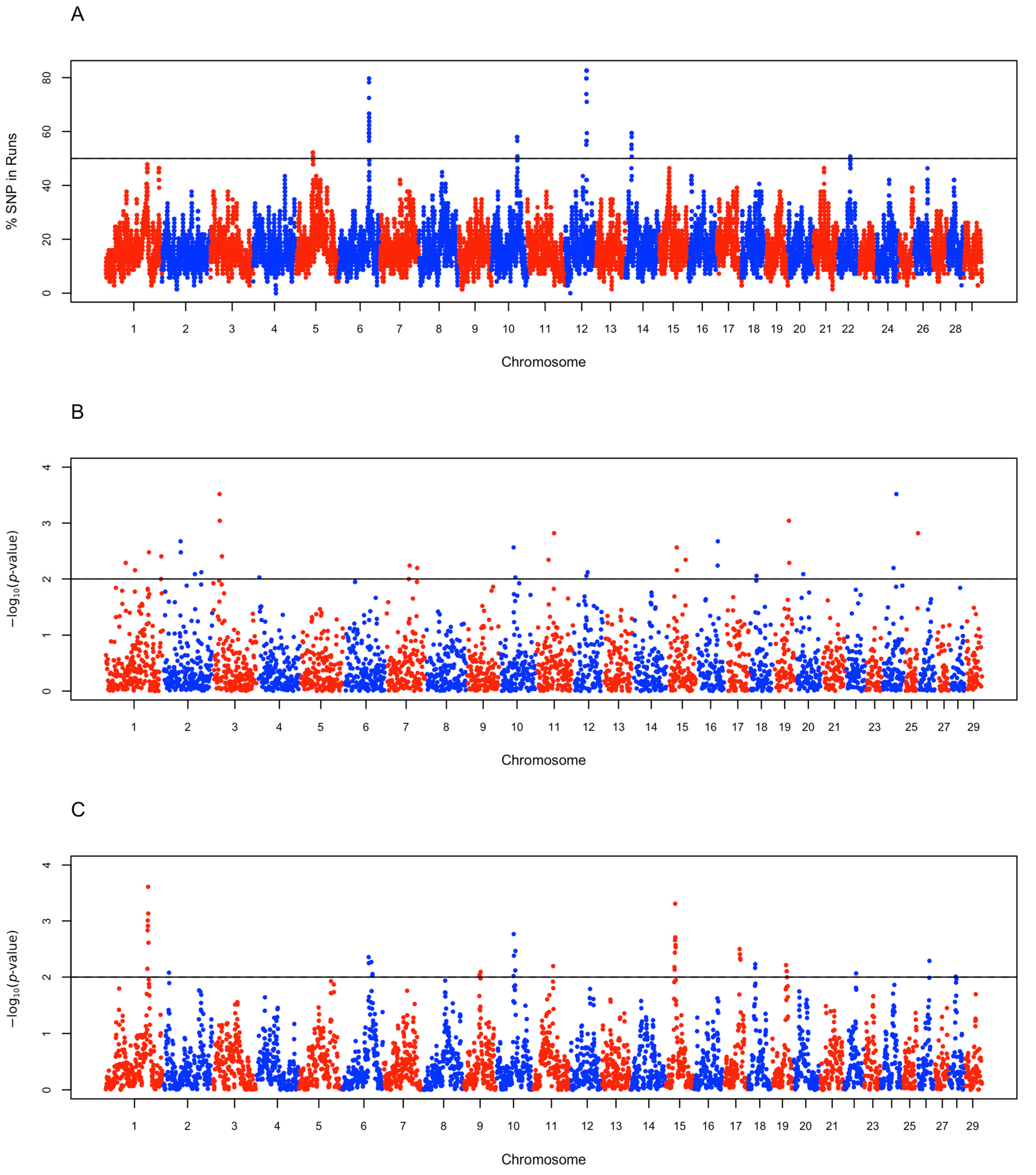
3.2. Detection of ROH Segments and Estimation of FROH
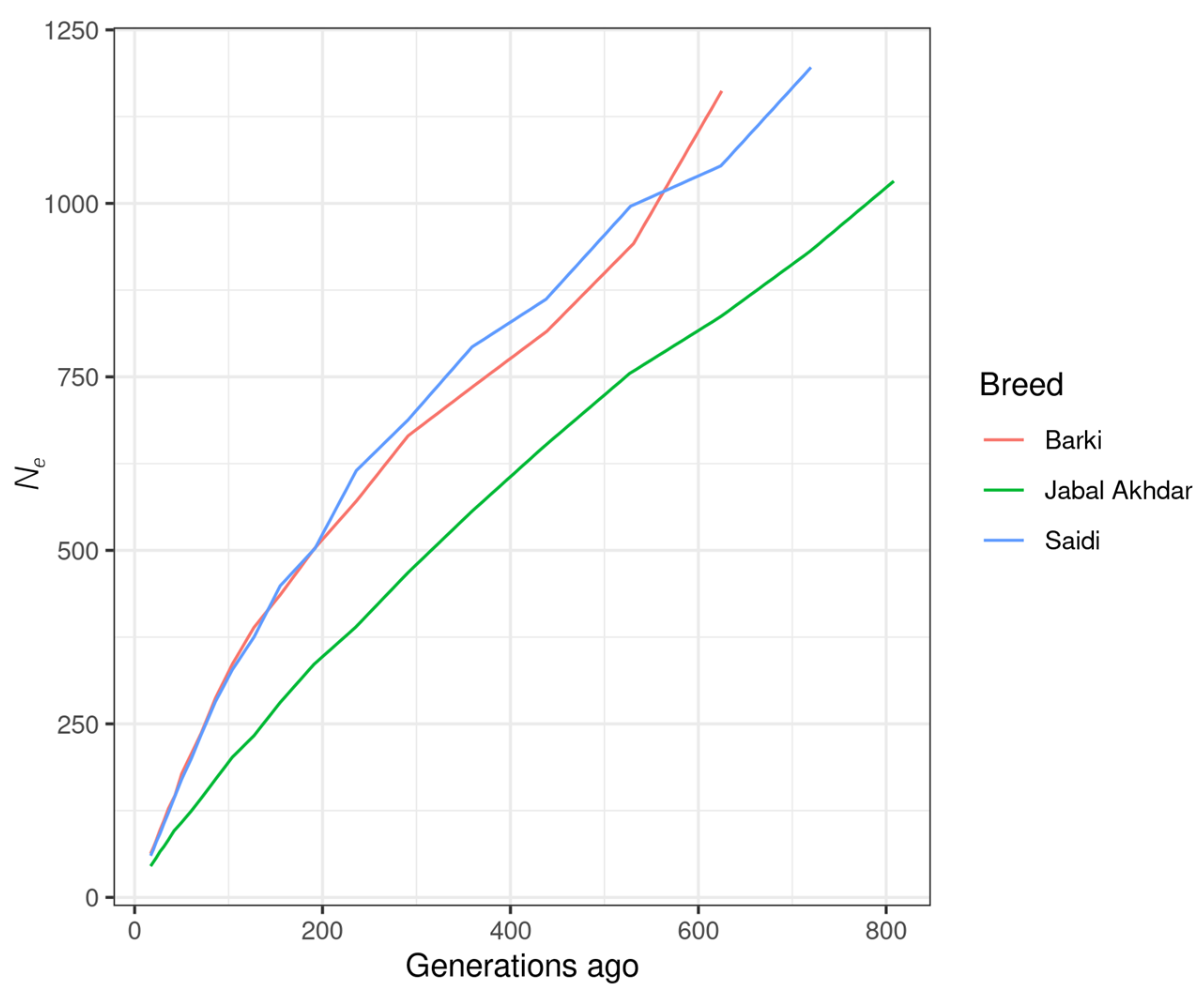
3.3. LD and Ne Analyses
3.4. Candidate Selection Regions and Functional Characterization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Utaaker, K.S.; Chaudhary, S.; Kifleyohannes, T.; Robertson, L.J. Global goat! Is the expanding goat population an important reservoir of Cryptosporidium? Front. Vet. Sci. 2021, 8, 648500. [Google Scholar] [CrossRef] [PubMed]
- Salem, A.Z.M.; Salem, M.Z.M.; El-Adawy, M.M.; Robinson, P.H. Nutritive evaluations of some browse tree foliages during the dry season: Secondary compounds, feed intake and in vivo digestibility in sheep and goats. Anim. Feed. Sci. Technol. 2006, 127, 251–267. [Google Scholar] [CrossRef]
- Silanikove, N. The struggle to maintain hydration and osmoregulation in animals experiencing severe dehydration and rapid rehydration: The story of ruminants. Exp. Physiol. 1994, 79, 281–300. [Google Scholar] [CrossRef]
- Silanikove, N. The physiological basis of adaptation in goats to harsh environments. Small Rumin. Res. 2000, 35, 181–193. [Google Scholar] [CrossRef]
- Aboul Naga, A.M.; Abdel Khalek, T.M.; Osman, M.; Elbeltagy, A.R.; Abdel-Aal, E.S.; Abou-Ammo, F.F.; El-Shafie, M.H. Physiological and genetic adaptation of desert sheep and goats to heat stress in the arid areas of Egypt. Small Rumin. Res. 2021, 203, 106499. [Google Scholar] [CrossRef]
- Aboulnaga, A.; Mohamed, S.; Ahmed, R.; Gamal, L.; Shafie, M. Physiological and genetic issues for tolerance to heat stress in subtropical sheep and goats raised in hot dry areas. Egypt. J. Anim. Prod. 2025, 62, 37–47. [Google Scholar] [CrossRef]
- Aboul-Naga, A.M.; Alsamman, A.M.; Nassar, A.E.; Mousa, K.H.; Osman, M.; Abdelsabour, T.H.; Mohamed, L.G.; Elshafie, M.H. Investigating genetic diversity and population structure of Egyptian goats across four breeds and seven regions. Small Rumin. Res. 2023, 226, 107017. [Google Scholar] [CrossRef]
- Al-Kalbani, M.; Coll, J. Recent trends in temperature and precipitation in Al Jabal Al Akhdar, Sultanate of Oman, and the implications for future climate change. J. Earth Sci. Clim. Change 2015, 6, 295. [Google Scholar] [CrossRef]
- Al-Abri, M.; Al Kharousi, K.; Al Hamrashdi, A.; Al Toobi, A.G.; Bahbahani, H.; Salem, M.M.I. Genome wide association analysis for body measurements in Jabal Akhdar Omani goats. Kuwait J. Sci. 2025, 52, 100372. [Google Scholar] [CrossRef]
- Mahgoub, O.; Kadim, I.T.; Al-Saqry, N.M.; Al-Busaidi, R.M. Potential of Omani Jebel Akhdar goat for meat production under feedlot conditions. Small Rumin. Res. 2005, 56, 223–230. [Google Scholar] [CrossRef]
- Shaat, I.; Al-Habsi, R. Current status of animal genetic resources in Oman. J. Agri. Sci. F. Tech. 2016, 2, 139–146. [Google Scholar]
- Al-Abri, M.; Al Kharousi, K.; Al Hamrashdi, A.; Al Toobi, A.G.; Salem, M.M.I. Genome wide association analysis for twinning ability in Jabal Akhdar Omani goats. Small Rumin. Res. 2023, 221, 106951. [Google Scholar] [CrossRef]
- Curik, I.; Ferenčaković, M.; Sölkner, J. Inbreeding and runs of homozygosity: A possible solution to an old problem. Livest. Sci. 2014, 166, 26–34. [Google Scholar] [CrossRef]
- Ouborg, N.J.; Pertoldi, C.; Loeschcke, V.; Bijlsma, R.K.; Hedrick, P.W. Conservation genetics in transition to conservation genomics. Trends Genet. 2010, 26, 177–187. [Google Scholar] [CrossRef]
- Dixit, S.P.; Singh, S.; Ganguly, I.; Bhatia, A.K.; Sharma, A.; Kumar, N.A.; Dang, A.K.; Jayakumar, S. Genome-wide runs of homozygosity revealed selection signatures in Bos indicus. Front. Genet. 2020, 11, 92. [Google Scholar] [CrossRef]
- Falchi, L.; Cesarani, A.; Criscione, A.; Hidalgo, J.; Garcia, A.; Mastrangelo, S.; Macciotta, N.P.P. Effect of genotyping density on the detection of runs of homozygosity and heterozygosity in cattle. J. Anim. Sci. 2024, 102, skae147. [Google Scholar] [CrossRef]
- Liu, J.; Shi, L.; Li, Y.; Chen, L.; Garrick, D.; Wang, L.; Zhao, F. Estimates of genomic inbreeding and identification of candidate regions that differ between Chinese indigenous sheep breeds. J. Anim. Sci. Biotechnol. 2021, 12, 95. [Google Scholar] [CrossRef]
- Shi, L.; Wang, L.; Liu, J.; Deng, T.; Yan, H.; Zhang, L.; Liu, X.; Gao, H.; Hou, X.; Wang, L.; et al. Estimation of inbreeding and identification of regions under heavy selection based on runs of homozygosity in a Large White pig population. J. Anim. Sci. Biotechnol. 2020, 11, 46. [Google Scholar] [CrossRef]
- Berihulay, H.; Islam, R.; Jiang, L.; Ma, Y. Genome-wide linkage disequilibrium and the extent of effective population sizes in six Chinese goat populations using a 50K single nucleotide polymorphism panel. Animals 2019, 9, 350. [Google Scholar] [CrossRef]
- Nandolo, W.; Mészáros, G.; Banda, L.J.; Gondwe, T.N.; Lamuno, D.; Mulindwa, H.A.; Nakimbugwe, H.N.; Wurzinger, M.; Utsunomiya, Y.T.; Woodward-Greene, M.J.; et al. Timing and extent of inbreeding in African goats. Front. Genet. 2019, 10, 537. [Google Scholar] [CrossRef]
- Zhao, Q.; Huang, C.; Chen, Q.; Su, Y.; Zhang, Y.; Wang, R.; Su, R.; Xu, H.; Liu, S.; Ma, Y.; et al. Genomic inbreeding and runs of homozygosity analysis of cashmere goat. Animals 2024, 14, 1246. [Google Scholar] [CrossRef]
- McQuillan, R.; Leutenegger, A.-L.; Abdel-Rahman, R.; Franklin, C.S.; Pericic, M.; Barac-Lauc, L.; Smolej-Narancic, N.; Janicijevic, B.; Polasek, O.; Tenesa, A.; et al. Runs of homozygosity in European populations. Am. J. Hum. Genet. 2008, 83, 359–372. [Google Scholar] [CrossRef] [PubMed]
- Bahbahani, H.; Clifford, H.; Wragg, D.; Mbole-Kariuki, M.N.; Van Tassell, C.; Sonstegard, T.; Woolhouse, M.; Hanotte, O. Signatures of positive selection in East African Shorthorn Zebu: A genome-wide single nucleotide polymorphism analysis. Sci. Rep. 2015, 5, 11729. [Google Scholar] [CrossRef]
- Bahbahani, H.; Salim, B.; Almathen, F.; Al Enezi, F.; Mwacharo, J.M.; Hanotte, O. Signatures of positive selection in African Butana and Kenana dairy zebu cattle. PLoS ONE 2018, 13, e0190446. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.Y.; Savegnago, R.P.; Lim, D.; Lee, S.H.; Gondro, C. Signatures of selection in Angus and Hanwoo beef cattle using imputed whole genome sequence data. Front. Genet. 2024, 15, 1368710. [Google Scholar] [CrossRef]
- Saravanan, K.A.; Panigrahi, M.; Kumar, H.; Bhushan, B.; Dutt, T.; Mishra, B.P. Genome-wide analysis of genetic diversity and selection signatures in three Indian sheep breeds. Livest. Sci. 2021, 243, 104367. [Google Scholar] [CrossRef]
- Li, T.; Jin, M.; Wang, H.; Zhang, W.; Yuan, Z.; Wei, C. Whole-Genome Scanning for Selection Signatures Reveals Candidate Genes Associated with Growth and Tail Length in Sheep. Animals 2024, 14, 687. [Google Scholar] [CrossRef]
- Almathen, F. Genomic signatures of positive selection in Awarik dromedary camels from southwestern of Saudi Arabia. Front. Vet. Sci. 2024, 11, 1443748. [Google Scholar] [CrossRef]
- Bahbahani, H.; Alfoudari, A.; Al-Ateeqi, A.; Al Abri, M.; Almathen, F. Positive selection footprints and haplotype distribution in the genome of dromedary camels. Animal 2024, 18, 101098. [Google Scholar] [CrossRef]
- Al Abri, M.; Alfoudari, A.; Mohammad, Z.; Almathen, F.; Al-Marzooqi, W.; Al-Hajri, S.; Al-Amri, M.; Bahbahani, H. Assessing genetic diversity and defining signatures of positive selection on the genome of dromedary camels from the southeast of the Arabian Peninsula. Front. Vet. Sci. 2023, 10, 1296610. [Google Scholar] [CrossRef]
- Brito, L.F.; Kijas, J.W.; Ventura, R.V.; Sargolzaei, M.; Porto-Neto, L.R.; Cánovas, A.; Feng, Z.; Jafarikia, M.; Schenkel, F.S. Genetic diversity and signatures of selection in various goat breeds revealed by genome-wide SNP markers. BMC Genom. 2017, 18, 229. [Google Scholar] [CrossRef]
- Waineina, R.W.; Okeno, T.O.; Ilatsia, E.D.; Ngeno, K. Selection signature analyses revealed genes associated with adaptation, production, and reproduction in selected goat breeds in Kenya. Front. Genet. 2022, 13, 858923. [Google Scholar] [CrossRef]
- Li, G.; Tang, J.; Huang, J.; Jiang, Y.; Fan, Y.; Wang, X.; Ren, J. Genome-wide estimates of runs of homozygosity, heterozygosity, and genetic load in two Chinese indigenous goat breeds. Front. Genet. 2022, 13, 774196. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.; Zhang, Y.; Gao, L.; Wang, S.; Liu, M.; Sun, E.; Lu, K.; Zhang, Y.; Li, B.; Li, G.; et al. Examination of homozygosity runs and selection signatures in native goat breeds of Henan, China. BMC Genom. 2024, 25, 1184. [Google Scholar] [CrossRef]
- Onzima, R.B.; Upadhyay, M.R.; Doekes, H.P.; Brito, L.F.; Bosse, M.; Kanis, E.; Groenen, M.A.M.; Crooijmans, R.P.M.A. Genome-wide characterization of selection signatures and runs of homozygosity in Ugandan goat breeds. Front. Genet. 2018, 9, 318. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Yao, N.; Yang, M.; Liu, X.; Dong, K.; Zhao, Q.; Pu, Y.; He, X.; Guan, W.; Yang, N.; et al. Exome sequencing reveals genetic differentiation due to high-altitude adaptation in the Tibetan cashmere goat (Capra hircus). BMC Genom. 2016, 17, 122. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Tao, H.; Li, P.; Li, L.; Zhong, T.; Wang, L.; Ma, J.; Chen, X.; Song, T.; Zhang, H. Whole-genome sequencing reveals selection signatures associated with important traits in six goat breeds. Sci. Rep. 2018, 8, 10405. [Google Scholar] [CrossRef]
- Jin, M.; Lu, J.; Fei, X.; Lu, Z.; Quan, K.; Liu, Y.; Chu, M.; Di, R.; Wei, C.; Wang, H. Selection signatures analysis reveals genes associated with high-altitude adaptation in Tibetan goats from Nagqu, Tibet. Animals 2020, 10, 1599. [Google Scholar] [CrossRef]
- Zhong, T.; Wang, X.; Huang, C.; Yang, L.; Zhao, Q.; Chen, X.; Freitas-De-Melo, A.; Zhan, S.; Wang, L.; Dai, D.; et al. A genome-wide perspective on the diversity and selection signatures in indigenous goats using 53 K single nucleotide polymorphism array. Animal 2023, 17, 100706. [Google Scholar] [CrossRef]
- Colli, L.; Milanesi, M.; Talenti, A.; Bertolini, F.; Chen, M.; Crisà, A.; Daly, K.G.; Del Corvo, M.; Guldbrandtsen, B.; Lenstra, J.A.; et al. Genome-wide SNP profiling of worldwide goat populations reveals strong partitioning of diversity and highlights post-domestication migration routes. Genet. Sel. Evol. 2018, 50, 58. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Aulchenko, Y.S.; Ripke, S.; Isaacs, A.; Van Duijn, C.M. GenABEL: An R library for genome-wide association analysis. Bioinformatics 2007, 23, 1294–1296. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2024. [Google Scholar]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Wickham, H.; Sievert, C. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009; Volume 10. [Google Scholar]
- Barbato, M.; Orozco-Terwengel, P.; Tapio, M.; Bruford, M.W. SNeP: A tool to estimate trends in recent effective population size trajectories using genome-wide SNP data. Front. Genet. 2015, 6, 109. [Google Scholar] [CrossRef]
- Voight, B.F.; Kudaravalli, S.; Wen, X.; Pritchard, J.K.; Hurst, L. A map of recent positive selection in the human genome. PLoS Biol. 2006, 4, e72. [Google Scholar]
- Tang, H.; Choudhry, S.; Mei, R.; Morgan, M.; Rodriguez-Cintron, W.; Burchard, E.G.; Risch, N.J. Recent genetic selection in the ancestral admixture of Puerto Ricans. Am. J. Hum. Genet. 2007, 81, 626–633. [Google Scholar] [CrossRef]
- Santos, W.B.; Schettini, G.P.; Maiorano, A.M.; Bussiman, F.O.; Balieiro, J.C.C.; Ferraz, G.C.; Pereira, G.L.; Baldassini, W.A.; Neto, O.R.M.; Oliveira, H.N.; et al. Genome-wide scans for signatures of selection in Mangalarga Marchador horses using high-throughput SNP genotyping. BMC Genom. 2021, 22, 737. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Browning, B.L.; Tian, X.; Zhou, Y.; Browning, S.R. Fast two-stage phasing of large-scale sequence data. Am. J. Hum. Genet. 2021, 108, 1880–1890. [Google Scholar] [CrossRef]
- Gautier, M.; Vitalis, R. rehh: An R package to detect footprints of selection in genome-wide SNP data from haplotype structure. Bioinformatics 2012, 28, 1176–1177. [Google Scholar] [CrossRef]
- Verity, R.; Collins, C.; Card, D.C.; Schaal, S.M.; Wang, L.; Lotterhos, K.E. minotaur: A platform for the analysis and visualization of multivariate results from genome scans with R Shiny. Mol. Ecol. Resour. 2017, 17, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Reimand, J.; Kull, M.; Peterson, H.; Hansen, J.; Vilo, J. g: Profiler—A web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007, 35 (Suppl. 2), W193–W200. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef] [PubMed]
- Kirin, M.; McQuillan, R.; Franklin, C.S.; Campbell, H.; McKeigue, P.M.; Wilson, J.F. Genomic runs of homozygosity record population history and consanguinity. PLoS ONE 2010, 5, e13996. [Google Scholar] [CrossRef]
- Cardoso, T.F.; Amills, M.; Bertolini, F.; Rothschild, M.; Marras, G.; Boink, G.; Jordana, J.; Capote, J.; Carolan, S.; Hallsson, J.H.; et al. Patterns of homozygosity in insular and continental goat breeds. Genet. Sel. Evol. 2018, 50, 56. [Google Scholar] [CrossRef]
- Oleksyk, T.K.; Smith, M.W.; O’Brien, S.J. Genome-wide scans for footprints of natural selection. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 185–205. [Google Scholar] [CrossRef]
- Beall, C.M.; Cavalleri, G.L.; Deng, L.; Elston, R.C.; Gao, Y.; Knight, J.; Li, C.; Li, J.C.; Liang, Y.; McCormack, M.; et al. Natural selection on EPAS1 (HIF2α) associated with low hemoglobin concentration in Tibetan highlanders. Proc. Natl. Acad. Sci. USA 2010, 107, 11459–11464. [Google Scholar] [CrossRef]
- Bigham, A.; Bauchet, M.; Pinto, D.; Mao, X.; Akey, J.M.; Mei, R.; Scherer, S.W.; Julian, C.G.; Wilson, M.J.; Herráez, D.L.; et al. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 2010, 6, e1001116. [Google Scholar] [CrossRef]
- Lukyanova, L.D.; Kirova, Y.I. Mitochondria-controlled signaling mechanisms of brain protection in hypoxia. Front. Neurosci. 2015, 9, 320. [Google Scholar] [CrossRef]
- Lukyanova, L.; Kirova, Y.I.; Germanova, E. Specific features of immediate expression of succinate-dependent receptor GPR91 in tissues during hypoxia. Bull. Exp. Biol. Med. 2016, 160, 742–747. [Google Scholar] [CrossRef]
- Abdulmalek, K.; Ashur, F.; Ezer, N.; Ye, F.; Magder, S.; Hussain, S.N.A. Differential expression of Tie-2 receptors and angiopoietins in response to in vivo hypoxia in rats. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2001, 281, L582–L590. [Google Scholar] [CrossRef]
- Louphrasitthiphol, P.; Ledaki, I.; Chauhan, J.; Falletta, P.; Siddaway, R.; Buffa, F.M.; Mole, D.R.; Soga, T.; Goding, C.R. MITF controls the TCA cycle to modulate the melanoma hypoxia response. Pigment. Cell Melanoma Res. 2019, 32, 792–808. [Google Scholar] [CrossRef] [PubMed]
- Du Puy, L.; Beqqali, A.; Monshouwer-Kloots, J.; Haagsman, H.P.; Roelen, B.A.J.; Passier, R. CAZIP, a novel protein expressed in the developing heart and nervous system. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2009, 238, 2903–2911. [Google Scholar] [CrossRef]
- Arora, R.; Kaur, M.; Kumar, A.; Chhabra, P.; Mir, M.A.; Ahlawat, S.; Singh, M.K.; Sharma, R.; Gera, R. Skeletal muscle transcriptomics of sheep acclimated to cold desert and tropical regions identifies genes and pathways accentuating their diversity. Int. J. Biometeorol. 2024, 68, 1811–1821. [Google Scholar] [CrossRef]
- Bahbahani, H.; Mohammad, Z.; Alfoudari, A.; Al Abri, M. Genomic insights into racing camels: Inbreeding levels and positive selection linked to athletic traits. Animal 2025, 19, 101467. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; He, S.; Wang, Q.; Li, F.; Kwak, M.-J.; Chen, S.; O’connell, D.; Zhang, T.; Pirooz, S.D.; Jeon, Y.H.; et al. Autophagic UVRAG Promotes UV-Induced Photolesion Repair by Activation of the CRL4(DDB2) E3 Ligase. Mol. Cell 2016, 62, 507–519. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Lei, B.; Liu, Y. An Analysis of a Transposable Element Expression Atlas during 27 Developmental Stages in Porcine Skeletal Muscle: Unveiling Molecular Insights into Pork Production Traits. Animals 2023, 13, 3581. [Google Scholar] [CrossRef]
- Hicks, M.R.; Saleh, K.K.; Clock, B.; Gibbs, D.E.; Yang, M.; Younesi, S.; Gane, L.; Gutierrez-Garcia, V.; Xi, H.; Pyle, A.D. Regenerating human skeletal muscle forms an emerging niche in vivo to support PAX7 cells. Nat. Cell Biol. 2023, 25, 1758–1773. [Google Scholar] [CrossRef]
- Morishima, N.; Ito, Y. Calpain-5 regulates muscle-specific protein expression and nuclear positioning during myoblast differentiation. J. Biol. Chem. 2024, 300, 107842. [Google Scholar] [CrossRef]
- Yang, L.; Wang, Y.; Sun, N.; Chen, J.; He, S. Genomic and functional evidence reveals convergent evolution in fishes on the Tibetan Plateau. Mol. Ecol. 2021, 30, 5752–5764. [Google Scholar] [CrossRef]
- Ji, K.; Jiao, D.; Yang, G.; Degen, A.A.; Zhou, J.; Liu, H.; Wang, W.; Cong, H. Transcriptome analysis revealed potential genes involved in thermogenesis in muscle tissue in cold-exposed lambs. Front. Genet. 2022, 13, 1017458. [Google Scholar] [CrossRef] [PubMed]
- Alhajeri, B.H.; Steppan, S.J. A phylogenetic test of adaptation to deserts and aridity in skull and dental morphology across rodents. J. Mammal. 2018, 99, 1197–1216. [Google Scholar] [CrossRef]
- Bahbahani, H.; Musa, H.H.; Wragg, D.; Shuiep, E.S.; Almathen, F.; Hanotte, O. Genome diversity and signatures of selection for production and performance traits in dromedary camels. Front. Genet. 2019, 10, 893. [Google Scholar] [CrossRef]
- Sebastian, A.; Loots, G.G. Transcriptional control of Sost in bone. Bone 2017, 96, 76–84. [Google Scholar] [CrossRef]
- Dauer, M.V.; Currie, P.D.; Berger, J. Skeletal malformations of Meox1-deficient zebrafish resemble human Klippel–Feil syndrome. J. Anat. 2018, 233, 687–695. [Google Scholar] [CrossRef]
- Gu, X.; Chang, X.; Yang, L.; Chamba, Y.; Geng, F. Quantitative Proteomic Analysis of Tibetan Pig Livers at Different Altitudes. Molecules 2023, 28, 1694. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, L.; Han, J.; Tang, X.; Ma, M.; Wang, K.; Zhang, X.; Ren, Q.; Chen, Q.; Qiu, Q. Comparative transcriptomic analysis revealed adaptation mechanism of Phrynocephalus erythrurus, the highest altitude Lizard living in the Qinghai-Tibet Plateau. BMC Evol. Biol. 2015, 15, 101. [Google Scholar] [CrossRef]
- Wu, H.; Guang, X.; Al-Fageeh, M.B.; Cao, J.; Pan, S.; Zhou, H.; Zhang, L.; AbuTarboush, M.H.; Xing, Y.; Xie, Z.; et al. Camelid genomes reveal evolution and adaptation to desert environments. Nat. Commun. 2014, 5, 5188. [Google Scholar] [CrossRef]
- Jin, Y.; McFie, P.J.; Banman, S.L.; Brandt, C.; Stone, S.J. Diacylglycerol acyltransferase-2 (DGAT2) and monoacylglycerol acyltransferase-2 (MGAT2) interact to promote triacylglycerol synthesis. J. Biol. Chem. 2014, 289, 28237–28248. [Google Scholar] [CrossRef]
- Zhang, J.-F.; Choi, S.-H.; Li, Q.; Wang, Y.; Sun, B.; Tang, L.; Wang, E.-Z.; Hua, H.; Li, X.-Z. Overexpression of DGAT2 stimulates lipid droplet formation and triacylglycerol accumulation in bovine satellite cells. Animals 2022, 12, 1847. [Google Scholar] [CrossRef]
- Liu, Q.; Li, J.; Zhang, W.; Xiao, C.; Zhang, S.; Nian, C.; Li, J.; Su, D.; Chen, L.; Zhao, Q.; et al. Glycogen accumulation and phase separation drives liver tumor initiation. Cell 2021, 184, 5559–5576.e19. [Google Scholar] [CrossRef] [PubMed]
- Velez-Irizarry, D.; Casiro, S.; Daza, K.R.; Bates, R.O.; Raney, N.E.; Steibel, J.P.; Ernst, C.W. Genetic control of longissimus dorsi muscle gene expression variation and joint analysis with phenotypic quantitative trait loci in pigs. BMC Genom. 2019, 20, 3. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Shi, B.; Xie, J.; Zhou, H.; Wang, J.; Liu, X.; Li, S.; Zhao, Z.; Luo, Y. Tissue Expression and Variation of the DGAT2 Gene and Its Effect on Carcass and Meat Quality Traits in Yak. Animals 2019, 9, 61. [Google Scholar] [CrossRef]
- Lu, H.; Wang, R.; Li, W.; Xie, H.; Wang, C.; Hao, Y.; Sun, Y.; Jia, Z. Plasma proteomic study of acute mountain sickness susceptible and resistant individuals. Sci. Rep. 2018, 8, 1265. [Google Scholar] [CrossRef]
- Blumthaler, M.; Ambach, W.; Ellinger, R. Increase in solar UV radiation with altitude. J. Photochem. Photobiol. B Biol. 1997, 39, 130–134. [Google Scholar] [CrossRef]
- Rocha, J.L.; Silva, P.; Santos, N.; Nakamura, M.; Afonso, S.; Qninba, A.; Boratynski, Z.; Sudmant, P.H.; Brito, J.C.; Nielsen, R.; et al. North African fox genomes show signatures of repeated introgression and adaptation to life in deserts. Nat. Ecol. Evol. 2023, 7, 1267–1286. [Google Scholar] [CrossRef]
- Pathania, S.; Nguyen, J.; Hill, S.J.; Scully, R.; Feunteun, J.; Livingston, D.M. BRCA1 is required for postreplication repair after UV-induced DNA damage. Mol. Cell 2011, 44, 235–251. [Google Scholar] [CrossRef]
- Perrett, R.M.; McArdle, C.A. Molecular mechanisms of gonadotropin-releasing hormone signaling: Integrating cyclic nucleotides into the network. Front. Endocrinol. 2013, 4, 180. [Google Scholar] [CrossRef]
- van der Meer, T.; Chan, W.I.; Palazon, L.S.; Nieduszynski, C.; Murphy, M.; Sobczak-Thépot, J.; Carrington, M.; Colledge, W.H. Cyclin A1 protein shows haplo-insufficiency for normal fertility in male mice. Reproduction 2004, 127, 503–511. [Google Scholar] [CrossRef]
- Huang, C.; Wu, D.; Khan, F.A.; Jiao, X.; Guan, K.; Huo, L. The GTPase SPAG-1 orchestrates meiotic program by dictating meiotic resumption and cytoskeleton architecture in mouse oocytes. Mol. Biol. Cell 2016, 27, 1776–1785. [Google Scholar] [CrossRef]
- Cao, M.; Wang, X.; Guo, S.; Kang, Y.; Pei, J.; Guo, X. F1 Male Sterility in Cattle-Yak Examined through Changes in Testis Tissue and Transcriptome Profiles. Animals 2022, 12, 2711. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Singh, S.B.; Sarkar, S. Genome Wide Expression Analysis Suggests Perturbation of Vascular Homeostasis during High Altitude Pulmonary Edema. PLoS ONE 2014, 9, e85902. [Google Scholar] [CrossRef] [PubMed]
- Ai, H.; Yang, B.; Li, J.; Xie, X.; Chen, H.; Ren, J. Population history and genomic signatures for high-altitude adaptation in Tibetan pigs. BMC Genom. 2014, 15, 834. [Google Scholar] [CrossRef]
- Edea, Z.; Dadi, H.; Dessie, T.; Kim, K.-S. Genomic signatures of high-altitude adaptation in Ethiopian sheep populations. Genes Genom. 2019, 41, 973–981. [Google Scholar] [CrossRef]
- Hou, L.; Pavan, W.J. Transcriptional and signaling regulation in neural crest stem cell-derived melanocyte development: Do all roads lead to Mitf? Cell Res. 2008, 18, 1163–1176. [Google Scholar] [CrossRef]
- Kawakami, A.; Fisher, D.E. The master role of microphthalmia-associated transcription factor in melanocyte and melanoma biology. Lab. Investig. 2017, 97, 649–656. [Google Scholar] [CrossRef]
- Tosser-Klopp, G.; Bardou, P.; Bouchez, O.; Cabau, C.; Crooijmans, R.; Dong, Y.; Donnadieu-Tonon, C.; Eggen, A.; Heuven, H.C.M.; Jamli, S.; et al. Design and characterization of a 52K SNP chip for goats. PLoS ONE 2014, 9, e86227. [Google Scholar] [CrossRef]
- Kardos, M.; Åkesson, M.; Fountain, T.; Flagstad, Ø.; Liberg, O.; Olason, P.; Sand, H.; Wabakken, P.; Wikenros, C.; Ellegren, H. Genomic consequences of intensive inbreeding in an isolated wolf population. Nat. Ecol. Evol. 2018, 2, 124–131. [Google Scholar] [CrossRef]


| Quality Control Criteria | Dataset 1 | Dataset 2 |
|---|---|---|
| Genotyping call rate | 19,059 | 2126 |
| Hardy-Weinberg Equilibrium | 121 | 29 |
| Minor allele frequency * | 998 | 5589 |
| Linkage disequilibrium ** | 584 | 7781 |
| Functional Category | Gene ID | Gene Name | Candidate Region Chr: Start-Stop (bp) | Identifying Statistic(s) |
|---|---|---|---|---|
| Hypoxia Tolerance | SUCNR1 | Succinate receptor 1 | 1: 115,626,920–115,636,639 | Rsb |
| ANGPTL1 | Angiopoietin-like 1 | 16: 58,825,678–58,849,019 | iHS | |
| MITF | Microphthalmia-Associated Transcription Factor | 22: 31,524,691–31,755,591 | ROH | |
| MTUS2 | Microtubule-Associated Scaffold Protein 2 | 12: 55,325,810–55,710,530 | ROH | |
| Muscle Development and Function | MBNL1 | Muscleblind-Like Protein 1 | 1: 114,974,326–115,192,808 | Rsb |
| ACTC1 | Actin Alpha Cardiac Muscle 1 | 10: 72,761,190–72,766,594 | ROH | |
| CAPN5 | Calpain-5 | 15: 26,421,548–26,478,072 | iHS | |
| Lipid Metabolism | DGAT2 | Diacylglycerol O-Acyltransferase 2 | 15: 27,720,879–27,753,736 | Rsb |
| G6PC | Glucose-6-Phosphatase | 19: 42,622,248–42,632,787 | iHS | |
| SUCLG2 | Succinate-CoA Ligase GDP-Forming Subunit Beta | 22: 33,808,508–34,098,836 | iHS and ROH | |
| UV Radiation Resistance | UVRAG | UV Radiation Resistance-Associated Gene | 15: 27,379,066–27,699,522 | Rsb |
| BRCA1 | BRCA1 DNA Repair Associated | 19: 42,742,613–42,809,204 | iHS | |
| Fertility | GNRHR | Gonadotrophin-Releasing Hormone Receptor | 6: 84,144,226–84,161,854 | Rsb |
| CCNA1 | Cyclin A1 | 12: 61,577,282–61,587,501 | ROH | |
| SPAG1 | Sperm-Associated Antigen 1 | 14: 17,990,033–18,077,090 | ROH | |
| Bone Development | SOST | Sclerostin | 19: 43,286,595–43,291,485 | iHS |
| MEOX1 | Mesenchyme Homeobox 1 | 19: 43,186,727–43,205,838 | iHS |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohammad, Z.; Bahbahani, H.; Alfoudari, A.; Al Kharousi, K.; Al Hamrashdi, A.A.; Al Toobi, A.G.; Al Abri, M. Genomic Analysis of Adaptability and Genetic Structure of Jabal Akhdar Goats: Evidence of Positive Selection in an Indigenous Omani Breed. Biology 2025, 14, 761. https://doi.org/10.3390/biology14070761
Mohammad Z, Bahbahani H, Alfoudari A, Al Kharousi K, Al Hamrashdi AA, Al Toobi AG, Al Abri M. Genomic Analysis of Adaptability and Genetic Structure of Jabal Akhdar Goats: Evidence of Positive Selection in an Indigenous Omani Breed. Biology. 2025; 14(7):761. https://doi.org/10.3390/biology14070761
Chicago/Turabian StyleMohammad, Zainab, Hussain Bahbahani, Ahmad Alfoudari, Kaadhia Al Kharousi, Al Abeer Al Hamrashdi, Al Ghalya Al Toobi, and Mohammad Al Abri. 2025. "Genomic Analysis of Adaptability and Genetic Structure of Jabal Akhdar Goats: Evidence of Positive Selection in an Indigenous Omani Breed" Biology 14, no. 7: 761. https://doi.org/10.3390/biology14070761
APA StyleMohammad, Z., Bahbahani, H., Alfoudari, A., Al Kharousi, K., Al Hamrashdi, A. A., Al Toobi, A. G., & Al Abri, M. (2025). Genomic Analysis of Adaptability and Genetic Structure of Jabal Akhdar Goats: Evidence of Positive Selection in an Indigenous Omani Breed. Biology, 14(7), 761. https://doi.org/10.3390/biology14070761






