The Identification of a Glucuronyltransferase-Related Gene, GlcAT-S, with Putative Mucus Protection and Anti-Inflammatory Effects from Gut-Damaged Drosophila by Dextran Sulfate Sodium (DSS)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Drosophila and Feeding Assay
2.2. cDNA Synthesis and Quantitative PCR Analysis
2.3. EC-Specific Knockdown of Selected Genes Using RNA Interference
2.4. Drosophila Gut Immunostaining and Imaging
2.5. Statistical Analysis
3. Results
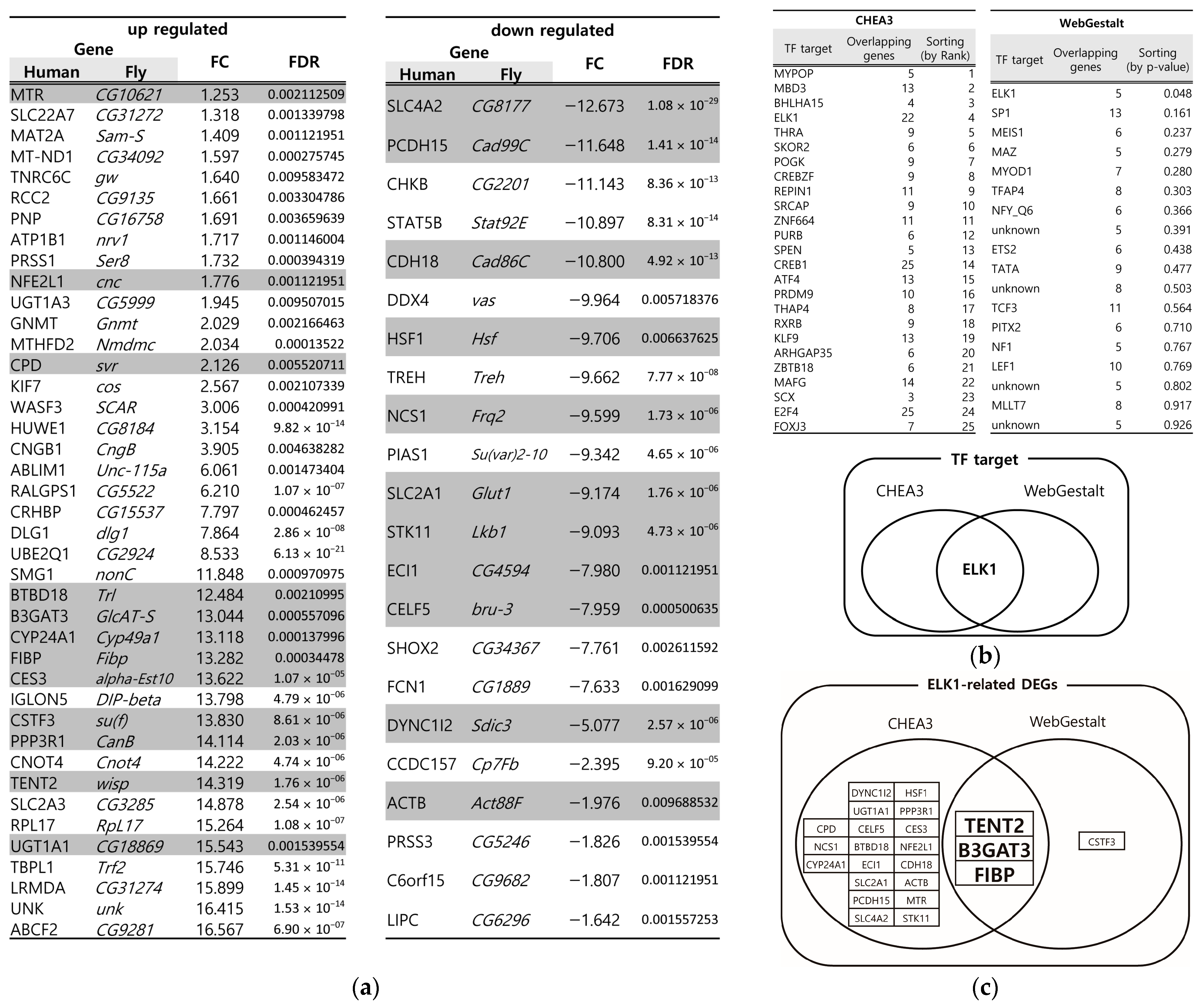
3.1. Identification of Key DEGs from the DSS-Induced Drosophila IBD Model Through Multiple Bioinformatics Tools
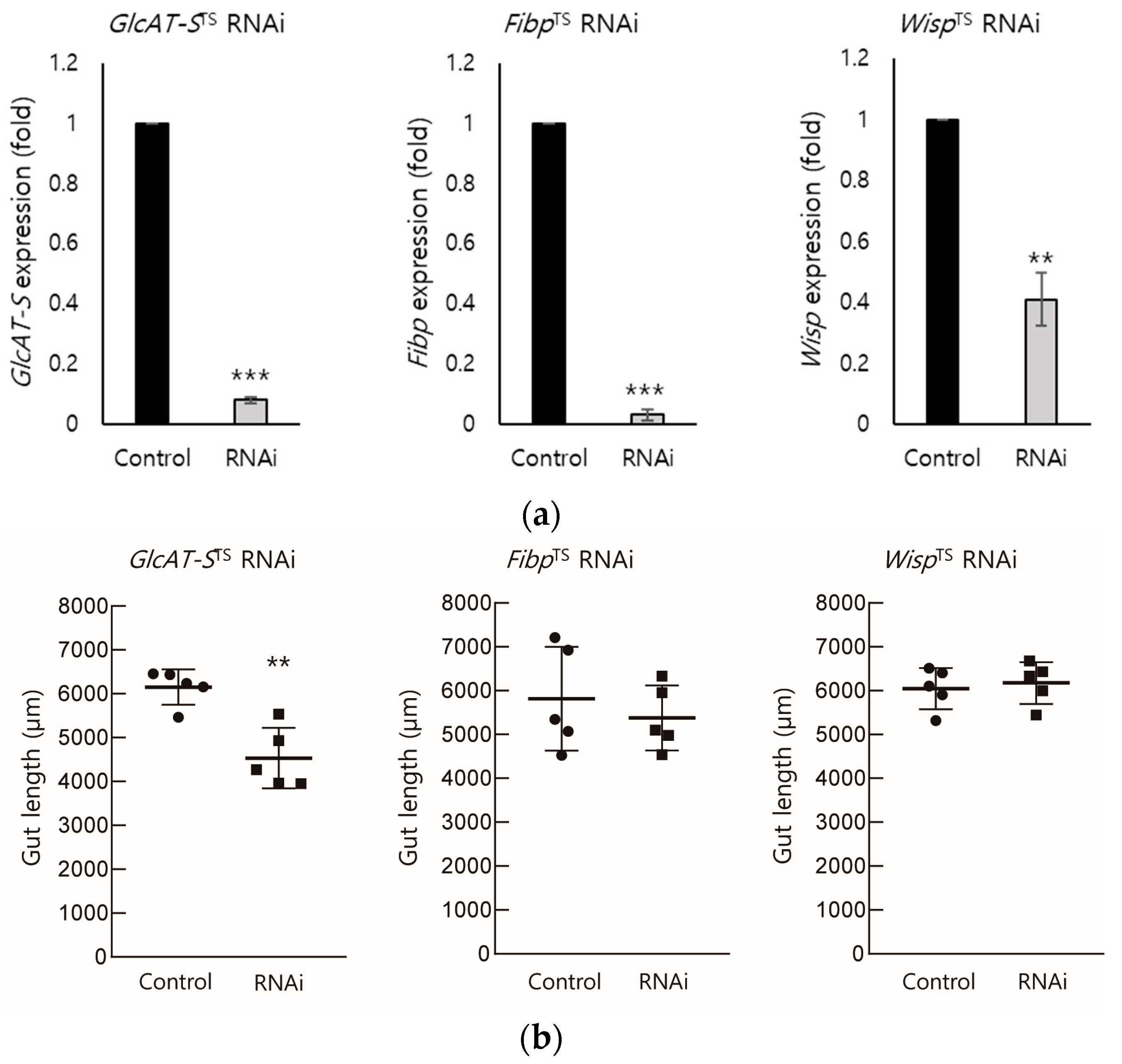
3.2. EC-Specific Knockdown of Screened DEGs
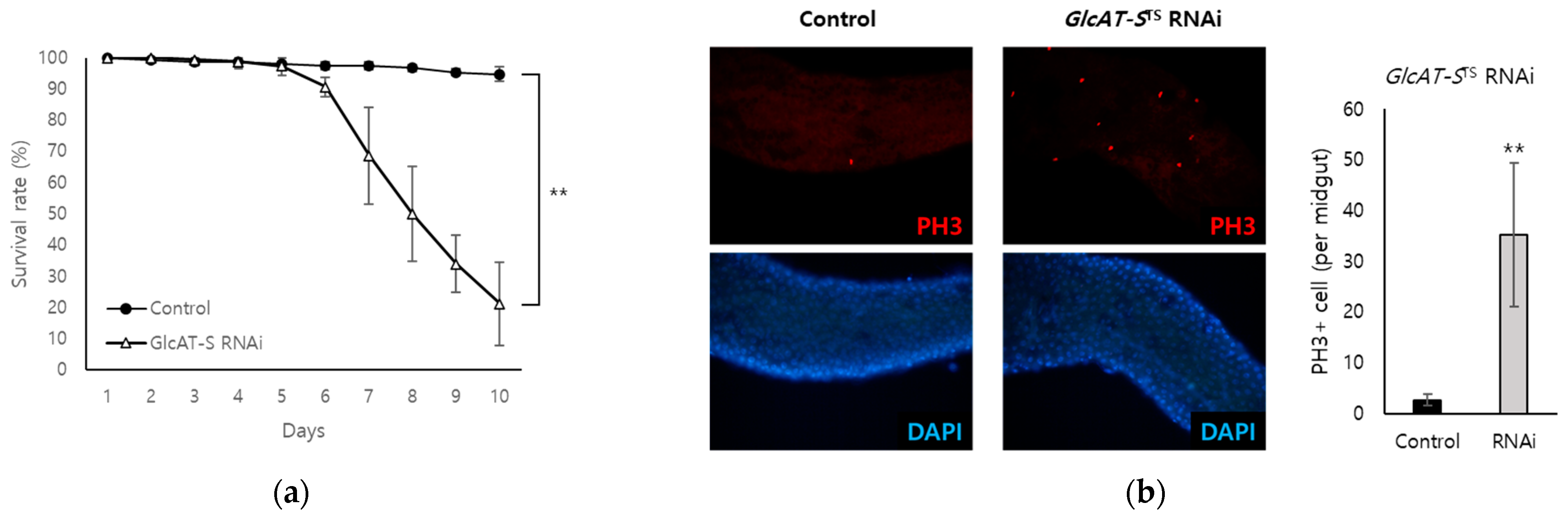
3.3. Confirmation of the Relationship Between GlcAT-S and Gut Tissue Damage Through EC-Specific Knockdown
3.4. Analysis of the Relationship Among GlcAT-S, Mucus Loss, and Inflammation in the Drosophila Gut-Damage Model
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Neurath, M.F. Cytokines in Inflammatory Bowel Disease. Nat. Rev. Immunol. 2014, 14, 329–342. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.R. Intestinal Mucosal Barrier Function in Health and Disease. Nat. Rev. Immunol. 2009, 9, 799–809. [Google Scholar] [CrossRef]
- Grondin, J.A.; Kwon, Y.H.; Far, P.M.; Haq, S.; Khan, W.I. Mucins in Intestinal Mucosal Defense and Inflammation: Learning from Clinical and Experimental Studies. Front. Immunol. 2020, 11, 2054. [Google Scholar] [CrossRef]
- Ananthakrishnan, A.N. Environmental Risk Factors for Inflammatory Bowel Disease. Gastroenterol. Hepatol. 2013, 9, 367–374. [Google Scholar] [CrossRef]
- Pandey, U.B.; Nichols, C.D. Human Disease Models in Drosophila melanogaster and the Role of the Fly in Therapeutic Drug Discovery. Pharmacol. Rev. 2011, 63, 411–436. [Google Scholar] [CrossRef]
- Apidianakis, Y.; Rahme, L.G. Drosophila melanogaster as a Model for Human Intestinal Infection and Pathology. Dis. Models Mech. 2011, 4, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Galeone, A.; Han, S.Y.; Story, B.A.; Consonni, G.; Mueller, W.F.; Steinmetz, L.M.; Vaccari, T.; Jafar-Nejad, H. Gut Barrier Defects, Intestinal Immune Hyperactivation and Enhanced Lipid Catabolism Drive Lethality in NGLY1-Deficient Drosophila. Nat. Commun. 2023, 14, 5667. [Google Scholar] [CrossRef] [PubMed]
- Roditi, I.; Lehane, M.J. Interactions between Trypanosomes and Tsetse Flies. Curr. Opin. Microbiol. 2008, 11, 345–351. [Google Scholar] [CrossRef]
- Chassaing, B.; Aitken, J.D.; Malleshappa, M.; Vijay-Kumar, M. Dextran Sulfate Sodium (DSS)-Induced Colitis in Mice. CP Immunol. 2014, 104, 15–25. [Google Scholar] [CrossRef]
- Katsandegwaza, B.; Horsnell, W.; Smith, K. Inflammatory Bowel Disease: A Review of Pre-Clinical Murine Models of Human Disease. Int. J. Mol. Sci. 2022, 23, 9344. [Google Scholar] [CrossRef]
- Zeng, C.; Liu, F.; Huang, Y.; Liang, Q.; He, X.; Li, L.; Xie, Y. Drosophila: An Important Model for Exploring the Pathways of Inflammatory Bowel Disease (IBD) in the Intestinal Tract. Int. J. Mol. Sci. 2024, 25, 12742. [Google Scholar] [CrossRef] [PubMed]
- Capo, F.; Wilson, A.; Di Cara, F. The Intestine of Drosophila Melanogaster: An Emerging Versatile Model System to Study Intestinal Epithelial Homeostasis and Host-Microbial Interactions in Humans. Microorganisms 2019, 7, 336. [Google Scholar] [CrossRef]
- Buchon, N.; Broderick, N.A.; Lemaitre, B. Gut Homeostasis in a Microbial World: Insights from Drosophila Melanogaster. Nat. Rev. Microbiol. 2013, 11, 615–626. [Google Scholar] [CrossRef]
- Dietzl, G.; Chen, D.; Schnorrer, F.; Su, K.-C.; Barinova, Y.; Fellner, M.; Gasser, B.; Kinsey, K.; Oppel, S.; Scheiblauer, S.; et al. A Genome-Wide Transgenic RNAi Library for Conditional Gene Inactivation in Drosophila. Nature 2007, 448, 151–156. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A Revolutionary Tool for Transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Tarca, A.L.; Carey, V.J.; Chen, X.; Romero, R.; Drăghici, S. Machine Learning and Its Applications to Biology. PLoS Comput. Biol. 2007, 3, e116. [Google Scholar] [CrossRef]
- Manzoni, C.; Kia, D.A.; Vandrovcova, J.; Hardy, J.; Wood, N.W.; Lewis, P.A.; Ferrari, R. Genome, Transcriptome and Proteome: The Rise of Omics Data and Their Integration in Biomedical Sciences. Brief. Bioinform. 2018, 19, 286–302. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Palmer, W.H.; Medd, N.C.; Beard, P.M.; Obbard, D.J. Isolation of a Natural DNA Virus of Drosophila Melanogaster, and Characterisation of Host Resistance and Immune Responses. PLoS Pathog. 2018, 14, e1007050. [Google Scholar] [CrossRef]
- Ewen-Campen, B.; Luan, H.; Xu, J.; Singh, R.; Joshi, N.; Thakkar, T.; Berger, B.; White, B.H.; Perrimon, N. Split-Intein Gal4 Provides Intersectional Genetic Labeling That Is Repressible by Gal80. Proc. Natl. Acad. Sci. USA 2023, 120, e2304730120. [Google Scholar] [CrossRef]
- Lee, S.H.; Hwang, D.; Goo, T.-W.; Yun, E.-Y. Prediction of Intestinal Stem Cell Regulatory Genes from Drosophila Gut Damage Model Created Using Multiple Inducers: Differential Gene Expression-Based Protein-Protein Interaction Network Analysis. Dev. Comp. Immunol. 2023, 138, 104539. [Google Scholar] [CrossRef]
- Park, D.; Singh, R.; Baym, M.; Liao, C.-S.; Berger, B. IsoBase: A Database of Functionally Related Proteins across PPI Networks. Nucleic Acids Res. 2011, 39, D295–D300. [Google Scholar] [CrossRef] [PubMed]
- Keenan, A.B.; Torre, D.; Lachmann, A.; Leong, A.K.; Wojciechowicz, M.L.; Utti, V.; Jagodnik, K.M.; Kropiwnicki, E.; Wang, Z.; Ma’ayan, A. ChEA3: Transcription Factor Enrichment Analysis by Orthogonal Omics Integration. Nucleic Acids Res. 2019, 47, W212–W224. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene Set Analysis Toolkit with Revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef] [PubMed]
- Osumi, R.; Sugihara, K.; Yoshimoto, M.; Tokumura, K.; Tanaka, Y.; Hinoi, E. Role of Proteoglycan Synthesis Genes in Osteosarcoma Stem Cells. Front. Oncol. 2024, 14, 1325794. [Google Scholar] [CrossRef]
- Huang, Y.-F.; Niu, W.-B.; Hu, R.; Wang, L.-J.; Huang, Z.-Y.; Ni, S.-H.; Wang, M.-Q.; Yang, Y.; Huang, Y.-S.; Feng, W.-J.; et al. FIBP Knockdown Attenuates Growth and Enhances Chemotherapy in Colorectal Cancer via Regulating GSK3β-Related Pathways. Oncogenesis 2018, 7, 77. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Bofill-De Ros, X.; Stanton, R.; Shao, T.-J.; Villanueva, P.; Gu, S. TENT2, TUT4, and TUT7 Selectively Regulate miRNA Sequence and Abundance. Nat. Commun. 2022, 13, 5260. [Google Scholar] [CrossRef]
- Kong, S.; Zhang, Y.H.; Zhang, W. Regulation of Intestinal Epithelial Cells Properties and Functions by Amino Acids. BioMed Res. Int. 2018, 2018, 2819154. [Google Scholar] [CrossRef]
- Frazer, L.C.; Good, M. Intestinal Epithelium in Early Life. Mucosal Immunol. 2022, 15, 1181–1187. [Google Scholar] [CrossRef]
- Weaver, L.T.; Austin, S.; Cole, T.J. Small Intestinal Length: A Factor Essential for Gut Adaptation. Gut 1991, 32, 1321–1323. [Google Scholar] [CrossRef]
- Viragova, S.; Li, D.; Klein, O.D. Activation of Fetal-like Molecular Programs during Regeneration in the Intestine and Beyond. Cell Stem Cell 2024, 31, 949–960. [Google Scholar] [CrossRef] [PubMed]
- Amcheslavsky, A.; Lindblad, J.L.; Bergmann, A. Transiently “Undead” Enterocytes Mediate Homeostatic Tissue Turnover in the Adult Drosophila Midgut. Cell Rep. 2020, 33, 108408. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Deng, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; Wang, X.; Zhao, L. Inflammatory Responses and Inflammation-Associated Diseases in Organs. Oncotarget 2018, 9, 7204–7218. [Google Scholar] [CrossRef]
- Oz, H.S.; Yeh, S.-L.; Neuman, M.G. Gastrointestinal Inflammation and Repair: Role of Microbiome, Infection, and Nutrition. Gastroenterol. Res. Pract. 2016, 2016, 6516708. [Google Scholar] [CrossRef]
- Cornick, S.; Tawiah, A.; Chadee, K. Roles and Regulation of the Mucus Barrier in the Gut. Tissue Barriers 2015, 3, e982426. [Google Scholar] [CrossRef]
- Saez, A.; Herrero-Fernandez, B.; Gomez-Bris, R.; Sánchez-Martinez, H.; Gonzalez-Granado, J.M. Pathophysiology of Inflammatory Bowel Disease: Innate Immune System. Int. J. Mol. Sci. 2023, 24, 1526. [Google Scholar] [CrossRef]
- Sun, J.; Shen, X.; Li, Y.; Guo, Z.; Zhu, W.; Zuo, L.; Zhao, J.; Gu, L.; Gong, J.; Li, J. Therapeutic Potential to Modify the Mucus Barrier in Inflammatory Bowel Disease. Nutrients 2016, 8, 44. [Google Scholar] [CrossRef]
- Rawat, P.S.; Seyed Hameed, A.S.; Meng, X.; Liu, W. Utilization of Glycosaminoglycans by the Human Gut Microbiota: Participating Bacteria and Their Enzymatic Machineries. Gut Microbes 2022, 14, 2068367. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, S.-H.; Yang, J.-Y. Mechanobiological Approach for Intestinal Mucosal Immunology. Biology 2025, 14, 110. [Google Scholar] [CrossRef]
- Murch, S.H.; MacDonald, T.T.; Walker-Smith, J.A.; Lionetti, P.; Levin, M.; Klein, N.J. Disruption of Sulphated Glycosaminoglycans in Intestinal Inflammation. Lancet 1993, 341, 711–714. [Google Scholar] [CrossRef]
- Kim, K.; Lane, E.A.; Saftien, A.; Wang, H.; Xu, Y.; Wirtz-Peitz, F.; Perrimon, N. Drosophila as a Model for Studying Cystic Fibrosis Pathophysiology of the Gastrointestinal System. Proc. Natl. Acad. Sci. USA 2020, 117, 10357–10367. [Google Scholar] [CrossRef]
- Kitagawa, H.; Uyama, T.; Sugahara, K. Molecular Cloning and Expression of a Human Chondroitin Synthase. J. Biol. Chem. 2001, 276, 38721–38726. [Google Scholar] [CrossRef]
- Pompili, S.; Latella, G.; Gaudio, E.; Sferra, R.; Vetuschi, A. The Charming World of the Extracellular Matrix: A Dynamic and Protective Network of the Intestinal Wall. Front. Med. 2021, 8, 610189. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.E.V.; Hansson, G.C. Immunological Aspects of Intestinal Mucus and Mucins. Nat. Rev. Immunol. 2016, 16, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Corfield, A.P. Mucins: A Biologically Relevant Glycan Barrier in Mucosal Protection. BBA Gen. Subj. 2015, 1850, 236–252. [Google Scholar] [CrossRef]
- Dasgupta, S.; Silva, J.; Wang, G.; Yu, R.K. Sulfoglucuronosyl Paragloboside Is a Ligand for T Cell Adhesion: Regulation of Sulfoglucuronosyl Paragloboside Expression via Nuclear Factor κB Signaling. J. Neurosci. Res. 2009, 87, 3591–3599. [Google Scholar] [CrossRef] [PubMed]
- Ren, F.; Wang, B.; Yue, T.; Yun, E.-Y.; Ip, Y.T.; Jiang, J. Hippo Signaling Regulates Drosophila Intestine Stem Cell Proliferation through Multiple Pathways. Proc. Natl. Acad. Sci. USA 2010, 107, 21064–21069. [Google Scholar] [CrossRef]
- Noah, T.K.; Donahue, B.; Shroyer, N.F. Intestinal Development and Differentiation. Exp. Cell Res. 2011, 317, 2702–2710. [Google Scholar] [CrossRef]
- Watnick, P.I.; Jugder, B.-E. Microbial Control of Intestinal Homeostasis via Enteroendocrine Cell Innate Immune Signaling. Trends Microbiol. 2020, 28, 141–149. [Google Scholar] [CrossRef]
- Pelaseyed, T.; Bergström, J.H.; Gustafsson, J.K.; Ermund, A.; Birchenough, G.M.H.; Schütte, A.; van der Post, S.; Svensson, F.; Rodríguez-Piñeiro, A.M.; Nyström, E.E.L.; et al. The Mucus and Mucins of the Goblet Cells and Enterocytes Provide the First Defense Line of the Gastrointestinal Tract and Interact with the Immune System. Immunol. Rev. 2014, 260, 8–20. [Google Scholar] [CrossRef]
- Hung, R.-J.; Hu, Y.; Kirchner, R.; Liu, Y.; Xu, C.; Comjean, A.; Tattikota, S.G.; Li, F.; Song, W.; Ho Sui, S.; et al. A Cell Atlas of the Adult Drosophila Midgut. Proc. Natl. Acad. Sci. USA 2020, 117, 1514–1523. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.H.; Hwang, D.; Lee, J.-W.; Goo, T.-W.; Yun, E.-Y. The Identification of a Glucuronyltransferase-Related Gene, GlcAT-S, with Putative Mucus Protection and Anti-Inflammatory Effects from Gut-Damaged Drosophila by Dextran Sulfate Sodium (DSS). Biology 2025, 14, 513. https://doi.org/10.3390/biology14050513
Lee SH, Hwang D, Lee J-W, Goo T-W, Yun E-Y. The Identification of a Glucuronyltransferase-Related Gene, GlcAT-S, with Putative Mucus Protection and Anti-Inflammatory Effects from Gut-Damaged Drosophila by Dextran Sulfate Sodium (DSS). Biology. 2025; 14(5):513. https://doi.org/10.3390/biology14050513
Chicago/Turabian StyleLee, Seung Hun, Dooseon Hwang, Jang-Won Lee, Tae-Won Goo, and Eun-Young Yun. 2025. "The Identification of a Glucuronyltransferase-Related Gene, GlcAT-S, with Putative Mucus Protection and Anti-Inflammatory Effects from Gut-Damaged Drosophila by Dextran Sulfate Sodium (DSS)" Biology 14, no. 5: 513. https://doi.org/10.3390/biology14050513
APA StyleLee, S. H., Hwang, D., Lee, J.-W., Goo, T.-W., & Yun, E.-Y. (2025). The Identification of a Glucuronyltransferase-Related Gene, GlcAT-S, with Putative Mucus Protection and Anti-Inflammatory Effects from Gut-Damaged Drosophila by Dextran Sulfate Sodium (DSS). Biology, 14(5), 513. https://doi.org/10.3390/biology14050513





