Functional and Genomic Evidence of L-Arginine-Dependent Bacterial Nitric Oxide Synthase Activity in Paenibacillus nitricinens sp. nov.
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Site, Soil Chemical Characterization, and Bacterial Isolation
2.2. 16S rRNA Sequencing of Isolates
2.3. Design and Amplification of bnos-Targeting Primers
2.4. Hybrid Whole-Genome Sequencing and Assembly
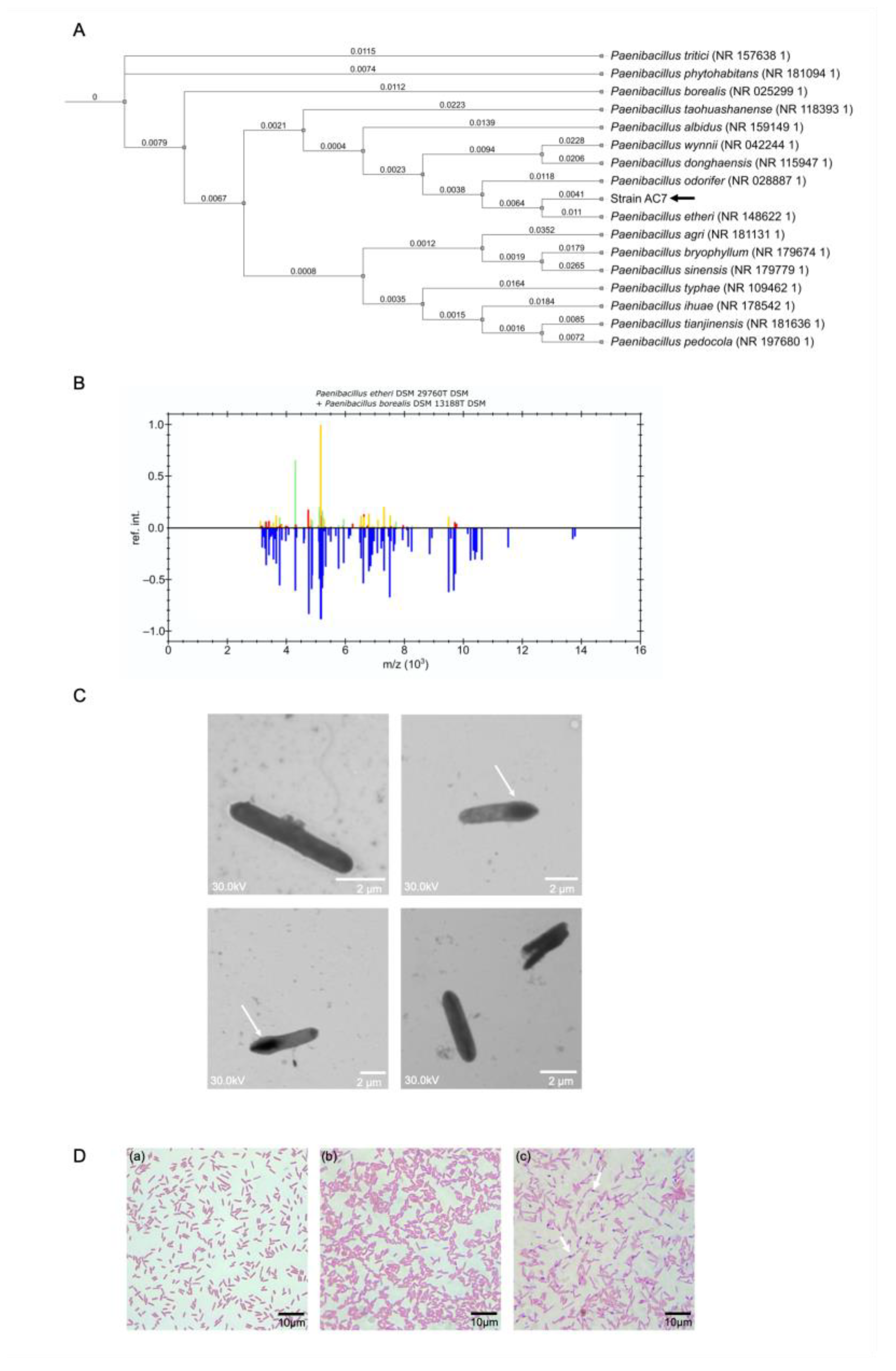
2.5. Phenotypic Characterization
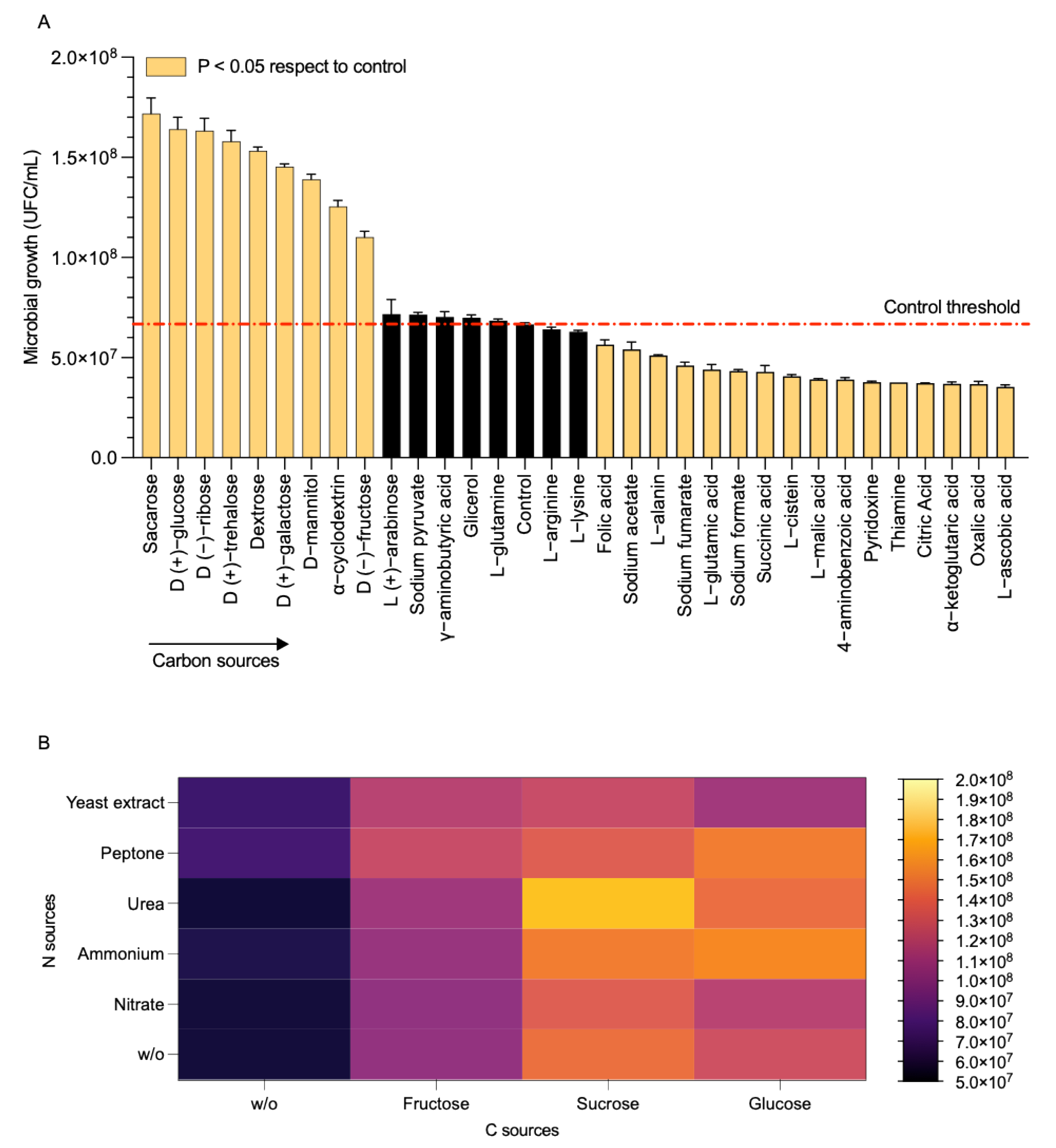
2.6. Growth and Nutrient Preference
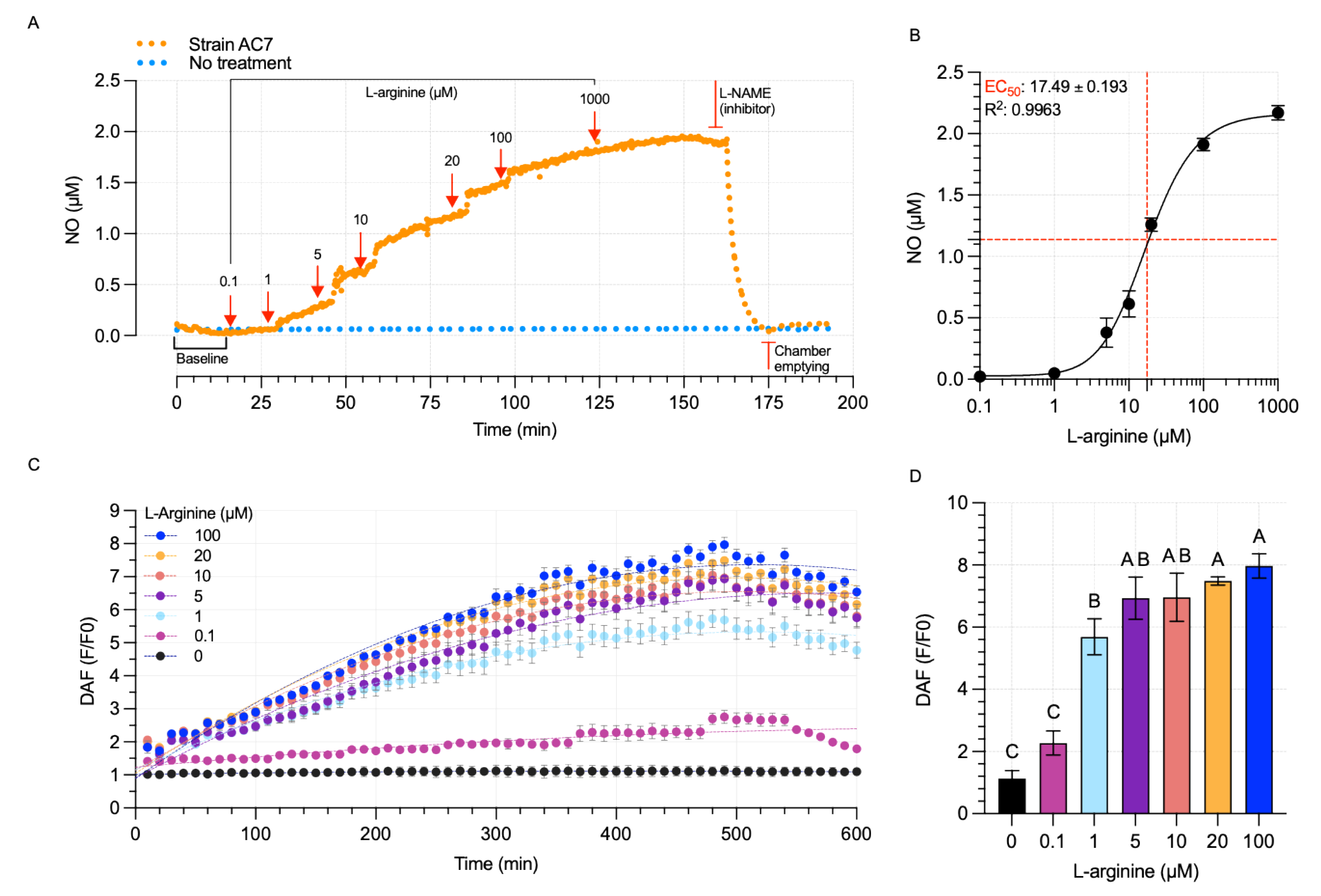
2.7. Extracellular NO Measurement
2.8. Intracellular NO Detection
2.9. Statistical Analysis
3. Results
3.1. Soil Characteristics and Preliminary Isolates
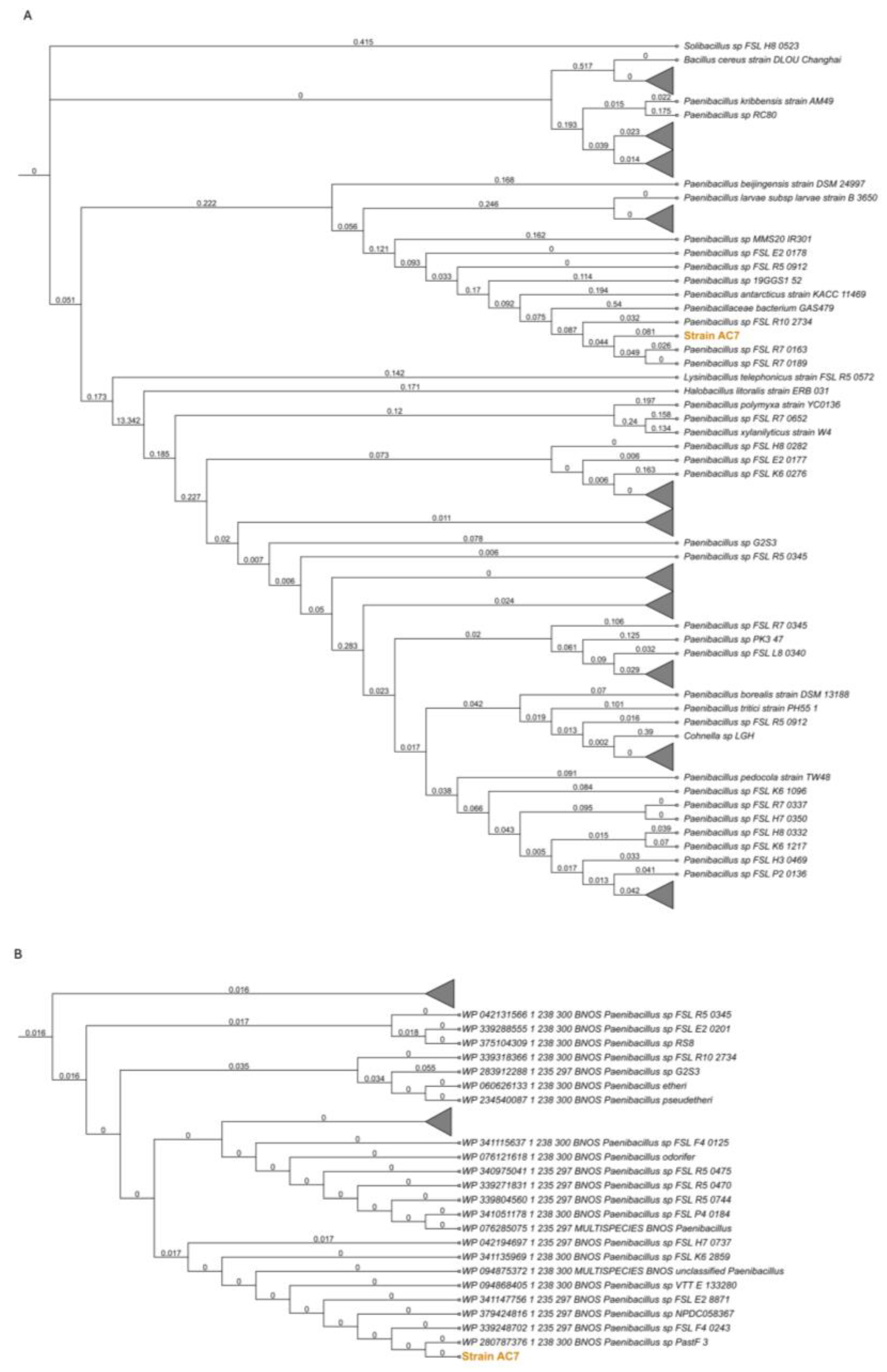
3.2. bnos Amplification and Phylogenetic Placement
3.3. Phylogenetic Analysis and Biochemical Characterization
3.4. Utilization of Carbon and Nitrogen Substrates
3.5. Bacterial NO Production
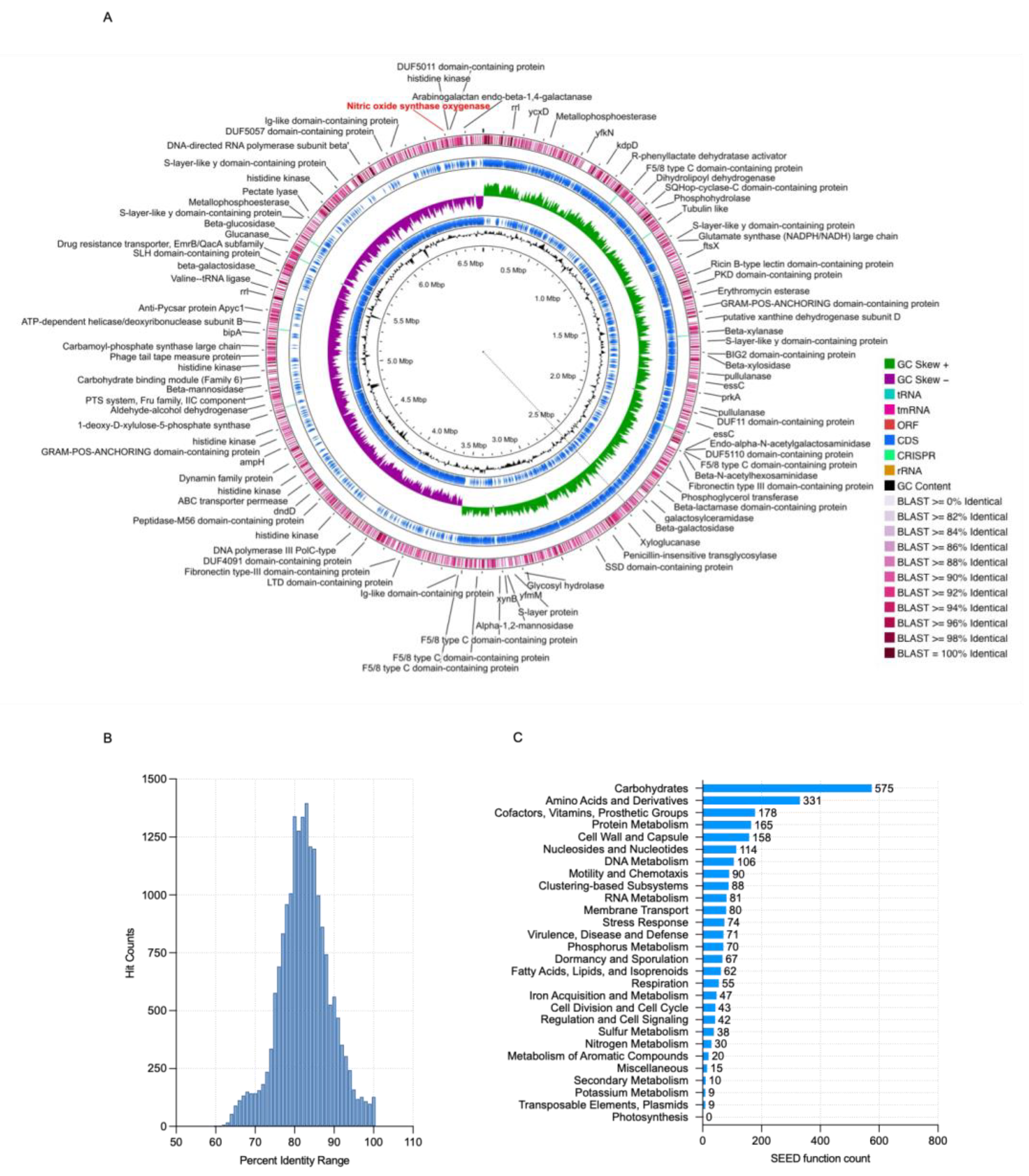
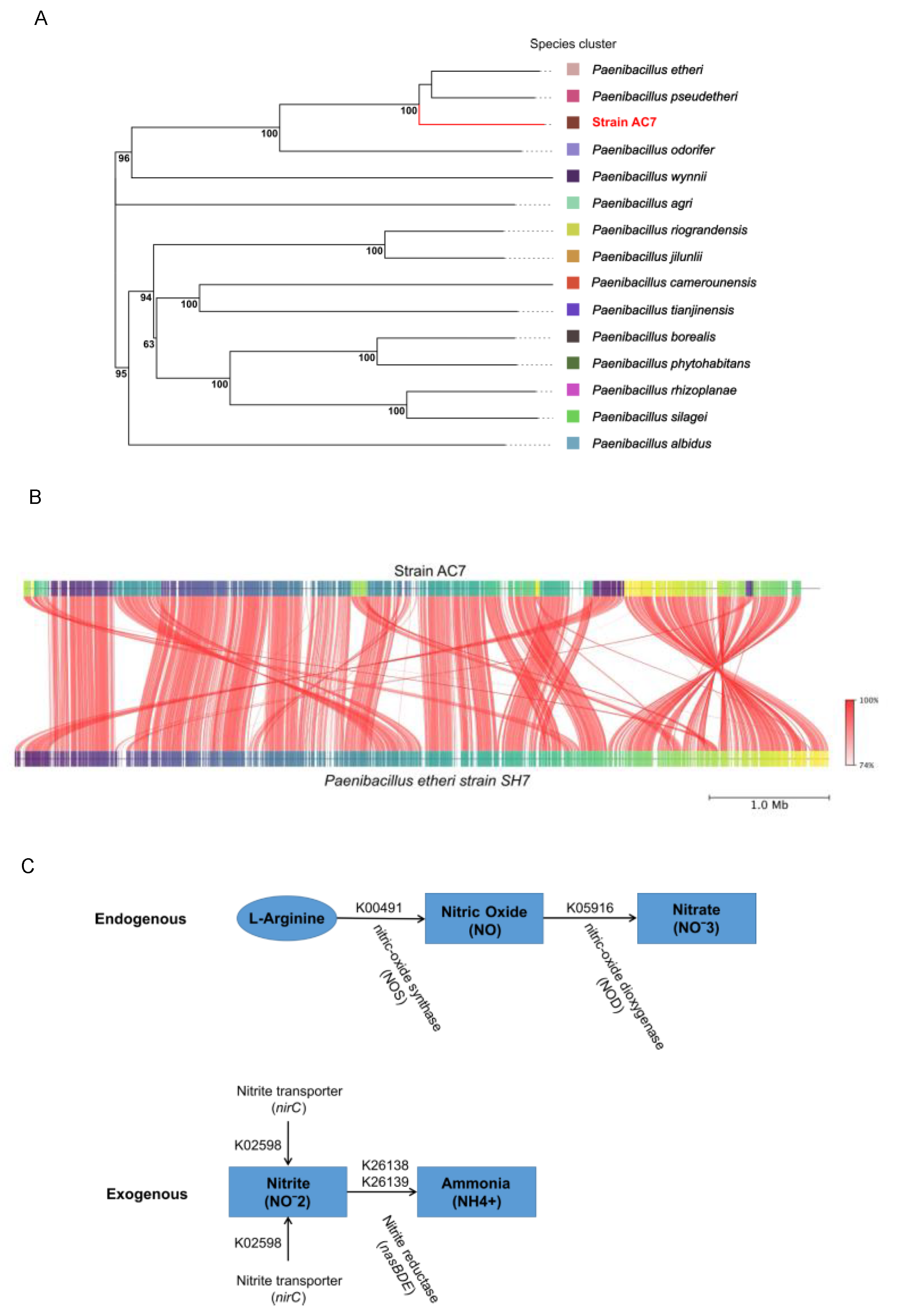
3.6. Genome Characterization and Phylogenetic Studies
4. Discussion
4.1. Aerobic NO Production via bNOS in Paenibacillus Nitricinens sp. nov.
4.2. Taxonomic Distinction and Functional Characterization
4.3. Metabolic Versatility and Environmental Adaptation
4.4. Implications for Soil Nitrogen Cycling
4.5. Potential Regulation of bNOS and Future Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ANI | Average nucleotide identity |
| CCCT | Colección Chilena de Cultivos Tipo |
| CFU | Colony-forming unit |
| dDDH | Digital DNA–DNA hybridization |
| FASTME | Fast Minimum Evolution |
| FBA | Flux balance analysis |
| GBDP | Genome BLAST Distance Phylogeny |
| GC skew | Guanine–cytosine skew |
| HSD | Tukey’s honest significant difference |
| MSA AA | Multiple-sequence alignment (amino acid) |
| OAT | OrthoANI Tool |
| OD600 | Optical density at 600 nanometers |
| PBS | Phosphate-buffered saline |
| pHMM | Profile hidden Markov model |
| rpm | Revolutions per minute |
| SNAP | S-nitroso-N-acetylpenicillamine |
| SRA | Sequence Read Archive |
| tmRNA | Transfer–messenger RNA |
| TYGS | Type (Strain) Genome Server |
| WGS | Whole-genome sequencing |
References
- Salas, A.; Cabrera, J.J.; Jiménez-Leiva, A.; Mesa, S.; Bedmar, E.J.; Richardson, D.J.; Gates, A.J.; Delgado, M.J. Bacterial nitric oxide metabolism: Recent insights in rhizobia. Adv. Microb. Physiol. 2021, 78, 259–315. [Google Scholar] [CrossRef] [PubMed]
- Nisbett, L.M.; Boon, E.M. Nitric Oxide Regulation of H-NOX Signaling Pathways in Bacteria. Biochemistry 2016, 55, 4873–4884. [Google Scholar] [CrossRef] [PubMed]
- Murali, R.; Pace, L.A.; Sanford, R.A.; Ward, L.M.; Lynes, M.M.; Hatzenpichler, R.; Lingappa, U.F.; Fischer, W.W.; Gennis, R.B.; Hemp, J. Diversity and evolution of nitric oxide reduction in bacteria and archaea. Proc. Natl. Acad. Sci. USA 2024, 121, e2316422121. [Google Scholar] [CrossRef] [PubMed]
- Blomberg, M.R.; Siegbahn, P.E. Mechanism for N2O generation in bacterial nitric oxide reductase: A quantum chemical study. Biochemistry 2012, 51, 5173–5186. [Google Scholar] [CrossRef]
- Torres, M.J.; Simon, J.; Rowley, G.; Bedmar, E.J.; Richardson, D.J.; Gates, A.J.; Delgado, M.J. Nitrous Oxide Metabolism in Nitrate-Reducing Bacteria: Physiology and Regulatory Mechanisms. Adv. Microb. Physiol. 2016, 68, 353–432. [Google Scholar] [CrossRef]
- Crane, B.R.; Sudhamsu, J.; Patel, B.A. Bacterial nitric oxide synthases. Annu. Rev. Biochem. 2010, 79, 445–470. [Google Scholar] [CrossRef]
- Sudhamsu, J.; Crane, B.R. Bacterial nitric oxide synthases: What are they good for? Trends Microbiol. 2009, 17, 212–218. [Google Scholar] [CrossRef]
- Correa-Aragunde, N.; Foresi, N.; Del Castello, F.; Lamattina, L. A singular nitric oxide synthase with a globin domain found in Synechococcus PCC 7335 mobilizes N from arginine to nitrate. Sci. Rep. 2018, 8, 12505. [Google Scholar] [CrossRef]
- Ras, G.; Zuliani, V.; Derkx, P.; Seibert, T.M.; Leroy, S.; Talon, R. Evidence for Nitric Oxide Synthase Activity in Staphylococcus xylosus Mediating Nitrosoheme Formation. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Adak, S.; Aulak, K.S.; Stuehr, D.J. Direct evidence for nitric oxide production by a nitric-oxide synthase-like protein from Bacillus subtilis. J. Biol. Chem. 2002, 277, 16167–16171. [Google Scholar] [CrossRef]
- Toyofuku, M.; Yoon, S.-S. Chapter Four—Nitric Oxide, an Old Molecule with Noble Functions in Pseudomonas aeruginosa Biology. In Advances in Microbial Physiology; Poole, R.K., Ed.; Academic Press: New York, NY, USA, 2018; Volume 72, pp. 117–145. [Google Scholar]
- Beno, S.M.; Cheng, R.A.; Orsi, R.H.; Duncan, D.R.; Guo, X.; Kovac, J.; Carroll, L.M.; Martin, N.H.; Wiedmann, M. Paenibacillus odorifer, the Predominant Paenibacillus Species Isolated from Milk in the United States, Demonstrates Genetic and Phenotypic Conservation of Psychrotolerance but Clade-Associated Differences in Nitrogen Metabolic Pathways. mSphere 2020, 5, e00739-19. [Google Scholar] [CrossRef] [PubMed]
- Grady, E.N.; MacDonald, J.; Liu, L.; Richman, A.; Yuan, Z.-C. Current knowledge and perspectives of Paenibacillus: A review. Microb. Cell Factories 2016, 15, 203. [Google Scholar] [CrossRef] [PubMed]
- Seldin, L. Paenibacillus, nitrogen fixation and soil fertility. In Endospore-Forming Soil Bacteria; Logan, N.A., Vos, P., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 287–307. [Google Scholar] [CrossRef]
- Crane, B.R. The enzymology of nitric oxide in bacterial pathogenesis and resistance. Biochem. Soc. Trans. 2008, 36, 1149–1154. [Google Scholar] [CrossRef] [PubMed]
- Gusarov, I.; Starodubtseva, M.; Wang, Z.Q.; McQuade, L.; Lippard, S.J.; Stuehr, D.J.; Nudler, E. Bacterial nitric-oxide synthases operate without a dedicated redox partner. J. Biol. Chem. 2008, 283, 13140–13147. [Google Scholar] [CrossRef]
- Pajares, S.; Bohannan, B.J. Ecology of nitrogen fixing, nitrifying, and denitrifying microorganisms in tropical forest soils. Front. Microbiol. 2016, 7, 1045. [Google Scholar] [CrossRef]
- Tang, Y.; Yu, G.; Zhang, X.; Wang, Q.; Ge, J.; Liu, S. Changes in nitrogen-cycling microbial communities with depth in temperate and subtropical forest soils. Appl. Soil Ecol. 2018, 124, 218–228. [Google Scholar] [CrossRef]
- Abdallah, K.; Stock, S.C.; Heeger, F.; Koester, M.; Nájera, F.; Matus, F.; Merino, C.; Spielvogel, S.; Gorbushina, A.A.; Kuzyakov, Y. Nitrogen Gain and Loss Along an Ecosystem Sequence: From Semi-desert to Rainforest. Front. Soil Sci. 2022, 2, 817641. [Google Scholar] [CrossRef]
- Glass, J.B.; Rousk, K. Microbial nitrogen transformation processes across environments: More than just a cycle. Trends Microbiol. 2024, 32, 519–521. [Google Scholar] [CrossRef]
- Luzio, L.W.; Sadzawka, R.A.; Besoain, M.E.; Lara, G.P. Influence of volcanic materials on red clay soil genesis. Rev. Cienc. Suelo Y Nutr. Veg. 2003, 3, 37–52. [Google Scholar]
- Weatherburn, M. Phenol-hypochlorite reaction for determination of ammonia. Anal. Chem. 1967, 39, 971–974. [Google Scholar] [CrossRef]
- Doane, T.A.; Horwáth, W.R. Spectrophotometric determination of nitrate with a single reagent. Anal. Lett. 2003, 36, 2713–2722. [Google Scholar] [CrossRef]
- Merino, C.; Kuzyakov, Y.; Godoy, K.; Cornejo, P.; Matus, F. Synergy effect of peroxidase enzymes and Fenton reactions greatly increase the anaerobic oxidation of soil organic matter. Sci. Rep. 2020, 10, 11289. [Google Scholar] [CrossRef] [PubMed]
- Agriculture, U.S.D.O.; Service, N.R.C.; Survey, S.; Staff, S.S. Keys to Soil Taxonomy; Books Express Publishing: Berkshire, UK, 2010. [Google Scholar]
- Sadzawka, A.; Carrasco, M.; Grez, R.; Mora, M.; Flores, H.; Neaman, A. Métodos de Análisis de Suelos Recomendados para los Suelos de Chile, Revision 2006. ed.; Serie Actas INIA N°34; Instituto de Investigaciones Agropecuarias: Santiago, Chile, 2006; Volume 1, pp. 33–50. [Google Scholar]
- Lemoine, F.; Correia, D.; Lefort, V.; Doppelt-Azeroual, O.; Mareuil, F.; Cohen-Boulakia, S.; Gascuel, O. NGPhylogeny.fr: New generation phylogenetic services for non-specialists. Nucleic Acids Res. 2019, 47, W260–W265. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Wu, Q.; Liu, Y.; Hu, S.; Huang, J.; Zhang, C. High-Quality Genome Assembly and Annotation Resource of Three Botryosphaeria Pathogens Causing Chinese Hickory Canker. Mol. Plant-Microbe Interact. 2022, 35, 941–943. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Gaete, A.; Andreani-Gerard, C.; Maldonado, J.E.; Muñoz-Torres, P.A.; Sepúlveda-Chavera, G.F.; González, M. Bioprospecting of Plant Growth-Promoting Traits of Pseudomonas sp. Strain C3 Isolated from the Atacama Desert: Molecular and Culture-Based Analysis. Diversity 2022, 14, 388. [Google Scholar] [CrossRef]
- Chauhan, A.; Jindal, T. Biochemical and Molecular Methods for Bacterial Identification. In Microbiological Methods for Environment, Food and Pharmaceutical Analysis; Chauhan, A., Jindal, T., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 425–468. [Google Scholar]
- Jofré, I.; Matus, F.; Mendoza, D.; Nájera, F.; Merino, C. Manganese-Oxidizing Antarctic Bacteria (Mn-Oxb) Release Reactive Oxygen Species (ROS) as Secondary Mn(II) Oxidation Mechanisms to Avoid Toxicity. Biology 2021, 10, 4. [Google Scholar] [CrossRef]
- Rao, N.S.S. Soil Microorganisms; CBS Publishers & Distributors: New Delhi, India, 2017; pp. 128–142. [Google Scholar]
- Blanco-Vargas, A.; Rodríguez-Gacha, L.M.; Sánchez-Castro, N.; Garzón-Jaramillo, R.; Pedroza-Camacho, L.D.; Poutou-Piñales, R.A.; Rivera-Hoyos, C.M.; Díaz-Ariza, L.A.; Pedroza-Rodríguez, A.M. Phosphate-solubilizing Pseudomonas sp., and Serratia sp., co-culture for Allium cepa L. growth promotion. Heliyon 2020, 6, e05218. [Google Scholar] [CrossRef]
- Sivaloganathan, D.M.; Brynildsen, M.P. Quantitative Modeling Extends the Antibacterial Activity of Nitric Oxide. Front. Physiol. 2020, 11, 330. [Google Scholar] [CrossRef] [PubMed]
- Kinkel, T.L.; Ramos-Montañez, S.; Pando, J.M.; Tadeo, D.V.; Strom, E.N.; Libby, S.J.; Fang, F.C. An essential role for bacterial nitric oxide synthase in Staphylococcus aureus electron transfer and colonization. Nat. Microbiol. 2016, 2, 16224. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; LoBue, A.; Heuser, S.K.; Leo, F.; Cortese-Krott, M.M. Using diaminofluoresceins (DAFs) in nitric oxide research. Nitric Oxide 2021, 115, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Lewis, A.M.; Matzdorf, S.S.; Rice, K.C. Fluorescent Detection of Intracellular Nitric Oxide in Staphylococcus aureus. Bio-Protoc. 2016, 6, e1878. [Google Scholar] [CrossRef]
- Holden, J.K.; Lewis, M.C.; Cinelli, M.A.; Abdullatif, Z.; Pensa, A.V.; Silverman, R.B.; Poulos, T.L. Targeting Bacterial Nitric Oxide Synthase with Aminoquinoline-Based Inhibitors. Biochemistry 2016, 55, 5587–5594. [Google Scholar] [CrossRef]
- Rinaldo, S.; Giardina, G.; Mantoni, F.; Paone, A.; Cutruzzolà, F. Beyond nitrogen metabolism: Nitric oxide, cyclic-di-GMP and bacterial biofilms. FEMS Microbiol. Lett. 2018, 365, fny029. [Google Scholar] [CrossRef]
- Cutruzzolà, F.; Frankenberg-Dinkel, N. Origin and impact of nitric oxide in Pseudomonas aeruginosa biofilms. J. Bacteriol. 2016, 198, 55–65. [Google Scholar] [CrossRef]
- Popova, T.G.; Teunis, A.; Vaseghi, H.; Zhou, W.; Espina, V.; Liotta, L.A.; Popov, S.G. Nitric oxide as a regulator of B. anthracis pathogenicity. Front. Microbiol. 2015, 6, 921. [Google Scholar] [CrossRef]
- Luhachack, L.; Nudler, E. Bacterial gasotransmitters: An innate defense against antibiotics. Curr. Opin. Microbiol. 2014, 21, 13–17. [Google Scholar] [CrossRef]
- Xu, H.; Qin, S.; Lan, Y.; Liu, M.; Cao, X.; Qiao, D.; Cao, Y.; Cao, Y. Comparative genomic analysis of Paenibacillus sp. SSG-1 and its closely related strains reveals the effect of glycometabolism on environmental adaptation. Sci. Rep. 2017, 7, 5720. [Google Scholar] [CrossRef]
- Rodriguez, R.L.M.; Castro Juan, C.; Kyrpides Nikos, C.; Cole James, R.; Tiedje James, M.; Konstantinidis Konstantinos, T. How Much Do rRNA Gene Surveys Underestimate Extant Bacterial Diversity? Appl. Environ. Microbiol. 2018, 84, e00014-18. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.; Guggiari, M.; Bravo, D.; Zopfi, J.; Cailleau, G.; Aragno, M.; Job, D.; Verrecchia, E.; Junier, P. Fungi, bacteria and soil pH: The oxalate–carbonate pathway as a model for metabolic interaction. Environ. Microbiol. 2012, 14, 2960–2970. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Yin, J.; Jin, M.; Gao, H. Distinct nitrite and nitric oxide physiologies in Escherichia coli and Shewanella oneidensis. Appl. Environ. Microbiol. 2018, 84, e00559-18. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.B.; Martiny, A.C.; Martiny, J.B. Global biogeography of microbial nitrogen-cycling traits in soil. Proc. Natl. Acad. Sci. USA 2016, 113, 8033–8040. [Google Scholar] [CrossRef]
- Richardson, A.R.; Payne, E.C.; Younger, N.; Karlinsey, J.E.; Thomas, V.C.; Becker, L.A.; Navarre, W.W.; Castor, M.E.; Libby, S.J.; Fang, F.C. Multiple targets of nitric oxide in the tricarboxylic acid cycle of Salmonella enterica serovar typhimurium. Cell Host Microbe 2011, 10, 33–43. [Google Scholar] [CrossRef]
- Wilbert, S.A.; Newman, D.K. The contrasting roles of nitric oxide drive microbial community organization as a function of oxygen presence. Curr. Biol. 2022, 32, 5221–5234.e5224. [Google Scholar] [CrossRef]
- Agapie, T.; Suseno, S.; Woodward, J.J.; Stoll, S.; Britt, R.D.; Marletta, M.A. NO formation by a catalytically self-sufficient bacterial nitric oxide synthase from Sorangium cellulosum. Proc. Natl. Acad. Sci. USA 2009, 106, 16221–16226. [Google Scholar] [CrossRef]
- Fuangthong, M.; Herbig, A.F.; Bsat, N.; Helmann, J.D. Regulation of the Bacillus subtilis fur and perR genes by PerR: Not all members of the PerR regulon are peroxide inducible. J. Bacteriol. 2002, 184, 3276–3286. [Google Scholar] [CrossRef]
- Picciano, A.L.; Crane, B.R. A nitric oxide synthase-like protein from Synechococcus produces NO/NO(3)(-) from l-arginine and NADPH in a tetrahydrobiopterin- and Ca(2+)-dependent manner. J. Biol. Chem. 2019, 294, 10708–10719. [Google Scholar] [CrossRef]
- Pilegaard, K. Processes regulating nitric oxide emissions from soils. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20130126. [Google Scholar] [CrossRef]
| Parameter | Units | Value | |
|---|---|---|---|
| Site characteristics | Coordinates | – | 40°12′ S–73°26′ W |
| Parent material | – | Metamorphic, mica schist | |
| Soil order 1 | – | Ultisol | |
| Clay type 2 | – | Q, I, K | |
| Texture 3 | – | CL | |
| MAT 4 | °C | 9.5 | |
| MAP 5 | mm a−1 | 4000 | |
| Elevation | m.a.s.l. | 1048 | |
| Vegetation 6 | – | DW, LP; NN, NP, PN, SC | |
| Soil chemical properties | Total C | % | 9.2 ± 2.1 |
| Total N | % | 0.47 ± 0.01 | |
| C:N ratio | – | 24.3 | |
| NO3− | % | 0.3 ± 0.0 | |
| NH4⁺ | % | 0.16 ± 0.02 | |
| pH (water) | – | 3.9 ± 0.2 | |
| Al saturation | % | 80 ± 9.2 | |
| Total Fe | g kg−1 soil | 10.4 ± 0.3 | |
| Fe2⁺ | g kg−1 soil | 5.3 ± 0.6 | |
| Fe3⁺ | g kg−1 soil | 5.1 ± 0.3 |
| Strain | Closest Accession |
Closest Organism | Identity (%) | Coverage (%) | E-Value |
Length (bp) | bnos -PCR |
Accession Number |
| AC1 | NR_104919.1 | Bacillus tequilensis | 99.86 | 100 | 0.0 | 1531 | - | PV535646 |
| AC2 | NR_179598.1 | Collimonas antrihumi | 99.31 | 100 | 0.0 | 1425 | - | PV529833 |
| AC5 | NR_074325.2 | Paraburkholderia bryophila | 98.98 | 100 | 0.0 | 1484 | - | PV535647 |
| AC7 | NR_025299.1 | Paenibacillus borealis | 97.98 | 100 | 0.0 | 1540 | + | PV248128 |
| Characteristics | Strain AC7 | P. etheri | P. odorifer | |
|---|---|---|---|---|
| Morphological characteristics | Gram Stain | - | + | - |
| Cell shape | Rod-shaped | Rod-shaped | Rod-shaped | |
| Spore formation | + | + | + | |
| Motility | - | + | + | |
| Physiological traits | Oxygen tolerance | Facultative anaerobe | Facultative anaerobe | Facultative anaerobe |
| pH range | 6.0–8.5 | 7.0–8.0 | 6.0–8.0 | |
| Temperature range (°C) | 12–30 | 5–35 | 5–35 | |
| Biochemical tests and enzymatic activity | Catalase | + | + | + |
| Oxidase | - | - | - | |
| Citrate | - | - | + | |
| Urease | - | - | - | |
| Starch utilization | + | + | + | |
| Indole | - | - | - | |
| Voges–Proskauer | + | + | + | |
| Methyl Red | - | - | - | |
| Fermentation | Glucose | + | + | + |
| Lactose | - | - | + | |
| Mannitol | + | + | + | |
| Functional capabilities | P solubilization | - | ND | ND |
| Free-nitrogen culture | + | ND | ND | |
| Genomic information | DNA G+C content | 43.79% | 44.30% | 44.35% |
| Category | Feature | Value |
|---|---|---|
| Annotation and gene prediction | Genome size | 6,790,125 bp |
| Number of contigs | 1 | |
| Largest contig | 6,790,125 bp | |
| N50/L50 | 6,790,125 bp/1 | |
| Genome topology | Putative circular | |
| GC content | 43.80% | |
| Sequencing coverage | 147.8× | |
| Ambiguous bases (Ns) | 0 | |
| Misassemblies | 0 | |
| Mismatches/100 kbp | 7697.61 | |
| Indels/100 kbp | 104 | |
| CDS (protein-coding genes) | 6080 (Prokka annotation) | |
| rRNA genes | 30 (10 × 5S, 10 × 16S, 10 × 23S) | |
| tRNA genes | 93 | |
| tmRNA gene | 1 | |
| Completeness metrics | BUSCO completeness (bacillales_odb10) | 99.8% (98.9% single-copy, 0.9% duplicated) |
| Fragmented BUSCOs | 0.20% | |
| Missing BUSCOs | 0% | |
| Comparative alignment | Genome fraction aligned to reference | 41.80% (Paenibacillus etheri) |
| 100% identity hits | 121 | |
| Hit range with >96% identity | 577–19,391 | |
| Other genomic features | Putative plasmids | None detected (PlasFlow classification) |
| MLST markers | 5 (glpK, gmk, tpiA, and 2× acat) | |
| Biosynthetic gene clusters | 4 (3 RiPPs, 1 terpene; antiSMASH v6.1.1) | |
| Antimicrobial resistance genes | None detected (staramr analysis) | |
| Submission details | BioProject ID | PRJNA1233252 |
| BioSample ID | SAMN47264457 | |
| Taxonomy ID | 3367691 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saavedra-Tralma, D.; Gaete, A.; Merino-Guzmán, C.; Parada-Ibáñez, M.; Nájera-de Ferrari, F.; Jofré-Fernández, I. Functional and Genomic Evidence of L-Arginine-Dependent Bacterial Nitric Oxide Synthase Activity in Paenibacillus nitricinens sp. nov. Biology 2025, 14, 733. https://doi.org/10.3390/biology14060733
Saavedra-Tralma D, Gaete A, Merino-Guzmán C, Parada-Ibáñez M, Nájera-de Ferrari F, Jofré-Fernández I. Functional and Genomic Evidence of L-Arginine-Dependent Bacterial Nitric Oxide Synthase Activity in Paenibacillus nitricinens sp. nov. Biology. 2025; 14(6):733. https://doi.org/10.3390/biology14060733
Chicago/Turabian StyleSaavedra-Tralma, Diego, Alexis Gaete, Carolina Merino-Guzmán, Maribel Parada-Ibáñez, Francisco Nájera-de Ferrari, and Ignacio Jofré-Fernández. 2025. "Functional and Genomic Evidence of L-Arginine-Dependent Bacterial Nitric Oxide Synthase Activity in Paenibacillus nitricinens sp. nov." Biology 14, no. 6: 733. https://doi.org/10.3390/biology14060733
APA StyleSaavedra-Tralma, D., Gaete, A., Merino-Guzmán, C., Parada-Ibáñez, M., Nájera-de Ferrari, F., & Jofré-Fernández, I. (2025). Functional and Genomic Evidence of L-Arginine-Dependent Bacterial Nitric Oxide Synthase Activity in Paenibacillus nitricinens sp. nov. Biology, 14(6), 733. https://doi.org/10.3390/biology14060733








