Genomic Diversity and Selection Signatures for Zaosheng Cattle
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation and DNA Sequencing
2.2. Read Mapping and Variant Calling
2.3. Population Genetic Analysis
2.4. Detection of Selection Signals
2.5. Local Ancestry Inference
3. Results
3.1. Data Collection, Sequencing, and Identification of SNPs
3.2. Population Structure and Genetic Diversity
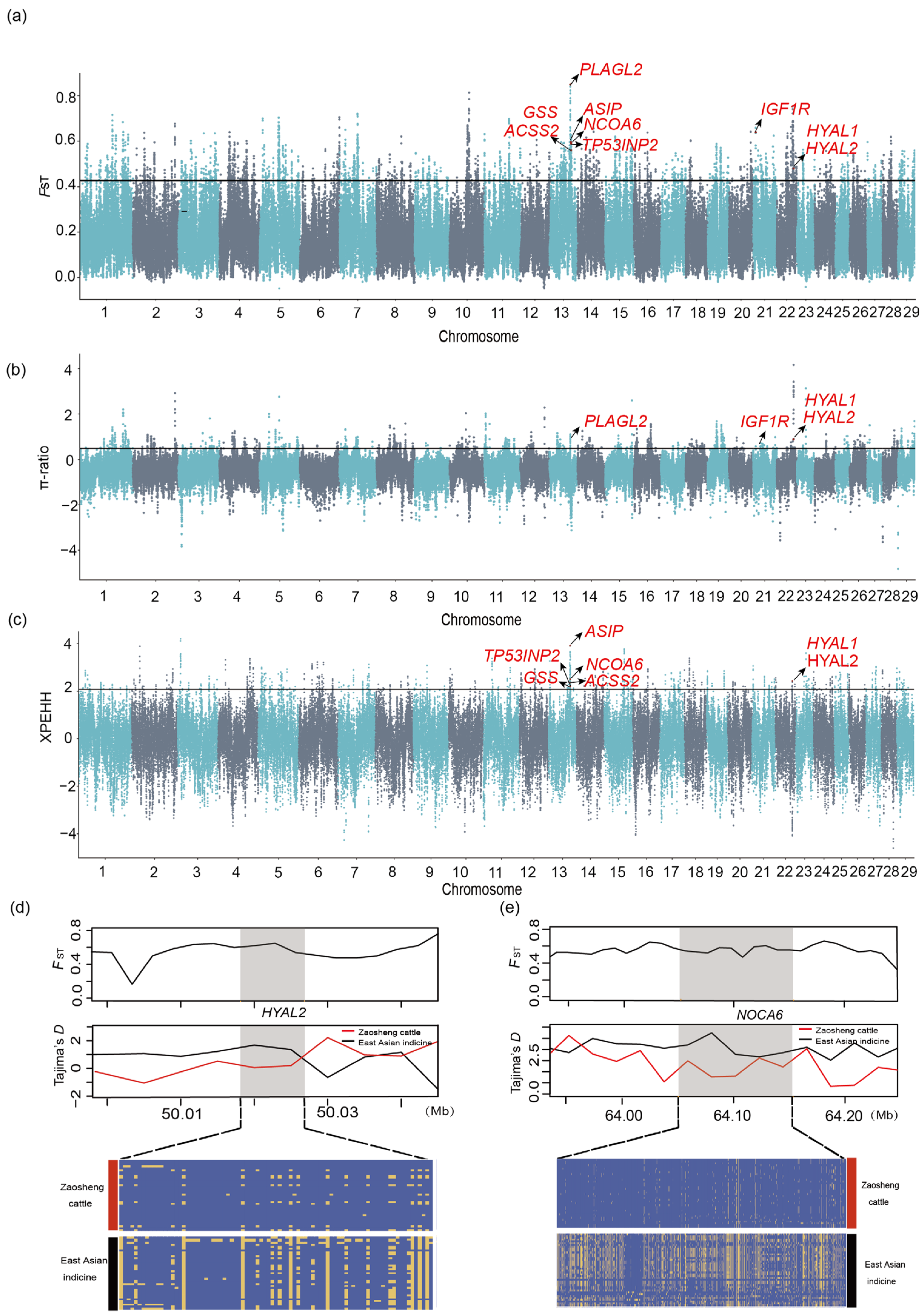
3.3. Genome-Wide Selective Scanning Signals from Zaosheng Cattle
3.3.1. Genetic Signature of Selection in Zaosheng Cattle
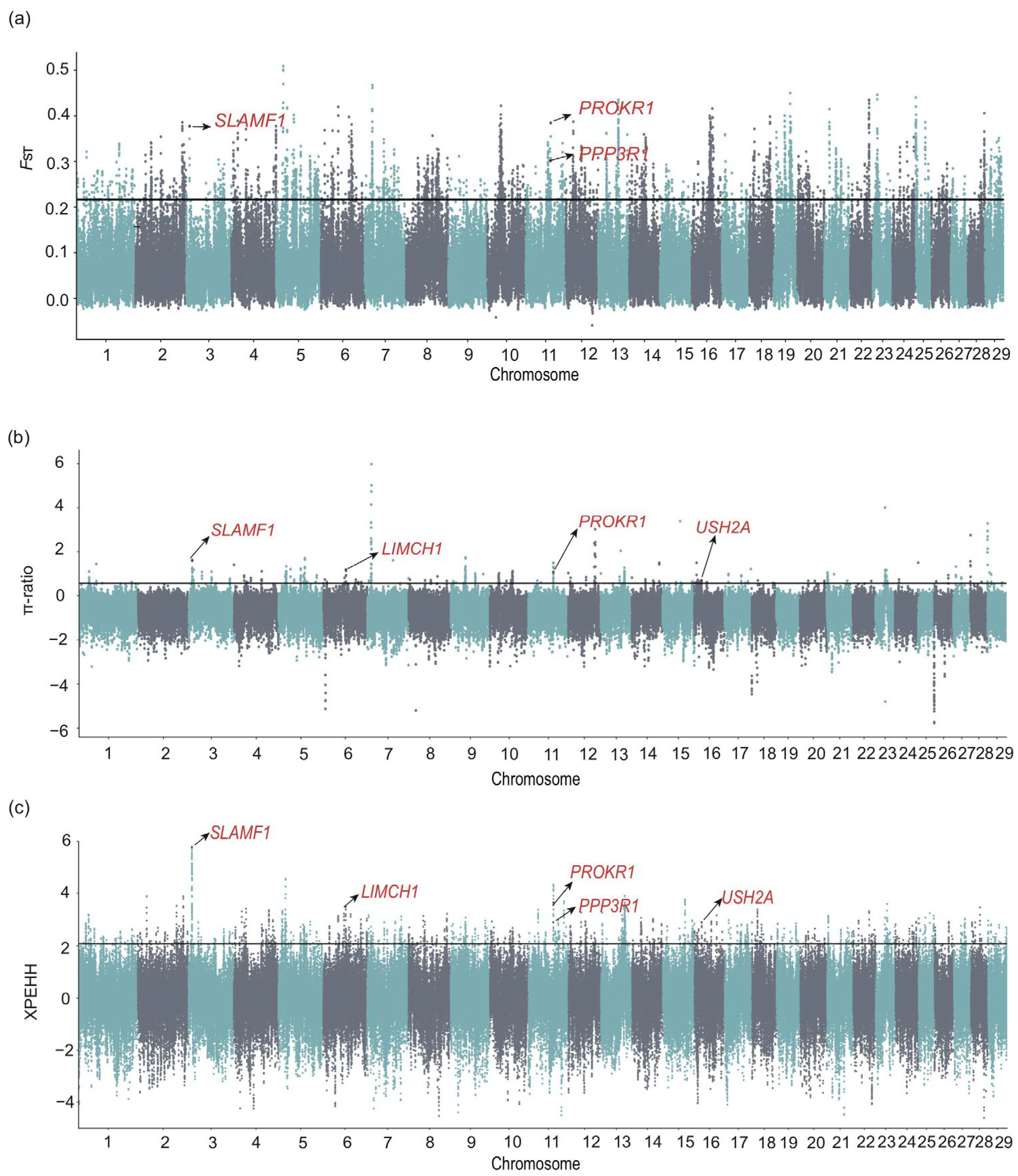
3.3.2. Selective Signals Between Zaosheng Cattle and East Asian Indicine Cattle
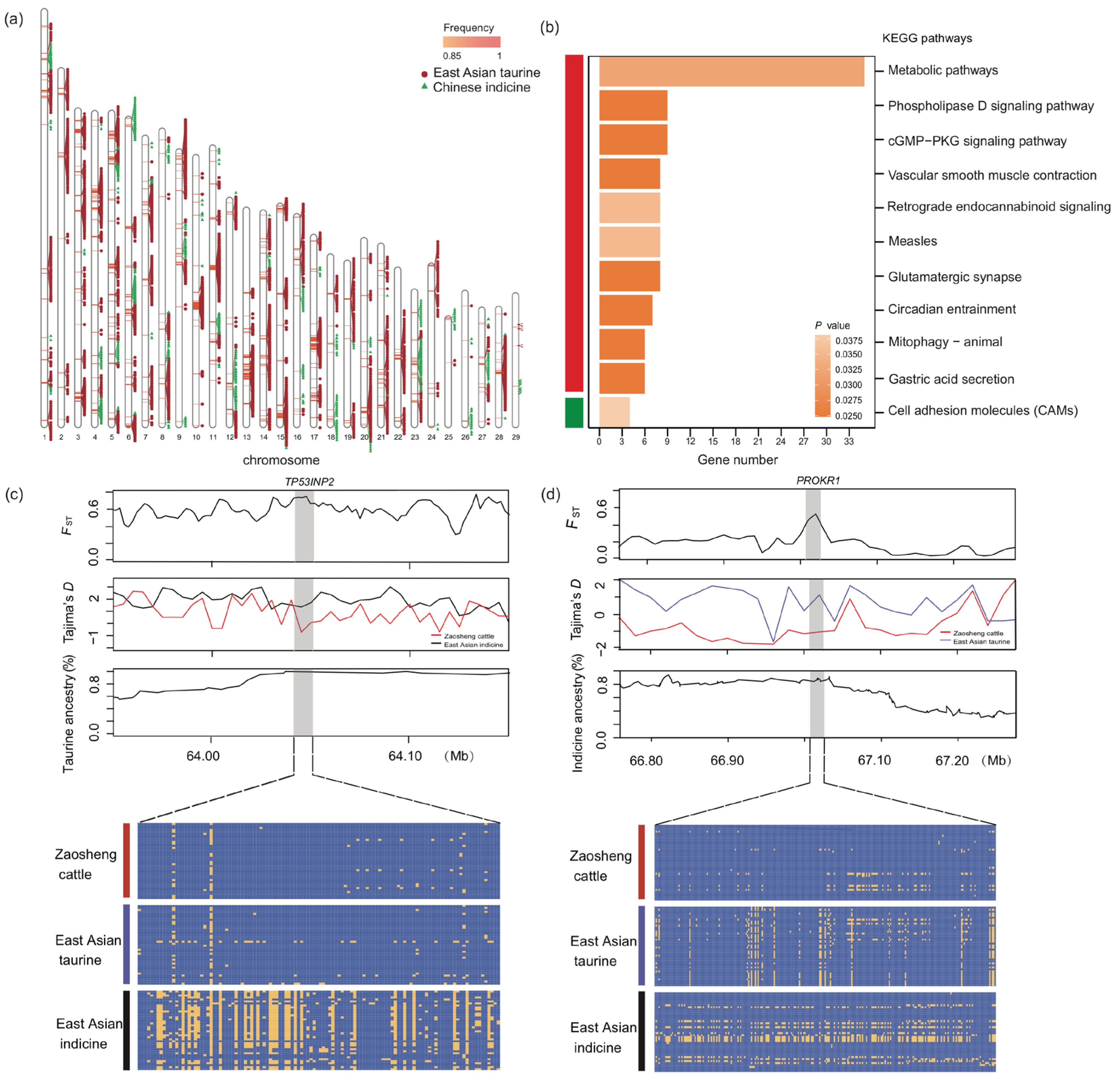
3.3.3. Selection Signature Between Zaosheng Cattle and East Asian Taurine Cattle
3.3.4. Local Ancestry Inference of Zaosheng Cattle
4. Discussion
4.1. Genetic Ancestry and Population Structure of Zaosheng Cattle
4.2. Selection Signatures and Environmental Adaptation
4.3. Local Ancestry Inference of Zaosheng Cattle
4.4. Conservation and Breeding Implications
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Decker, J.E.; McKay, S.D.; Rolf, M.M.; Kim, J.; Molina Alcala, A.; Sonstegard, T.S.; Hanotte, O.; Gotherstrom, A.; Seabury, C.M.; Praharani, L.; et al. Worldwide Patterns of Ancestry, Divergence, and Admixture in Domesticated Cattle. PLoS Genet. 2014, 10, e1004254. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Cai, Y.; Chen, Q.; Li, R.; Wang, K.; Huang, Y.; Hu, S.; Huang, S.; Zhang, H.; Zheng, Z.; et al. Whole-genome resequencing reveals world-wide ancestry and adaptive introgression events of domesticated cattle in East Asia. Nat. Commun. 2018, 9, 2337. [Google Scholar] [CrossRef] [PubMed]
- Gurgul, A.; Jasielczuk, I.; Ropka-Molik, K.; Semik-Gurgul, E.; Pawlina-Tyszko, K.; Szmatola, T.; Szyndler-Nedza, M.; Bugno-Poniewierska, M.; Blicharski, T.; Szulc, K.; et al. A genome-wide detection of selection signatures in conserved and commercial pig breeds maintained in Poland. BMC Genet. 2018, 19, 95. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Y.; Liu, W.; Li, Y.; Pan, J.; Pu, Y.; Han, J.; Orlando, L.; Ma, Y.; Jiang, L. A single-nucleotide mutation within the TBX3 enhancer increased body size in Chinese horses. Curr. Biol. 2022, 32, 480–487.e6. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Kon, T.; Chen, C.; Ichikawa, R.; Zheng, Q.; Pei, L.; Takemura, I.; Nsobi, L.H.; Tabata, H.; Pan, H.; et al. Whole-genome sequencing of endangered Zhoushan cattle suggests its origin and the association of MC1R with black coat colour. Sci. Rep. 2021, 11, 17359. [Google Scholar] [CrossRef]
- Yang, J.; Li, W.-R.; Lv, F.-H.; He, S.-G.; Tian, S.-L.; Peng, W.-F.; Sun, Y.-W.; Zhao, Y.-X.; Tu, X.-L.; Zhang, M.; et al. Whole-Genome Sequencing of Native Sheep Provides Insights into Rapid Adaptations to Extreme Environments. Mol. Biol. Evol. 2016, 33, 2576–2592. [Google Scholar] [CrossRef]
- Guo, X.; Xing, C.-H.; Wei, W.; Zhang, X.-F.; Wei, Z.-Y.; Ren, L.-L.; Jiang, J.-J.; Li, M.; Wang, J.-X.; He, X.-X.; et al. Genome-wide scan for selection signatures and genes related to heat tolerance in domestic chickens in the tropical and temperate regions in Asia. Poult. Sci. 2022, 101, 101821. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Yaxuan, R.; Kaixing, Q.; Suolang, Q.; Basang, Z.; Chuzhao, L.; Ningbo, C. Local ancestry and selection in admixed Sanjiang cattle. Stress Biol. 2023, 3, 30. [Google Scholar]
- Hengwei, Y.; Ke, Z.; Gong, C.; Chugang, M.; Hongbao, W.; Linsen, Z. Genome-wide analysis reveals genomic diversity and signatures of selection in Qinchuan beef cattle. BMC Genom. 2024, 25, 558. [Google Scholar]
- Wang, Y.Q.; Ma, J.; Wang, J.; Zhang, L.P.; Hu, J.W.; Ma, M.H.; Xu, L.Y.; Chen, Y.; Zhu, B.; Wang, Z.Z.; et al. Genetic Origin and Introgression Pattern of Pingliang Red Cattle Revealed Using Genome-Wide SNP Analyses. Genes 2023, 14, 2198. [Google Scholar] [CrossRef]
- Vitti, J.J.; Grossman, S.R.; Sabeti, P.C. Detecting Natural Selection in Genomic Data. Annu. Rev. Genet. 2013, 47, 97–120. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Heng, L.; Bob, H.; Alec, W.; Tim, F.; Jue, R.; Nils, H.; Gabor, M.; Goncalo, A.; Richard, D. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Alexander, D.H.; Lange, K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. Bmc Bioinform. 2011, 12, 246. [Google Scholar] [CrossRef] [PubMed]
- Patterson, N.; Price, A.L.; Reich, D. Population structure and eigenanalysis. PLoS Genet. 2006, 2, 2074–2093. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Nielsen, R.; Williamson, S.; Kim, Y.; Hubisz, M.J.; Clark, A.G.; Bustamante, C. Genomic scans for selective sweeps using SNP data. Genome Res. 2005, 15, 1566–1575. [Google Scholar] [CrossRef]
- DeGiorgio, M.; Huber, C.D.; Hubisz, M.J.; Hellmann, I.; Nielsen, R. Sweepfinder2: Increased sensitivity, robustness and flexibility. Bioinformatics 2016, 32, 1895–1897. [Google Scholar] [CrossRef] [PubMed]
- Hudson, R.R.; Slatkin, M.; Maddison, W.P. Estimation of Levels of Gene Flow from Dna-Sequence Data. Genetics 1992, 132, 583–589. [Google Scholar] [CrossRef]
- Sabeti, P.C.; Reich, D.E.; Higgins, J.M.; Levine, H.Z.P.; Richter, D.J.; Schaffner, S.F.; Gabriel, S.B.; Platko, J.V.; Patterson, N.J.; McDonald, G.J.; et al. Detecting recent positive selection in the human genome from haplotype structure. Nature 2002, 419, 832–837. [Google Scholar] [CrossRef] [PubMed]
- Szpiech, Z.A.; Hernandez, R.D. selscan: An Efficient Multithreaded Program to Perform EHH-Based Scans for Positive Selection. Mol. Biol. Evol. 2014, 31, 2824–2827. [Google Scholar] [CrossRef]
- Dias-Alves, T.; Mairal, J.; Blum, M.G.B. Loter: A Software Package to Infer Local Ancestry for a Wide Range of Species. Mol. Biol. Evol. 2018, 35, 2318–2326. [Google Scholar] [CrossRef]
- Hao, Z.D.; Lv, D.K.; Ge, Y.; Shi, J.S.; Weijers, D.; Yu, G.C.; Chen, J.H. RIdeogram: Drawing SVG graphics to visualize and map genome-wide data on the idiograms. Peerj Comput. Sci. 2020, 6, e251. [Google Scholar] [CrossRef] [PubMed]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef]
- Huang, Z.; Zhang, M.; Plec, A.A.; Estill, S.J.; Cai, L.; Repa, J.J.; McKnight, S.L.; Tu, B.P. ACSS2 promotes systemic fat storage and utilization through selective regulation of genes involved in lipid metabolism. Proc. Natl. Acad. Sci. USA 2018, 115, E9499–E9506. [Google Scholar] [CrossRef]
- Oh, G.-S.; Kim, S.-R.; Lee, E.-S.; Yoon, J.; Shin, M.-K.; Ryu, H.K.; Kim, D.S.; Kim, S.-W. Regulation of Hepatic Gluconeogenesis by Nuclear Receptor Coactivator 6. Mol. Cells 2022, 45, 180–192. [Google Scholar] [CrossRef]
- Liu, Y.; Fang, X.; Zhao, Z.; Li, J.; Albrecht, E.; Schering, L.; Maak, S.; Yang, R. Polymorphisms of the ASIP gene and the haplotype are associated with fat deposition traits and fatty acid composition in Chinese Simmental steers. Arch. Tierz. 2019, 62, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Cheng, H.; Liu, Y.; Sun, L.; Chen, N.; Jiang, F.; You, W.; Yang, Z.; Zhang, B.; Song, E.; et al. Assessing Genomic Diversity and Selective Pressures in Bohai Black Cattle Using Whole-Genome Sequencing Data. Animals 2022, 12, 665. [Google Scholar] [CrossRef] [PubMed]
- Tsoneva, E.; Vasileva-Slaveva, M.B.; Kostov, S.G.; Yordanov, A.D. ROMO1-a potential immunohistochemical prognostic marker for cancer development. Oncologie 2023, 25, 753–758. [Google Scholar] [CrossRef]
- Ortega, M.S.; Denicol, A.C.; Cole, J.B.; Null, D.J.; Hansen, P.J. Use of single nucleotide polymorphisms in candidate genes associated with daughter pregnancy rate for prediction of genetic merit for reproduction in Holstein cows. Anim. Genet. 2016, 47, 288–297. [Google Scholar] [CrossRef]
- Sekar, M.; Thirumurugan, K. Autophagic Regulation of Adipogenesis Through TP53INP2: Insights from In Silico and In Vitro Analysis. Mol. Biotechnol. 2024, 66, 1188–1205. [Google Scholar] [CrossRef] [PubMed]
- Galvan, L.; Francelle, L.; Gaillard, M.-C.; de Longprez, L.; Carrillo-de Sauvage, M.-A.; Liot, G.; Cambon, K.; Stimmer, L.; Luccantoni, S.; Flament, J.; et al. The striatal kinase DCLK3 produces neuroprotection against mutant huntingtin. Brain 2018, 141, 1434–1454. [Google Scholar] [CrossRef]
- Jin, Z.; Zhang, G.; Liu, Y.; He, Y.; Yang, C.; Du, Y.; Gao, F. The suppressive role of HYAL1 and HYAL2 in the metastasis of colorectal cancer. J. Gastroenterol. Hepatol. 2019, 34, 1766–1776. [Google Scholar] [CrossRef]
- Ardicli, S.; Samli, H.; Vatansever, B.; Soyudal, B.; Dincel, D.; Balci, F. Comprehensive assessment of candidate genes associated with fattening performance in Holstein-Friesian bulls. Arch. Anim. Breed. 2019, 62, 9–32. [Google Scholar] [CrossRef]
- Arikawa, L.M.; Mota, L.F.M.; Schmidt, P.I.; Frezarim, G.B.; Fonseca, L.F.S.; Magalhaes, A.F.B.; Silva, D.A.; Carvalheiro, R.; Chardulo, L.A.L.; de Albuquerque, L.G. Genome-wide scans identify biological and metabolic pathways regulating carcass and meat quality traits in beef cattle. Meat Sci. 2024, 209, 109402. [Google Scholar] [CrossRef]
- Huang, J.; Jia, Y.; Li, Q.; Son, K.; Hamilton, C.; Burris, W.R.; Bridges, P.J.; Stromberg, A.J.; Matthews, J.C. Glutathione content and expression of proteins involved with glutathione metabolism differs in longissimus dorsi, subcutaneous adipose, and liver tissues of finished vs. growing beef steers. J. Anim. Sci. 2018, 96, 5152–5165. [Google Scholar] [CrossRef]
- Engel, P. SLAMF receptors and disease. Clin. Immunol. 2019, 204, 1–2. [Google Scholar] [CrossRef]
- Santos Silva, D.B.d.; Fonseca, L.F.S.; Magalhães, A.F.B.; Muniz, M.M.M.; Baldi, F.; Ferro, J.A.; Chardulo, L.A.L.; Pinheiro, D.G.; Albuquerque, L.G.d. Transcriptome profiling of muscle in Nelore cattle phenotypically divergent for the ribeye muscle area. Genomics 2020, 112, 1257–1263. [Google Scholar] [CrossRef] [PubMed]
- Baryla, M.; Goryszewska-Szczurek, E.; Kaczynski, P.; Balboni, G.; Waclawik, A. Prokineticin 1 is a novel factor regulating porcine corpus luteum function. Sci. Rep. 2023, 13, 5085. [Google Scholar] [CrossRef] [PubMed]
- Birch, D.G.; Samarakoon, L.; Melia, M.; Duncan, J.L.; Ayala, A.R.; Audo, I.; Cheetham, J.K.; Durham, T.A.; Iannaccone, A.; Pennesi, M.E.; et al. The RUSH2A Study: Dark-Adapted Visual Fields in Patients with Retinal Degeneration Associated with Biallelic Variants in the USH2A Gene. Investig. Ophthalmol. Vis. Sci. 2022, 63, 17. [Google Scholar] [CrossRef]
- Marom, A.; Barak, A.F.; Kramer, M.P.; Lewinsky, H.; Binsky-Ehrenreich, I.; Cohen, S.; Tsitsou-Kampeli, A.; Kalchenko, V.; Kuznetsov, Y.; Mirkin, V.; et al. CD84 mediates CLL-microenvironment interactions. Oncogene 2017, 36, 628–638. [Google Scholar] [CrossRef]
- McArdel, S.L.; Terhorst, C.; Sharpe, A.H. Roles of CD48 in regulating immunity and tolerance. Clin. Immunol. 2016, 164, 10–20. [Google Scholar] [CrossRef]
- Carre, G.-A.; Couty, I.; Hennequet-Antier, C.; Govoroun, M.S. Gene Expression Profiling Reveals New Potential Players of Gonad Differentiation in the Chicken Embryo. PLoS ONE 2011, 6, e23959. [Google Scholar] [CrossRef]
- Chu, T.; Dufort, I.; Sirard, M.A. Effect of ovarian stimulation on oocyte gene expression in cattle. Theriogenology 2012, 77, 1928–1938. [Google Scholar] [CrossRef]
- Colpan, M.; Moroz, N.A.; Gray, K.T.; Cooper, D.A.; Diaz, C.A.; Kostyukova, A.S. Tropomyosin-binding properties modulate competition between tropomodulin isoforms. Arch. Biochem. Biophys. 2016, 600, 23–32. [Google Scholar] [CrossRef]
- Hitsumoto, T.; Tsukamoto, O.; Matsuoka, K.; Li, J.; Liu, L.; Kuramoto, Y.; Higo, S.; Ogawa, S.; Fujino, N.; Yoshida, S.; et al. Restoration of Cardiac Myosin Light Chain Kinase Ameliorates Systolic Dysfunction by Reducing Superrelaxed Myosin. Circulation 2023, 147, 1902–1918. [Google Scholar] [CrossRef]
- Shrestha, M.M.; Lim, C.-Y.; Bi, X.; Robinson, R.C.; Han, W. Tmod3 Phosphorylation Mediates AMPK-Dependent GLUT4 Plasma Membrane Insertion in Myoblasts. Front. Endocrinol. 2021, 12, 653557. [Google Scholar] [CrossRef] [PubMed]
- Doran, A.G.; Berry, D.P.; Creevey, C.J. Whole genome association study identifies regions of the bovine genome and biological pathways involved in carcass trait performance in Holstein-Friesian cattle. BMC Genom. 2014, 15, 837. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.-F.; Lyu, S.; Wang, X.; Zhang, Z.; Qu, K.; Xu, J.; Cai, C.; Li, Z.; Xie, J.; Ru, B.; et al. The combination between NCSTN gene copy number variation and growth traits in Chinese cattle. Anim. Biotechnol. 2021, 32, 683–687. [Google Scholar] [CrossRef]
- Cao, X.-K.; Huang, Y.-Z.; Ma, Y.-L.; Cheng, J.; Qu, Z.-X.; Ma, Y.; Bai, Y.-Y.; Tian, F.; Lin, F.-P.; Ma, Y.-L.; et al. Integrating CNVs into meta-QTL identified GBP4 as positional candidate for adult cattle stature. Funct. Integr. Genom. 2018, 18, 559–567. [Google Scholar] [CrossRef]
- Yu, X.; Zhai, C.; Fan, Y.; Zhang, J.; Liang, N.; Liu, F.; Cao, L.; Wang, J.; Du, J. TUSC3: A novel tumour suppressor gene and its functional implications. J. Cell. Mol. Med. 2017, 21, 1711–1718. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.M.; Kim, J.S.; Yoo, Y.D. A novel protein, Romo1, induces ROS production in the mitochondria. Biochem. Biophys. Res. Commun. 2006, 347, 649–655. [Google Scholar] [CrossRef]
- Sun, G.; Cao, Y.; Qian, C.; Wan, Z.; Zhu, J.; Guo, J.; Shi, L. Romo1 is involved in the immune response of glioblastoma by regulating the function of macrophages. Aging 2020, 12, 1114–1127. [Google Scholar] [CrossRef]
- Guo, S.S.; Lv, C.Y.; Ouyang, S.J.; Wang, X.L.; Liao, A.H.; Yuan, S.Q. GOLGA4, A Golgi matrix protein, is dispensable for spermatogenesis and male fertility in mice. Biochem. Biophys. Res. Commun. 2020, 529, 642–646. [Google Scholar] [CrossRef]
- Zhang, F.; Qu, K.; Chen, N.; Hanif, Q.; Jia, Y.; Huang, Y.; Dang, R.; Zhang, J.; Lan, X.; Chen, H.; et al. Genome-Wide SNPs and InDels Characteristics of Three Chinese Cattle Breeds. Animals 2019, 9, 596. [Google Scholar] [CrossRef]
- Xia, X.; Zhang, F.; Li, S.; Luo, X.; Peng, L.; Dong, Z.; Pausch, H.; Leonard, A.S.; Crysnanto, D.; Wang, S.; et al. Structural variation and introgression from wild populations in East Asian cattle genomes confer adaptation to local environment. Genome Biol. 2023, 24, 211. [Google Scholar] [CrossRef]
- Midgley, A.C.; Woods, E.L.; Jenkins, R.H.; Brown, C.; Khalid, U.; Chavez, R.; Hascall, V.; Steadman, R.; Phillips, A.O.; Meran, S. Hyaluronidase-2 Regulates RhoA Signaling, Myofibroblast Contractility, and Other Key Profibrotic Myofibroblast Functions. Am. J. Pathol. 2020, 190, 1236–1255. [Google Scholar] [CrossRef] [PubMed]
- Donelan, W.; Dominguez-Gutierrez, P.R.; Kusmartsev, S. Deregulated hyaluronan metabolism in the tumor microenvironment drives cancer inflammation and tumor-associated immune suppression. Front. Immunol. 2022, 13, 971278. [Google Scholar] [CrossRef]
- Mahajan, M.A.; Samuels, H.H. Nuclear receptor coactivator/coregulator NCoA6(NRC) is a pleiotropic coregulator involved in transcription, cell survival, growth and development. Nucl. Recept. Signal. 2008, 6, e002. [Google Scholar] [CrossRef]
- Wang, S.; Raza, S.H.A.; Zhang, K.; Mei, C.; Alamoudi, M.O.; Aloufi, B.H.; Alshammari, A.M.; Zan, L. Selection signatures of Qinchuan cattle based on whole-genome sequences. Anim. Biotechnol. 2023, 34, 1483–1491. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Y.; Xu, H. LIMCH1 suppress the growth of lung cancer by interacting with HUWE1 to sustain p53 stability. Gene 2019, 712, 143963. [Google Scholar] [CrossRef]
- Chen, Q.; Huang, B.; Zhan, J.; Wang, J.; Qu, K.; Zhang, F.; Shen, J.; Jia, P.; Ning, Q.; Zhang, J.; et al. Whole-genome analyses identify loci and selective signals associated with body size in cattle. J. Anim. Sci. 2020, 98, skaa068. [Google Scholar] [CrossRef] [PubMed]
- Harjunpaa, H.; Asens, M.L.; Guenther, C.; Fagerholm, S.C. Cell Adhesion Molecules and Their Roles and Regulation in the Immune and Tumor Microenvironment. Front. Immunol. 2019, 10, 1078. [Google Scholar] [CrossRef]
- Shen, J.; Hanif, Q.; Cao, Y.; Yu, Y.; Lei, C.; Zhang, G.; Zhao, Y. Whole Genome Scan and Selection Signatures for Climate Adaption in Yanbian Cattle. Front. Genet. 2020, 11, 94. [Google Scholar] [CrossRef]
- Baryla, M.; Kaczynski, P.; Goryszewska-Szczurek, E.; Waclawik, A. The regulation of the expression of prokineticin 1 and its receptors and its mechanism of action in the porcine corpus luteum. Theriogenology 2024, 226, 39–48. [Google Scholar] [CrossRef]
- Dong, S.; Li, J.; Zhang, X. Tumor protein p53-induced nuclear protein 2 modulates osteogenic differentiation of human adipose derived stem/stromal cells by activating Wnt/beta-catenin signaling. Am. J. Transl. Res. 2020, 12, 6853–6867. [Google Scholar]
- Sala, D.; Ivanova, S.; Plana, N.; Ribas, V.; Duran, J.; Bach, D.; Turkseven, S.; Laville, M.; Vida, H.; Karczewska-Kupczewska, M.; et al. Autophagy-regulating TP53INP2 mediates muscle wasting and is repressed in diabetes. J. Clin. Investig. 2014, 124, 1914–1927. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, J.; Wang, Y.; Shi, F.; Guo, H.; Gao, B.; Yang, J.; Gu, L.; Yang, D.; Zhang, F.; Gao, D.; et al. Genomic Diversity and Selection Signatures for Zaosheng Cattle. Biology 2025, 14, 623. https://doi.org/10.3390/biology14060623
Xu J, Wang Y, Shi F, Guo H, Gao B, Yang J, Gu L, Yang D, Zhang F, Gao D, et al. Genomic Diversity and Selection Signatures for Zaosheng Cattle. Biology. 2025; 14(6):623. https://doi.org/10.3390/biology14060623
Chicago/Turabian StyleXu, Jianfeng, Yanyan Wang, Fuyue Shi, Hailong Guo, Bo Gao, Junxiang Yang, Lingrong Gu, Dezhi Yang, Fengtao Zhang, Dengwei Gao, and et al. 2025. "Genomic Diversity and Selection Signatures for Zaosheng Cattle" Biology 14, no. 6: 623. https://doi.org/10.3390/biology14060623
APA StyleXu, J., Wang, Y., Shi, F., Guo, H., Gao, B., Yang, J., Gu, L., Yang, D., Zhang, F., Gao, D., Gao, Z., Wang, S., & Wang, J. (2025). Genomic Diversity and Selection Signatures for Zaosheng Cattle. Biology, 14(6), 623. https://doi.org/10.3390/biology14060623




