Halfway Through Ex Situ Population Genetic Lifespan: The Case of Cochlearia polonica
Simple Summary
Abstract
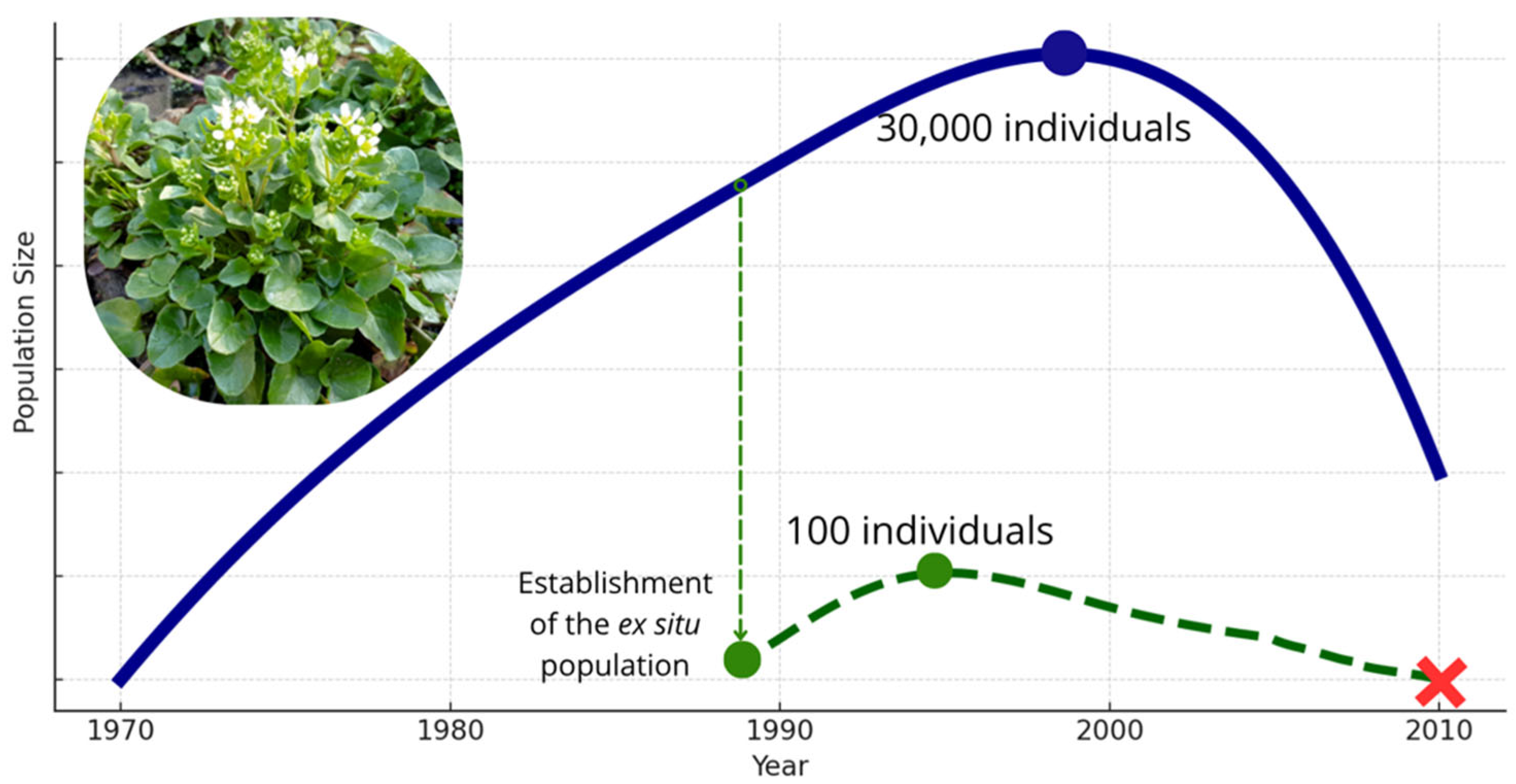
1. Introduction
2. Materials and Methods
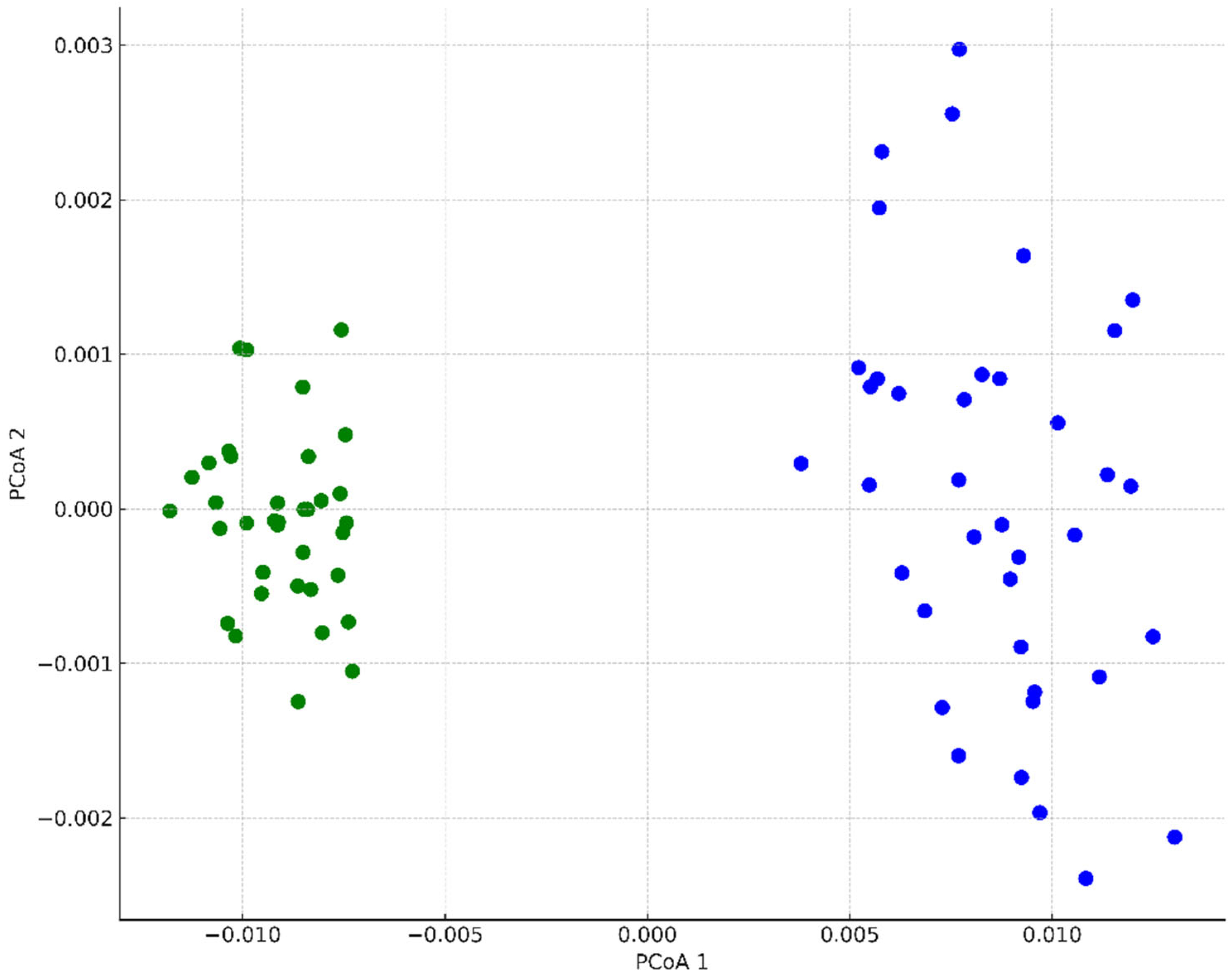
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.G.; da Fonseca, G.A.B.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Kruckeberg, A.R.; Rabinowitz, D. Biological aspects of endemism in higher plants. Annu. Rev. Ecol. Syst. 1985, 16, 447–479. [Google Scholar] [CrossRef]
- Brooks, T.M.; Mittermeier, R.A.; Mittermeier, C.G.; da Fonseca, G.A.B.; Rylands, A.B.; Konstant, W.R.; Hilton-Taylor, C. Habitat loss and extinction in the hotspots of biodiversity. Conserv. Biol. 2002, 16, 909–923. [Google Scholar] [CrossRef]
- IUCN. The IUCN Red List of Threatened Species; Version 2022-2; IUCN: Gland, Switzerland, 2022. [CrossRef]
- Frankham, R.; Ballou, J.D.; Briscoe, D.A. Introduction to Conservation Genetics; Cambridge University Press: Cambridge, UK, 2002. [Google Scholar] [CrossRef]
- Mace, G.M.; Gittleman, J.L.; Purvis, A. Preserving the tree of life. Science 2003, 300, 1707–1709. [Google Scholar] [CrossRef]
- Antonelli, A.; Fry, C.; Smith, R.J.; Simmonds, M.S.J.; Kersey, P.J.; Pritchard, H.W.; Abbo, M.S.; Acedo, C.; Adams, J.; Ainsworth, A.M.; et al. State of the World’s Plants and Fungi 2020; Royal Botanic Gardens, Kew: London, UK, 2020. [Google Scholar] [CrossRef]
- Higgins, K.; Lynch, M. Metapopulation extinction caused by mutation accumulation. Proc. Natl. Acad. Sci. USA 2001, 98, 2928–2933. [Google Scholar] [CrossRef]
- Panahi, P.; Jamzad, Z.; Jalili, A.; Talebi, K.S.; Pourhashemi, M. The role of the National Botanical Garden of Iran in ex situ conservation of Buxus hyrcana Pojark.; An endangered species. Urban For. Urban Green. 2021, 57, 126951. [Google Scholar] [CrossRef]
- Pritchard, D.J.; Fa, J.E.; Oldfiels, S.; Harrop, S.R. Bring the captive closer to the wild: Redefining the role of ex situ conservation. Oryx 2012, 46, 18–23. [Google Scholar] [CrossRef]
- Li, Z.; Pritchard, H.W. The science and economics of ex situ plant conservation. Trends Plant Sci. 2009, 14, 614–621. [Google Scholar] [CrossRef]
- Havens, K.; Kramer, A.T.; Guerrant, E.O. Ex situ plant conservation and beyond. BioScience 2014, 64, 735–745. [Google Scholar]
- Chwedorzewska, K.J.; Bednarek, P.T.; Puchalski, J. Studies on changes in specific rye genome regions due to seed ageing and regeneration. Cell. Mol. Biol. Lett. 2002, 7b, 569–576. [Google Scholar]
- Havens, K.; Vitt, P.; Maunder, M.; Guerrant, E.O.; Dixon, K. Ex situ plant conservation and beyond. Bioscience 2006, 56, 525–531. [Google Scholar] [CrossRef]
- Ellstrand, N.C.; Elam, D.R. Population genetic consequences of small population size: Implications for plant conservation. Annu. Rev. Ecol. Syst. 1993, 24, 217–242. [Google Scholar] [CrossRef]
- Aguilar, R.; Cristobal-Perez, E.J.; Balvino-Olvera, F.J.; Aguilar-Aguilar, M.D.; Aguirre-Acosta, N.; Ashworth, L.; Lobo, J.A.; Marten-Rodriguez, S.; Fuchs, E.J.; Sanchez-Montoya, G.; et al. Habitat fragmentation reduces plant progeny quality: A global synthesis. Ecol. Lett. 2019, 22, 1163–1173. [Google Scholar] [CrossRef]
- Young, A.; Boyle, T.; Brown, T. The population genetic consequences of habitat fragmentation from plants. Trends Ecol. Evol. 1996, 11, 413–418. [Google Scholar] [CrossRef]
- Cieślak, E.; Cieślak, J.; Ronikier, M. Phylogeographical structure of a narrow-endemic plant in an isolated high-mountain range: The case of Cochlearia tatrae in the Tatra Mts (Western Carpathians). Preslia 2021, 93, 125–148. [Google Scholar] [CrossRef]
- Kwiatkowska, A. Rozmieszczenie warzuchy polskiej (Cochlearia polonica E. Fröhlich) w okolicy Olkusza. Fragm. Florist. Geobot. 1957, 27, 11–20. [Google Scholar]
- Piech, K. Doronicum austriacum Jacq. i Cochlearia officinalis L. w okolicy Olkusza. Acta Soc. Bot. Pol. 1924, 2, 216–221. [Google Scholar] [CrossRef][Green Version]
- Fröhlich, E. Systematische Studien über polnische Esslöffel (Cochlearia L.) unter Berücksichtigung der verwandten europäischen Arten. Bull. Acad. Pol. Sci. Lettr. Cl. Math.-Nat. Ser. B 1937, 1, 129–146. [Google Scholar]
- Bajer, A. Cytological studies on Cochlearia polonica Fröhl. Acta Soc. Bot. Pol. 1950, 20, 635–646. [Google Scholar] [CrossRef][Green Version]
- Koch, M.; Dobeš, C.; Bernhardt, K.; Kochjarová, J. Cochlearia macrorrhiza (Brassicaceae): A bridging species between Cochlearia taxa from the Eastern Alps and the Carpathians? Plant Syst. Evol. 2003, 242, 137–147. [Google Scholar] [CrossRef]
- Kochjarová, J.; Valachovič, M.; Bureš, P.; Mráz, P. The genus Cochlearia L. (Brassicaceae) in the Eastern Carpathians and adjacent area. Bot. J. Linn. Soc. 2006, 151, 355–364. [Google Scholar] [CrossRef]
- Kwiatkowska, A. Cochlearia polonica Fröhlich–warzucha polska. In Polish Red Data Book of Plants; Kaźmierczakowa, R., Zarzycki, K., Eds.; Instytut Botaniki PAN, Instytut Ochrony Przyrody PAN: Kraków, Poland, 2001; pp. 166–167. [Google Scholar]
- Cieślak, E.; Korbecka, G.; Ronikier, M. Genetic structure of the critically endangered endemic Cochlearia polonica (Brassicaceae): Efficiency of the last-chance transplantation. Bot. J. Linn. Soc. 2007, 155, 527–532. [Google Scholar] [CrossRef][Green Version]
- Rucińska, A.; Puchalski, J. Comparative molecular studies on genetic diversity of ex situ garden collection and its source population of Cochlearia polonica. Biodivers. Conserv. 2011, 20, 401–413. [Google Scholar] [CrossRef]
- Burska, A. Cultivation of Cochlearia polonica Fröhlich in Botanical Garden of the Polish Academy of Sciences in Powsin. Biul. Ogr. Bot. 1995, 4, 3–4. (In Polish) [Google Scholar]
- Enßlin, A.; Sandner, T.M.; Matthies, D. Consequences of ex situ cultivation of plants: Genetic diversity, fitness and adaptation of the monocarpic Cynoglossum officinale L. in botanic gardens. Biol. Conserv. 2011, 144, 272–278. [Google Scholar] [CrossRef]
- Lauterbach, D.; Burkart, M.; Gemeinholzer, B. Rapid genetic differentiation between ex situ and their in-situ source populations: An example of the endangered Silene otitis (Caryophyllaceae). Bot. J. Linn. Soc. 2011, 168, 64–75. [Google Scholar] [CrossRef]
- Guerrant, E.O. Experimental reintroduction of Stephanomeria malheurensis. In Restoring Diversity. Strategies for Reintroduction of Endangered Plants; Falk, D.A., Millar, C.I., Olwell, M., Eds.; Island Press: Washington, DC, USA, 1996; pp. 399–402. [Google Scholar]
- Guerrant, E.O., Jr.; Pavlik, B. Reintroduction of rare plants: Genetics, demography, and the role of ex situ conservation methods. In Conservation Biology for the Coming Decade, 2nd ed.; Fiedler, P.L., Kareiva, P.M., Eds.; Chapman and Hall: New York, NY, USA, 1998; pp. 80–108. [Google Scholar]
- Husband, B.C.; Campbell, L.G. Population responses to novel environments: Implications for ex situ plant conservation. In Ex Situ Plant Conservation: Supporting Species Survival in the Wild; Guerrant, E.O., Jr., Havens, K., Maunder, M., Eds.; Island Press: Washington, DC, USA, 2004; pp. 231–266. [Google Scholar]
- Guerrant, E.O.; Havens, K.; Vitt, P. Sampling for effective ex situ plant conservation. Int. J. Plant Sci. 2014, 175, 11–20. [Google Scholar] [CrossRef]
- Mounce, R.; Smith, P.; Brockington, S. Ex situ conservation of plant diversity in the world’s botanic gardens. Nat. Plants 2017, 3, 795–802. [Google Scholar] [CrossRef]
- Guerrant, E.O.; Havens, K.; Maunder, M. Ex Situ Plant Conservation: Supporting Species Survival in the Wild; Island Press: Washington, DC, USA, 2004. [Google Scholar]
- Crane, P.R.; Hopper, S.D.; Raven, P.H.; Stevenson, D.W. Plant science research in botanic gardens. Trends Plant Sci. 2009, 14, 575–577. [Google Scholar] [CrossRef]
- Cibrian-Jaramillo, A.; Hird, A.; Oleas, N.; Ma, H.; Meerow, A.W.; Francisco-Ortega, J.; Griffith, M.P. What is the conservation value of a plant in a botanic garden? Using indicators to improve management of ex situ collections. Bot. Rev. 2013, 79, 559–577. [Google Scholar] [CrossRef]
- Cavender, N.; Westwood, M.; Bechtoldt, C.; Donnelly, G.; Oldfield, S.; Gardner, M.; Rae, D.; McNamara, W. Strengthening the conservation value of ex situ tree collections. Oryx 2015, 49, 416–424. [Google Scholar] [CrossRef]
- BGCI. Garden Search Database. Available online: https://www.bgci.org/resources/bgci-databases/gardensearch/ (accessed on 6 April 2025).
- Chen, G.; Sun, W. The role of botanical gardens in scientific research, conservation, and citizen science. Plant Divers. 2018, 40, 181–188. [Google Scholar] [CrossRef] [PubMed]
- McGlaughlin, M.E.; Riley, L.; Brandsrud, M.; Arcibal, E.; Helenurm, M.K.; Helenurm, K. How much is enough? Minimum sampling intensity required to capture extant genetic diversity in ex situ seed collections: Examples from the endangered plant Sibara filifolia (Brassicaceae). Conserv. Genet. 2015, 16, 253–266. [Google Scholar] [CrossRef]
- Griffith, M.P.; Calonje, M.; Meerow, A.W.; Francisco-Ortega, J.; Knowles, L.; Aguilar, R.; Tut, F.; Sánchez, V.; Meyer, A.; Noblick, L.R.; et al. Will the same ex situ protocols give similar results for closely related species? Biodivers. Conserv. 2017, 26, 2951–2966. [Google Scholar] [CrossRef]
- Hoban, S. New guidance for ex situ gene conservation: Sampling realistic population systems and accounting for collection attrition. Biol Conserv. 2019, 235, 199–208. [Google Scholar] [CrossRef]
- Hoban, S.; Callicrate, T.; Clark, J.; Deans, S.; Dosmann, M.; Fant, J.; Gailing, O.; Havens, K.; Hipp, A.L.; Kadav, P.; et al. Taxonomic similarity does not predict necessary sample size for ex situ conservation: A comparison among five genera. Proc. R. Soc. B. Biol Sci. 2020, 287, 20200102. [Google Scholar] [CrossRef]
- Vos, P.; Hogers, R.; Bleeke, M.; van de Lee, T.; Hornes, M.; Frijters, A.; Zauber, M. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef] [PubMed]
- Chwedorzewska, K.J.; Bednarek, P.T.; Puchalski, J.; Krajewski, P. AFLP profiling of long-term stored and regenerated rye genebank samples. Cell. Mol. Biol. Lett. 2002, 7a, 457–463. [Google Scholar]
- Duchesne, P.; Bernatchez, L. AFLPOP: A computer program for simulated and real population allocation based on AFLP data. Mol. Ecol. Notes 2002, 2, 380–383. [Google Scholar] [CrossRef][Green Version]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research- an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Nei, M. Analysis of Gene Diversity in Subdivided Populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef] [PubMed]
- Bensch, S.; Ĺkesson, M. Ten years of AFLP in ecology and evolution: Why so few animals? Mol. Ecol. 2005, 14, 2899–2914. [Google Scholar] [CrossRef]
- Excoffier, L. Patterns of DNA sequence diversity and genetic structure after a range expansion: Lessons from the infinite–island model. Mol. Ecol. 2004, 13, 853–864. [Google Scholar] [CrossRef]
- Excoffier, L.; Smouse, E.P.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction sites. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.X. Statistical tests of neutrality of mutations against population growth, hitchhiking, and background selection. Genetics 1997, 147, 915–925. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; Vonholdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol. Ecol. Notes 2007, 7, 574–578. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P. PAST: Paleontological statistics software packagefor education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Cornuet, J.M.; Luikart, G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 1996, 144, 2001–2014. [Google Scholar] [CrossRef] [PubMed]
- Tero, N.; Aspi, J.; Siikamäki, P.; Jäkäläniemi, A.; Tuomi, J. Genetic structure and gene flow in a metapopulation of an endangered plant species, Silene tatarica. Mol. Ecol. 2003, 12, 2073–2085. [Google Scholar] [CrossRef]
- Schneider, S.; Excoffier, L. Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: Application to human mitochondrial DNA. Genetics 1999, 152, 1079–1089. [Google Scholar] [CrossRef]
- Harpending, H.C. Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum. Biol. 1994, 66, 591–600. [Google Scholar]
- Rosche, C.; Baasch, A.; Runge, K.; Brade, P.; Träger, S.; Parisod, C.; Hensen, I. Tracking population genetic signatures of local extinction with herbarium specimens. Ann. Bot. 2022, 129, 857–868. [Google Scholar] [CrossRef]
- Rutherford, S.; Wilson, T.C.; Yap, J.-Y.S.; Lee, E.; Errington, G.; Rossetto, M. Evolutionary processes in an undescribed eucalypt: Implications for the translocation of a critically endangered species. Ann. Bot. 2022, 130, 491–508. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Conery, J.; Bürger, R. Mutational meltdowns in sexual populations. Evolution 1995, 49, 1067–1080. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.; Bürger, R. Mutation Accumulation and the Extinction of Small Populations. Am. Nat. 1995, 146, 489–518. [Google Scholar] [CrossRef]
- Koch, M.A.; Hurka, H.; Mummenhoff, K. Chloroplast DNA restriction site variation and RAPD-analyses in Cochlearia (Brassicaceae). Biosystematics and speciation processes. Nord. J. Bot. 1996, 16, 585–604. [Google Scholar] [CrossRef]
- Ramsay, M.M.; Dixon, K.W. Propagation Science, Recovery and Translocation of Terrestrial Orchids. In Orchid Conservation; Dixon, K.W., Kell, S.P., Barrett, R.L., Cribb, P.J., Eds.; Natural History Publications: Kota Kinabalu, Malaysia, 2003; pp. 259–288. [Google Scholar]
- Bijlsma, R.; Bundgaard, J.; Boerema, A.C. Does inbreeding affect the extinction risk of small populations?: Predictions from Drosophila. J. Evol. Biol. 2000, 13, 502–514. [Google Scholar] [CrossRef]
- Fox, C.W.; Reed, D.H. Inbreeding depression increases with environmental stress: An experimental study and meta-analysis. Evolution 2011, 65, 246–258. [Google Scholar] [CrossRef] [PubMed]
- Orr, H.A.; Unckless, R.L. Population Extinction and the Genetics of Adaptation. Am. Nat. 2008, 172, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Bijlsma, R.; Loeschcke, V. Genetic erosion impedes adaptive responses to stressful environments. Evol. Appl. 2012, 5, 117–129. [Google Scholar] [CrossRef]
- Puchalski, J.; Niemczyk, M.; Walerowski, P.; Podyma, W.; Kapler, A. Seed banking of Polish endangered plants–the FlorNatur Project. Biodivers. Res. Conserv. 2014, 34, 65–72. [Google Scholar] [CrossRef]
- Lande, R. Risks of population extinction from demographic and environmental stochasticity and random catastrophes. Am. Nat. 1993, 142, 911–927. [Google Scholar] [CrossRef]
- Volis, S. Conservation utility of botanic garden living collections: Setting a strategy and appropriate methodology. Plant Divers. 2017, 39, 365–372. [Google Scholar] [CrossRef]
- Ensslinn, A.; Godefroid, S. How the cultivation of wild plants in botanic gardens can change their genetic and phenotypic status and what this means for their conservation value. Sibbaldia J. Bot. Gard. Hortic. 2019, 17, 51–69. [Google Scholar] [CrossRef]
| Population | N | Ne | Nu | Np | P [%] | I | He |
|---|---|---|---|---|---|---|---|
| Botanical Garden | 36 | 1.044 | 1 | 27 | 7.42% | 0.038 | * 0.026 |
| Centuria | 38 | 1.078 | 2 | 42 | 11.54% | 0.063 | ** 0.043 |
| Source of Variation | d.f. | SS | Variance Components | Percentage of Variation |
|---|---|---|---|---|
| Among populations | 1 | 275.810 | 7.312 | 57.19 |
| Within populations | 72 | 394.149 | 5.474 | 42.81 |
| Total | 73 | 669.956 | 12.786 |
| Pairwise FST | ΦPT | Nei GD | Nei Unbiased Genetic Identity |
|---|---|---|---|
| 0.572 | 0.572 | 0.059 | 0.943 |
| Test | Description | Populations | Mean | SD | |
|---|---|---|---|---|---|
| BG | C | ||||
| Tajima’s D test | S | 27 | 42 | 34.50 | 10.607 |
| Pi | 7.962 | 13.774 | 10.868 | 4.110 | |
| Tajima’s D | 0.7733 | 1.343 | 1.058 | 0.403 | |
| Tajima’s D p-value | 0.835 | 0.947 | 0.891 | 0.079 | |
| Fu’s FS test | Theta pi | 7.962 | 13.774 | 10.868 | 4.110 |
| Exp. no. of alleles | 14.024 | 18.611 | 16.317 | 3.244 | |
| FS | −24.921 | −24.379 | −24.650 | 0.383 | |
| FS p-value | 0.000 | 0.000 | 0.000 | 0.000 | |
| Model | Statistic | Population | Mean | SD | |
|---|---|---|---|---|---|
| BG | C | ||||
| Demographic expansion | SSD | 0.005 | 0.001 | 0.003 | 0.003 |
| Model (SSD) p-value | 0.150 | 0.740 | 0.445 | 0.417 | |
| Raggedness index | 0.013 | 0.005 | 0.009 | 0.006 | |
| Raggedness p-value | 0.230 | 0.600 | 0.415 | 0.262 | |
| Spatial expansion | SSD | 0.005 | 0.001 | 0.003 | 0.003 |
| Model (SSD) p-value | 0.140 | 0.620 | 0.380 | 0.339 | |
| Raggedness index | 0.013 | 0.005 | 0.009 | 0.006 | |
| Raggedness p-value | 0.340 | 0.560 | 0.450 | 0.156 | |
| Population | SIGN Test | Standardised Test | Wilcoxon Test |
|---|---|---|---|
| Botanical Garden | Heex = 11.96 Hd = 5 Hex = 22 p = 0.00009 | T2 = 3.801 p = 0.00007 | One tail for heterozygosity deficiency: 0.99976 One tail for heterozygosity excess: 0.00027 Two tails for heterozygosity excess and deficiency: 0.00054 |
| Centuria | Heex = 18.66 Hd = 7 Hex = 35 p = 0.0000 | T2 = 5.713 p = 0.0000 | One tail for heterozygosity deficiency: 1.0000 One tail for heterozygosity excess: 0.0000 Two tails for heterozygosity excess and deficiency: 0.0000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rucińska, A.; Chwedorzewska, K.J.; Bednarek, P.T.; Boczkowska, M.; Puchalski, J.; Androsiuk, P.; Czaplicka, E. Halfway Through Ex Situ Population Genetic Lifespan: The Case of Cochlearia polonica. Biology 2025, 14, 681. https://doi.org/10.3390/biology14060681
Rucińska A, Chwedorzewska KJ, Bednarek PT, Boczkowska M, Puchalski J, Androsiuk P, Czaplicka E. Halfway Through Ex Situ Population Genetic Lifespan: The Case of Cochlearia polonica. Biology. 2025; 14(6):681. https://doi.org/10.3390/biology14060681
Chicago/Turabian StyleRucińska, Anna, Katarzyna Joanna Chwedorzewska, Piotr Tomasz Bednarek, Maja Boczkowska, Jerzy Puchalski, Piotr Androsiuk, and Ewa Czaplicka. 2025. "Halfway Through Ex Situ Population Genetic Lifespan: The Case of Cochlearia polonica" Biology 14, no. 6: 681. https://doi.org/10.3390/biology14060681
APA StyleRucińska, A., Chwedorzewska, K. J., Bednarek, P. T., Boczkowska, M., Puchalski, J., Androsiuk, P., & Czaplicka, E. (2025). Halfway Through Ex Situ Population Genetic Lifespan: The Case of Cochlearia polonica. Biology, 14(6), 681. https://doi.org/10.3390/biology14060681








