Exploring the Habitat Distribution of Decapterus macarellus in the South China Sea Under Varying Spatial Resolutions: A Combined Approach Using Multiple Machine Learning and the MaxEnt Model
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
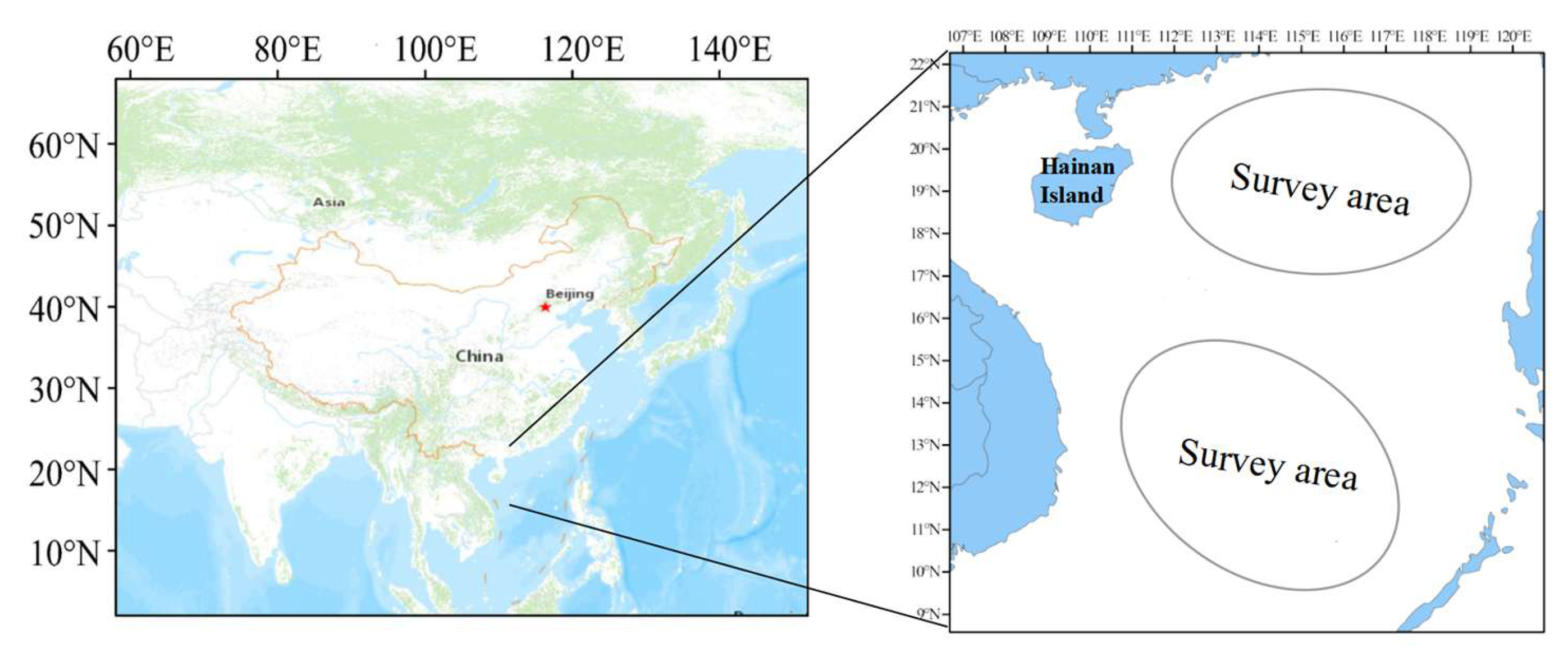
2.1. Sampling Information of D. macarellus
2.2. Screening of Environmental Data
2.3. Model Construction and Performance Evaluation
2.4. Model Parameter Settings
3. Results
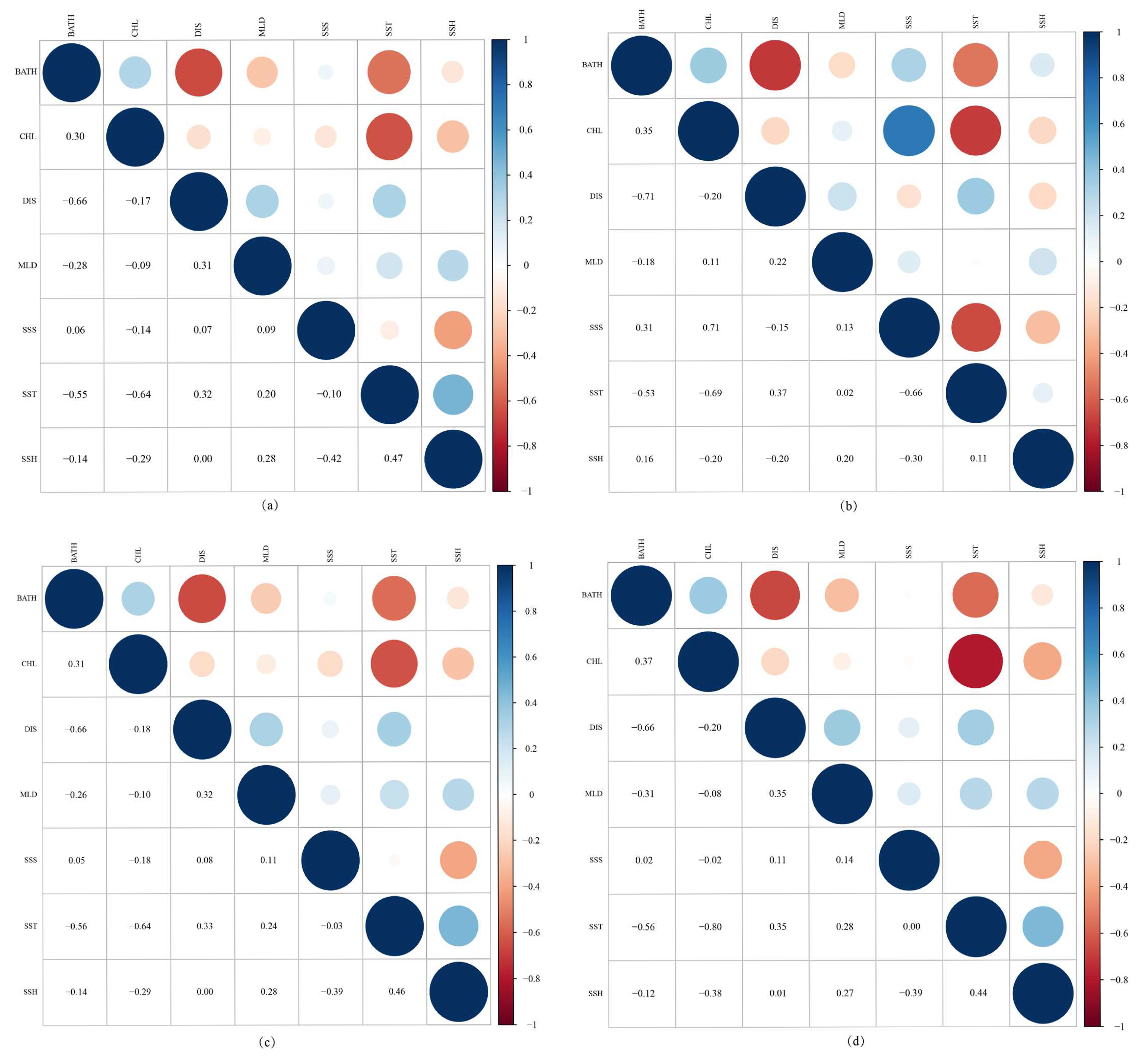
3.1. Screening Factors
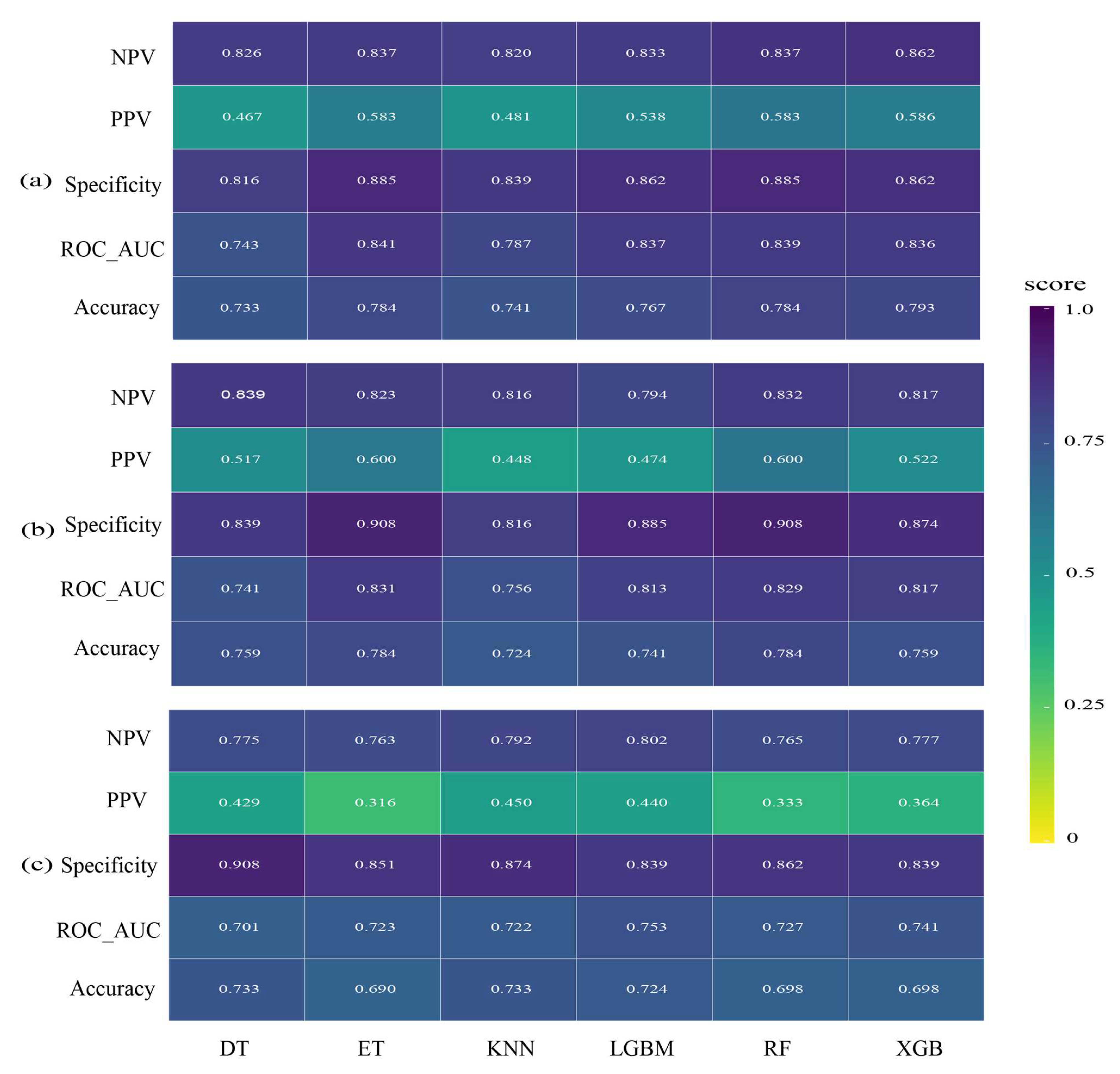
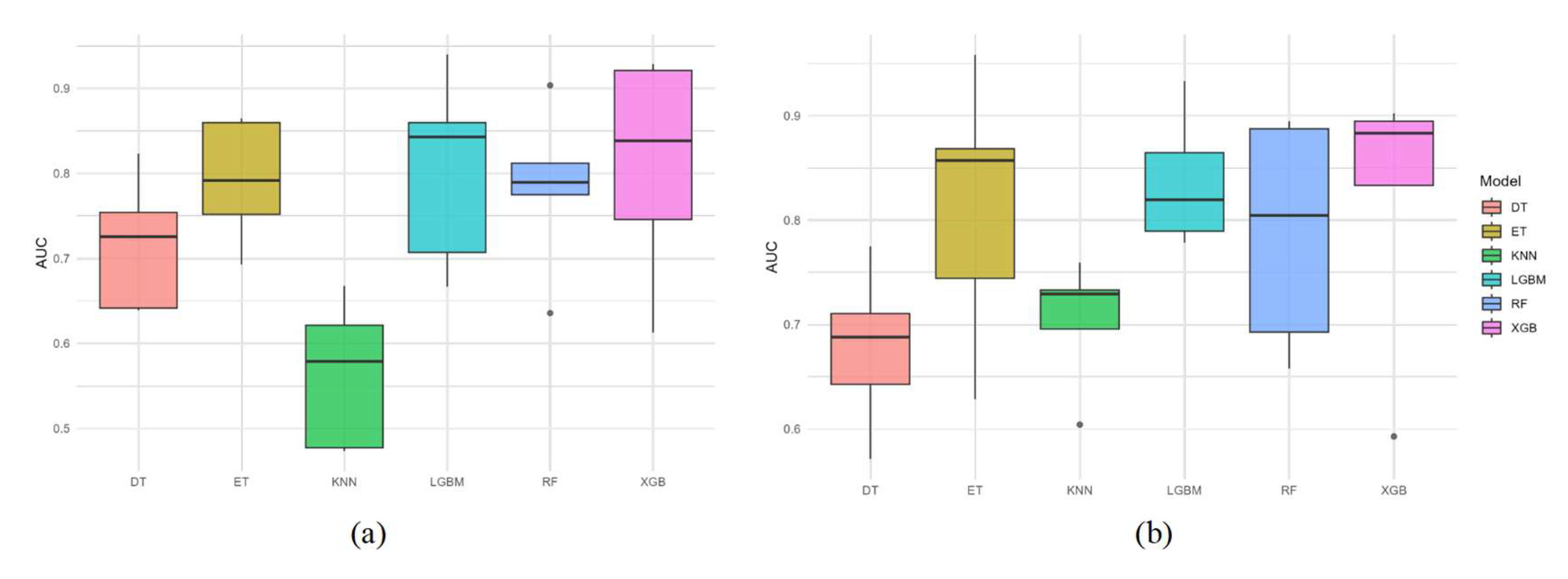
3.2. Model Performance
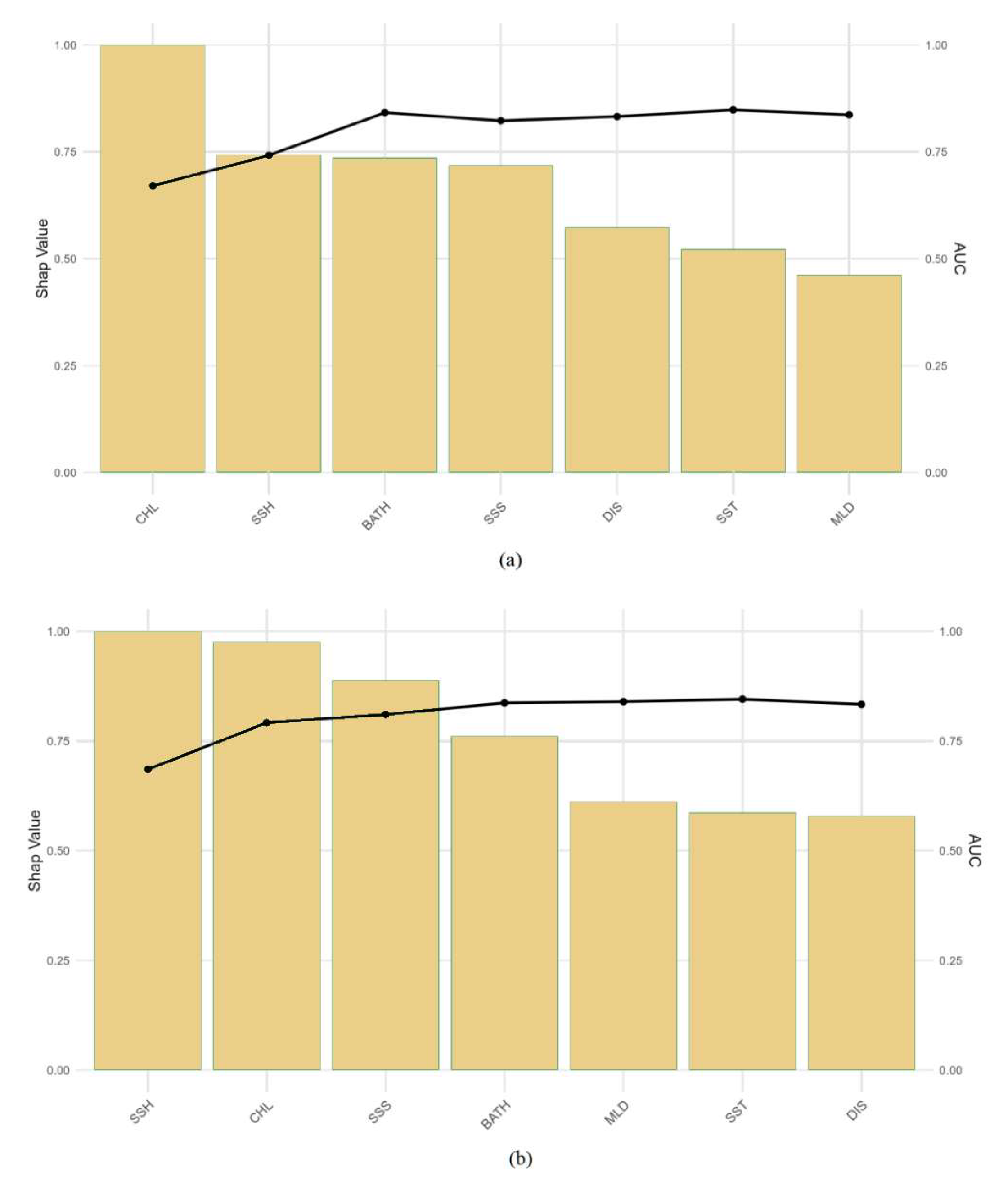
3.3. Analysis of Variable Importance
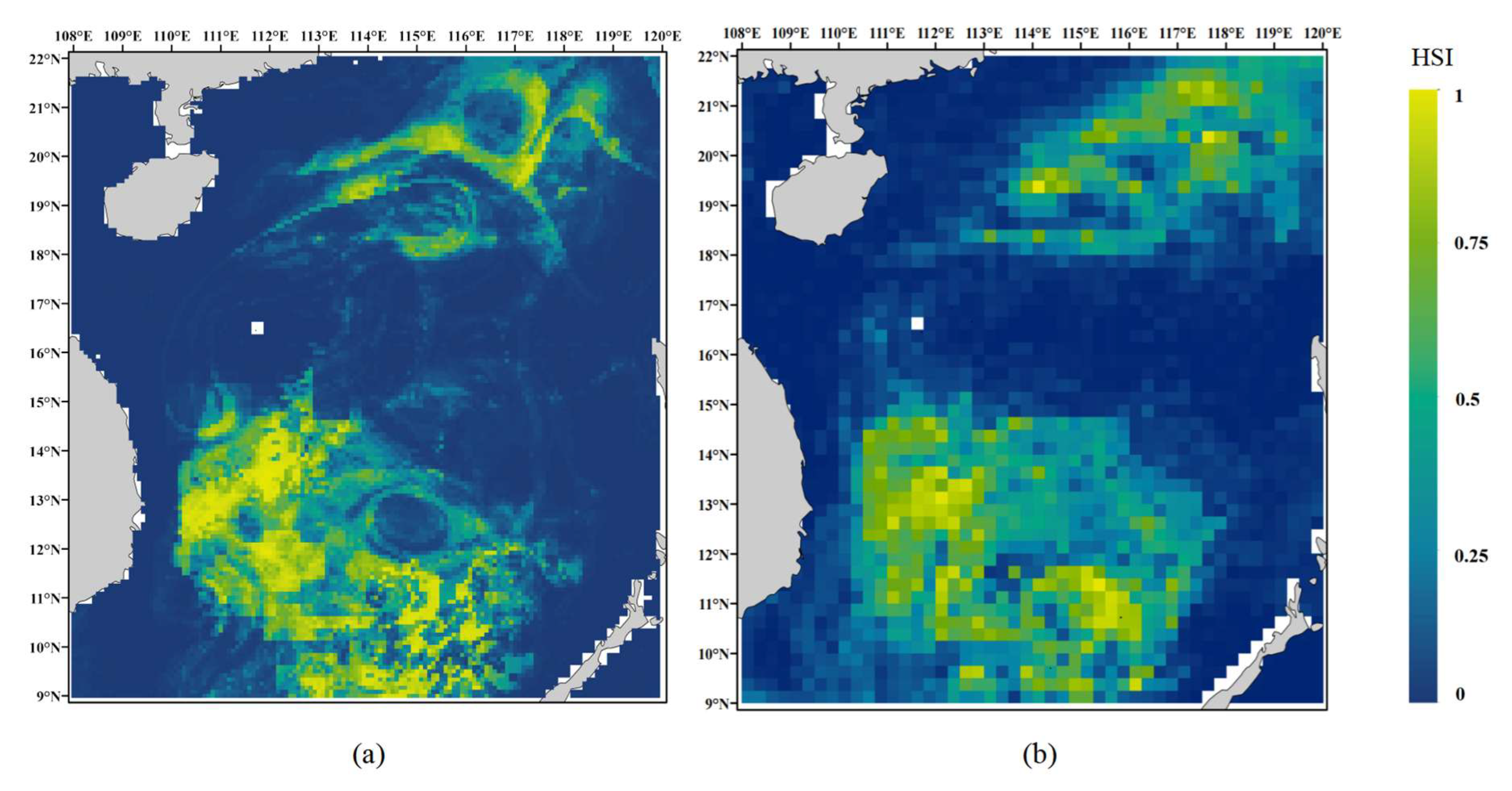
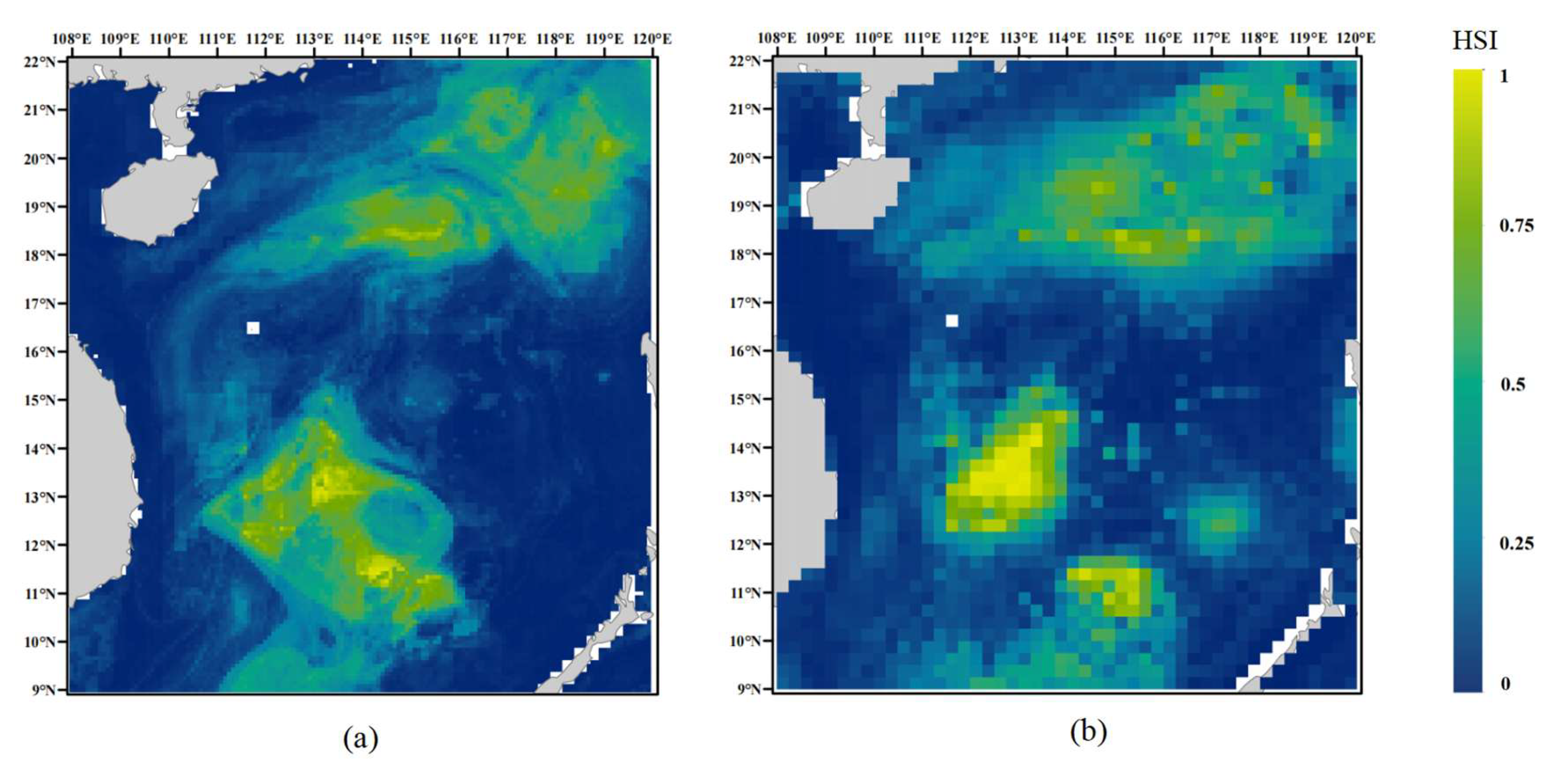
3.4. Habitat Suitability Prediction for D. macarellus in the South China Sea
3.5. External Validation
4. Discussion
4.1. Justification for the Selection of Environmental Variables
4.2. Model Performance Comparison
4.3. Comparison of Spatial Resolution Effects
4.4. Ecological Rationality of Predicted Potential Habitats
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kang, B.; Pecl, G.T.; Lin, L.; Sun, P.; Zhang, P.; Li, Y.; Zhao, L.; Peng, X.; Yan, Y.; Shen, C. Climate change impacts on China’s marine ecosystems. Rev. Fish Biol. Fish. 2021, 31, 599–629. [Google Scholar] [CrossRef]
- Li, Y.; Sun, M.; Yang, X.; Yang, M.; Kleisner, K.M.; Mills, K.E.; Tang, Y.; Du, F.; Qiu, Y.; Ren, Y. Social–ecological vulnerability and risk of China’s marine capture fisheries to climate change. Proc. Natl. Acad. Sci. USA 2024, 121, e2313773120. [Google Scholar] [CrossRef] [PubMed]
- Moustahfid, H.; Hendrickson, L.C.; Arkhipkin, A.; Pierce, G.J.; Gangopadhyay, A.; Kidokoro, H.; Markaida, U.; Nigmatullin, C.; Sauer, W.H.; Jereb, P. Ecological-fishery forecasting of squid stock dynamics under climate variability and change: Review, challenges, and recommendations. Rev. Fish. Sci. Aquac. 2021, 29, 682–705. [Google Scholar] [CrossRef]
- Mondal, S.; Punt, A.E.; Mendes, D.; Osuka, K.E.; Lee, M.A. Teleconnection Impacts of Climatic Variability on Tuna and Billfish Fisheries of the South Atlantic and Indian Ocean: A Study Towards Sustainable Fisheries Management. Fish Fish. 2025, 26, 240–256. [Google Scholar] [CrossRef]
- Yati, E.; Sadiyah, L.; Satria, F.; Alabia, I.D.; Sulma, S.; Prayogo, T.; Marpaung, S.; Harsa, H.; Kushardono, D.; Lumban-Gaol, J. Spatial distribution models for the four commercial tuna in the sea of maritime continent using multi-sensor remote sensing and maximum entropy. Mar. Environ. Res. 2024, 198, 106540. [Google Scholar] [CrossRef]
- Belkin, I.M. Remote sensing of ocean fronts in marine ecology and fisheries. Remote Sens. 2021, 13, 883. [Google Scholar] [CrossRef]
- Xue, M.; Tong, J.; Ma, W.; Zhu, Z.; Wang, W.; Lyu, S.; Chen, X. Reimagining habitat suitability modeling for Pacific saury (Cololabis saira) in the Northwest Pacific Ocean through acoustic data analysis from fishing vessels. Ecol. Inform. 2025, 85, 102971. [Google Scholar] [CrossRef]
- Shi, Y.; Kang, B.; Fan, W.; Xu, L.; Zhang, S.; Cui, X.; Dai, Y. Spatio-temporal variations in the potential habitat distribution of pacific sardine (Sardinops sagax) in the Northwest Pacific Ocean. Fishes 2023, 8, 86. [Google Scholar] [CrossRef]
- Braun, C.D.; Lezama-Ochoa, N.; Farchadi, N.; Arostegui, M.C.; Alexander, M.; Allyn, A.; Bograd, S.J.; Brodie, S.; Crear, D.P.; Curtis, T.H. Widespread habitat loss and redistribution of marine top predators in a changing ocean. Sci. Adv. 2023, 9, eadi2718. [Google Scholar] [CrossRef]
- Yang, S.; Chen, D.; Deng, K. Global effects of climate change in the South China Sea and its surrounding areas. Ocean-Land-Atmos. Res. 2024, 3, 0038. [Google Scholar] [CrossRef]
- Deng, L.; Zhao, J.; Sun, S.; Ai, B.; Zhou, W.; Cao, W. Two-decade satellite observations reveal variability in size-fractionated phytoplankton primary production in the South China Sea. Deep Sea Res. Part I Oceanogr. Res. Pap. 2024, 206, 104258. [Google Scholar] [CrossRef]
- Peng, R.; Chen, X.; Wu, Q.; Yan, Z.; Fu, Y.; Qin, B.; Hao, R.; Yu, K. Wildfire particulates enhance phytoplankton growth and alter communities in the South China Sea under wind-driven upwelling. J. Geophys. Res. Biogeosciences 2024, 129, e2024JG008066. [Google Scholar] [CrossRef]
- Wang, D.K.-H. Fisheries management in the South China Sea. In Routledge Handbook of the South China Sea; Routledge: London, UK, 2021; pp. 243–261. [Google Scholar]
- Song, J.; Yao, L.; Guo, J.; Fu, Y.; Cai, Y.; Wang, M. The Seasonal Correlation Between El Niño and Southern Oscillation Events and Sea Surface Temperature Anomalies in the South China Sea from 1958 to 2024. J. Mar. Sci. Eng. 2025, 13, 153. [Google Scholar] [CrossRef]
- Wang, Y.; Su, C.; Zhang, J.; He, J.; Liu, W.; Wang, T. Causation, impacts and mitigation strategies of typical marine ecological disasters in coasts of the South China Sea. Reg. Stud. Mar. Sci. 2025, 85, 104167. [Google Scholar] [CrossRef]
- Xiao, J.; Wang, W.; Wang, X.; Tian, P.; Niu, W. Recent deterioration of coral reefs in the South China Sea due to multiple disturbances. PeerJ 2022, 10, e13634. [Google Scholar] [CrossRef]
- Shi, J.; Li, C.; Wang, T.; Zhao, J.; Liu, Y.; Xiao, Y. Distribution pattern of coral reef fishes in China. Sustainability 2022, 14, 15107. [Google Scholar] [CrossRef]
- Zuo, X.; Su, F.; Yu, K.; Wang, Y.; Wang, Q.; Wu, H. Spatially modeling the synergistic impacts of global warming and sea-level rise on coral reefs in the South China Sea. Remote Sens. 2021, 13, 2626. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, P.; Panhwar, S.K.; Li, J.; Yan, L.; Chen, Z.; Zhang, K. The initial assessment of an important pelagic fish, Mackerel Scad, in the South China Sea using data-poor length-based methods. Mar. Coast. Fish. 2023, 15, e210258. [Google Scholar] [CrossRef]
- Hodapp, D.; Roca, I.T.; Fiorentino, D.; Garilao, C.; Kaschner, K.; Kesner-Reyes, K.; Schneider, B.; Segschneider, J.; Kocsis, Á.T.; Kiessling, W. Climate change disrupts core habitats of marine species. Glob. Change Biol. 2023, 29, 3304–3317. [Google Scholar] [CrossRef]
- Retnoningtyas, H.; Agustina, S.; Natsir, M.; Ningtias, P.; Hakim, A.; Dhani, A.K.; Hartati, I.D.; Pingkan, J.; Simanjuntak, C.P.; Wiryawan, B. Reproductive biology of the mackerel scad, Decapterus macarellus (Cuvier, 1833), in the Sulawesi Sea, Indonesia. Reg. Stud. Mar. Sci. 2024, 69, 103300. [Google Scholar] [CrossRef]
- TENRIWARE, T.; Nur, M.; Nasyrah, A.F.A. Biological reproduction of mackerel scad, Decapterus macarellus (Cuvier, 1833) caught by purse-seine net in Majene Waters, West Sulawesi, Indonesia. Biodiversitas J. Biol. Divers. 2023, 24, 3012–3018. [Google Scholar] [CrossRef]
- Silooy, F.; Tupamahu, A.; Ongkers, O.; Matrutty, D.; Pattikawa, J. Sex ratio, age group and length at first maturity of Mackerel Scad (Decapterus macarellus Cuvier, 1833) in the southern waters of Ambon, eastern Indonesia. IOP Conf. Ser. Earth Environ. Sci. 2021, 777, 012008. [Google Scholar] [CrossRef]
- Shen, Q.; Zhang, P.; Yu, W.; Xiong, P.; Cai, Y.; Li, J.; Chen, Z.; Fan, J. Impact of Climate Change on the Habitat Distribution of Decapterus macarellus in the South China Sea. J. Mar. Sci. Eng. 2025, 13, 156. [Google Scholar] [CrossRef]
- Rufino, M.M.; Mendo, T.; Samarão, J.; Gaspar, M.B. Estimating fishing effort in small-scale fisheries using high-resolution spatio-temporal tracking data (an implementation framework illustrated with case studies from Portugal). Ecol. Indic. 2023, 154, 110628. [Google Scholar] [CrossRef]
- Meeanan, C.; Noranarttragoon, P.; Sinanun, P.; Takahashi, Y.; Kaewnern, M.; Matsuishi, T.F. Estimation of the spatiotemporal distribution of fish and fishing grounds from surveillance information using machine learning: The case of short mackerel (Rastrelliger brachysoma) in the Andaman Sea, Thailand. Reg. Stud. Mar. Sci. 2023, 62, 102914. [Google Scholar] [CrossRef]
- Han, H.; Yang, C.; Jiang, B.; Shang, C.; Sun, Y.; Zhao, X.; Xiang, D.; Zhang, H.; Shi, Y. Construction of chub mackerel (Scomber japonicus) fishing ground prediction model in the northwestern Pacific Ocean based on deep learning and marine environmental variables. Mar. Pollut. Bull. 2023, 193, 115158. [Google Scholar] [CrossRef]
- Austin, M.P. Spatial prediction of species distribution: An interface between ecological theory and statistical modelling. Ecol. Model. 2002, 157, 101–118. [Google Scholar] [CrossRef]
- Segurado, P.; Araujo, M.B. An evaluation of methods for modelling species distributions. J. Biogeogr. 2004, 31, 1555–1568. [Google Scholar] [CrossRef]
- Araújo, M.B.; Luoto, M. The importance of biotic interactions for modelling species distributions under climate change. Glob. Ecol. Biogeogr. 2007, 16, 743–753. [Google Scholar] [CrossRef]
- Silva, L.D.; Costa, H.; de Azevedo, E.B.; Medeiros, V.; Alves, M.; Elias, R.B.; Silva, L. Modelling native and invasive woody species: A comparison of ENFA and MaxEnt applied to the Azorean forest. In Modeling, Dynamics, Optimization and Bioeconomics II: DGS III, Porto, Portugal, February 2014, and Bioeconomy VII, Berkeley, USA, March 2014-Selected Contributions 3; Springer: Berlin/Heidelberg, Germany, 2017; pp. 415–444. [Google Scholar]
- Gladju, J.; Kamalam, B.S.; Kanagaraj, A. Applications of data mining and machine learning framework in aquaculture and fisheries: A review. Smart Agric. Technol. 2022, 2, 100061. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- De Ville, B. Decision trees. Wiley Interdiscip. Rev. Comput. Stat. 2013, 5, 448–455. [Google Scholar] [CrossRef]
- Geurts, P.; Ernst, D.; Wehenkel, L. Extremely randomized trees. Mach. Learn. 2006, 63, 3–42. [Google Scholar] [CrossRef]
- Peterson, L.E. K-nearest neighbor. Scholarpedia 2009, 4, 1883. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd acm sigkdd international conference on knowledge discovery and data mining, San Francisco, CA, USA, 13 August 2016; pp. 785–794. [Google Scholar]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.-Y. Lightgbm: A highly efficient gradient boosting decision tree. Adv. Neural Inf. Process. Syst. 2017, 30, 3149–3157. [Google Scholar]
- Barbet-Massin, M.; Jiguet, F.; Albert, C.H.; Thuiller, W. Selecting pseudo-absences for species distribution models: How, where and how many? Methods Ecol. Evol. 2012, 3, 327–338. [Google Scholar] [CrossRef]
- Carrington, A.M.; Manuel, D.G.; Fieguth, P.W.; Ramsay, T.; Osmani, V.; Wernly, B.; Bennett, C.; Hawken, S.; Magwood, O.; Sheikh, Y. Deep ROC analysis and AUC as balanced average accuracy, for improved classifier selection, audit and explanation. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 45, 329–341. [Google Scholar] [CrossRef]
- Šimundić, A.-M. Measures of diagnostic accuracy: Basic definitions. Ejifcc 2009, 19, 203. [Google Scholar]
- Kessler, N.; Cyteval, C.; Gallix, B.t.; Lesnik, A.; Blayac, P.-M.; Pujol, J.; Bruel, J.-M.; Taourel, P. Appendicitis: Evaluation of sensitivity, specificity, and predictive values of US, Doppler US, and laboratory findings. Radiology 2004, 230, 472–478. [Google Scholar] [CrossRef]
- Sommer, U.; Stibor, H.; Katechakis, A.; Sommer, F.; Hansen, T. Pelagic food web configurations at different levels of nutrient richness and their implications for the ratio fish production: Primary production. In Sustainable Increase of Marine Harvesting: Fundamental Mechanisms and New Concepts: Proceedings of the 1st Maricult Conference Held in Trondheim, Norway, 25–28 June 2000; Springer: Berlin, Germany, 2002; pp. 11–20. [Google Scholar]
- Claireaux, G.; Lagardère, J.-P. Influence of temperature, oxygen and salinity on the metabolism of the European sea bass. J. Sea Res. 1999, 42, 157–168. [Google Scholar] [CrossRef]
- Britton-Simmons, K.H.; Rhoades, A.L.; Pacunski, R.E.; Galloway, A.W.; Lowe, A.T.; Sosik, E.A.; Dethier, M.N.; Duggins, D.O. Habitat and bathymetry influence the landscape-scale distribution and abundance of drift macrophytes and associated invertebrates. Limnol. Oceanogr. 2012, 57, 176–184. [Google Scholar] [CrossRef]
- Telesh, I.; Schubert, H.; Skarlato, S. Life in the salinity gradient: Discovering mechanisms behind a new biodiversity pattern. Estuar. Coast. Shelf Sci. 2013, 135, 317–327. [Google Scholar] [CrossRef]
- Garrido, S.; Ben-Hamadou, R.; Oliveira, P.B.; Cunha, M.E.; Chícharo, M.A.; van der Lingen, C.D. Diet and feeding intensity of sardine Sardina pilchardus: Correlation with satellite-derived chlorophyll data. Mar. Ecol. Prog. Ser. 2008, 354, 245–256. [Google Scholar] [CrossRef]
- Hou, G.; Wang, J.; Chen, Z.; Zhou, J.; Huang, W.; Zhang, H. Molecular and morphological identification and seasonal distribution of eggs of four Decapterus fish species in the northern South China Sea: A key to conservation of spawning ground. Front. Mar. Sci. 2020, 7, 590564. [Google Scholar] [CrossRef]
- Ekanayake, I.; Meddage, D.; Rathnayake, U. A novel approach to explain the black-box nature of machine learning in compressive strength predictions of concrete using Shapley additive explanations (SHAP). Case Stud. Constr. Mater. 2022, 16, e01059. [Google Scholar] [CrossRef]
- Brugere, L.; Kwon, Y.; Frazier, A.E.; Kedron, P. Improved prediction of tree species richness and interpretability of environmental drivers using a machine learning approach. For. Ecol. Manag. 2023, 539, 120972. [Google Scholar] [CrossRef]
- Shehadeh, A.; Alshboul, O.; Al Mamlook, R.E.; Hamedat, O. Machine learning models for predicting the residual value of heavy construction equipment: An evaluation of modified decision tree, LightGBM, and XGBoost regression. Autom. Constr. 2021, 129, 103827. [Google Scholar] [CrossRef]
- Zhang, P.; Jia, Y.; Shang, Y. Research and application of XGBoost in imbalanced data. Int. J. Distrib. Sens. Netw. 2022, 18, 15501329221106935. [Google Scholar] [CrossRef]
- Bellamy, C.; Scott, C.; Altringham, J. Multiscale, presence-only habitat suitability models: Fine-resolution maps for eight bat species. J. Appl. Ecol. 2013, 50, 892–901. [Google Scholar] [CrossRef]
- Ely, B.; Viñas, J.; Alvarado Bremer, J.R.; Black, D.; Lucas, L.; Covello, K.; Labrie, A.V.; Thelen, E. Consequences of the historical demography on the global population structure of two highly migratory cosmopolitan marine fishes: The yellowfin tuna (Thunnus albacares) and the skipjack tuna (Katsuwonus pelamis). BMC Evol. Biol. 2005, 5, 19. [Google Scholar] [CrossRef]
- Erauskin-Extramiana, M.; Arrizabalaga, H.; Hobday, A.J.; Cabré, A.; Ibaibarriaga, L.; Arregui, I.; Murua, H.; Chust, G. Large-scale distribution of tuna species in a warming ocean. Glob. Change Biol. 2019, 25, 2043–2060. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Song, L.; Zhang, T.; Li, Y.; Shen, J.; Zhang, M.; Li, K. Effects of different spatial resolutions on prediction accuracy of Thunnus alalunga fishing ground in waters near the Cook Islands Based on Long Short-Term Memory (LSTM) Neural Network Model. J. Ocean Univ. China 2023, 22, 1427–1438. [Google Scholar] [CrossRef]
- Buenafe, K.C.V.; Everett, J.D.; Dunn, D.C.; Mercer, J.; Suthers, I.M.; Schilling, H.T.; Hinchliffe, C.; Dabalà, A.; Richardson, A.J. A global, historical database of tuna, billfish, and saury larval distributions. Sci. Data 2022, 9, 423. [Google Scholar] [CrossRef] [PubMed]
- Moëzzi, F.; Eagderi, S. Quantifying how spatial resolution affects fish distribution model performance and prediction: A case study of Caspian Kutum, Rutilus frisii. Int. J. Aquat. Biol. 2024, 12, 533–545. [Google Scholar]
- Núñez-Riboni, I.; Akimova, A.; Sell, A.F. Effect of data spatial scale on the performance of fish habitat models. Fish Fish. 2021, 22, 955–973. [Google Scholar] [CrossRef]
- Nagendra, H.; Lucas, R.; Honrado, J.P.; Jongman, R.H.; Tarantino, C.; Adamo, M.; Mairota, P. Remote sensing for conservation monitoring: Assessing protected areas, habitat extent, habitat condition, species diversity, and threats. Ecol. Indic. 2013, 33, 45–59. [Google Scholar] [CrossRef]
- Pettorelli, N.; Laurance, W.F.; O’Brien, T.G.; Wegmann, M.; Nagendra, H.; Turner, W. Satellite remote sensing for applied ecologists: Opportunities and challenges. J. Appl. Ecol. 2014, 51, 839–848. [Google Scholar] [CrossRef]
- Marvin, D.C.; Koh, L.P.; Lynam, A.J.; Wich, S.; Davies, A.B.; Krishnamurthy, R.; Stokes, E.; Starkey, R.; Asner, G.P. Integrating technologies for scalable ecology and conservation. Glob. Ecol. Conserv. 2016, 7, 262–275. [Google Scholar] [CrossRef]
- Zhao, Y.; Deng, X.; Xiang, W.; Chen, L.; Ouyang, S. Predicting potential suitable habitats of Chinese fir under current and future climatic scenarios based on Maxent model. Ecol. Inform. 2021, 64, 101393. [Google Scholar] [CrossRef]
- Phillips, S.J.; Dudík, M. Modeling of species distributions with Maxent: New extensions and a comprehensive evaluation. Ecography 2008, 31, 161–175. [Google Scholar] [CrossRef]
- Elith, J.; Phillips, S.J.; Hastie, T.; Dudík, M.; Chee, Y.E.; Yates, C.J. A statistical explanation of MaxEnt for ecologists. Divers. Distrib. 2011, 17, 43–57. [Google Scholar] [CrossRef]
- Wang, H.; Zhou, S.; Li, X.; Liu, H.; Chi, D.; Xu, K. The influence of climate change and human activities on ecosystem service value. Ecol. Eng. 2016, 87, 224–239. [Google Scholar] [CrossRef]

| Variable | Description | Source | Unit | Spatial Resolution |
|---|---|---|---|---|
| SSS | Seawater salinity | https://marine.copernicus.eu/ | ‰ | 0.083° and 0.25° |
| SSH | Sea surface height above geoid | https://marine.copernicus.eu/ | m | 0.083° and 0.25° |
| MLD | Ocean mixed layer thickness | https://marine.copernicus.eu/ | m | 0.083° and 0.25° |
| SST | Sea surface temperature | https://marine.copernicus.eu/ | °C | 0.083° and 0.25° |
| DIS | Distance from shore | https://globalfishingwatch.org/ | km | 0.083° |
| BATH | Bathymetry | https://globalfishingwatch.org/ | m | 0.083° |
| CHL | Mass concentration of chlorophyll-a in seawater | https://marine.copernicus.eu/ | mg⋅m−3 | 0.25° |
| Models | Parameter Settings |
|---|---|
| RF | tuneLength = 3; ntree = 500 |
| DT | rpart |
| XGB | Eta = 0.1; max_depth = 0.8; nrounds = 100 |
| KNN | K = 5 |
| ET | ntree = 500; mtry = 3; nodesize = 1 |
| LGBM | distribution = “bernoulli”; n.trees = 100; interaction.depth = 6; shrinkage = 0.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, Q.; Zhang, P.; Feng, X.; Chen, Z.; Fan, J. Exploring the Habitat Distribution of Decapterus macarellus in the South China Sea Under Varying Spatial Resolutions: A Combined Approach Using Multiple Machine Learning and the MaxEnt Model. Biology 2025, 14, 753. https://doi.org/10.3390/biology14070753
Shen Q, Zhang P, Feng X, Chen Z, Fan J. Exploring the Habitat Distribution of Decapterus macarellus in the South China Sea Under Varying Spatial Resolutions: A Combined Approach Using Multiple Machine Learning and the MaxEnt Model. Biology. 2025; 14(7):753. https://doi.org/10.3390/biology14070753
Chicago/Turabian StyleShen, Qikun, Peng Zhang, Xue Feng, Zuozhi Chen, and Jiangtao Fan. 2025. "Exploring the Habitat Distribution of Decapterus macarellus in the South China Sea Under Varying Spatial Resolutions: A Combined Approach Using Multiple Machine Learning and the MaxEnt Model" Biology 14, no. 7: 753. https://doi.org/10.3390/biology14070753
APA StyleShen, Q., Zhang, P., Feng, X., Chen, Z., & Fan, J. (2025). Exploring the Habitat Distribution of Decapterus macarellus in the South China Sea Under Varying Spatial Resolutions: A Combined Approach Using Multiple Machine Learning and the MaxEnt Model. Biology, 14(7), 753. https://doi.org/10.3390/biology14070753







