Genome-Wide Identification of the TIFY Family in Longan and Their Potential Functional Analysis in Anthocyanin Synthesis
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Sequence Analysis of DlTIFY Genes
2.2. Sequence Alignments and Phylogenetic Analyses
2.3. Chromosomal Location and Gene Duplication
2.4. Analysis of Cis-Acting Elements of DlTIFY Genes
2.5. Expression Patterns of DlTIFY Genes
2.6. Subcellular Location and Protein Interaction Network Analysis of DlTIFY7
2.7. Overexpression of DlTIFY7 in Tobacco
3. Results
3.1. Genome-Wide Identification of DlTIFY Genes and Chromosomal Location
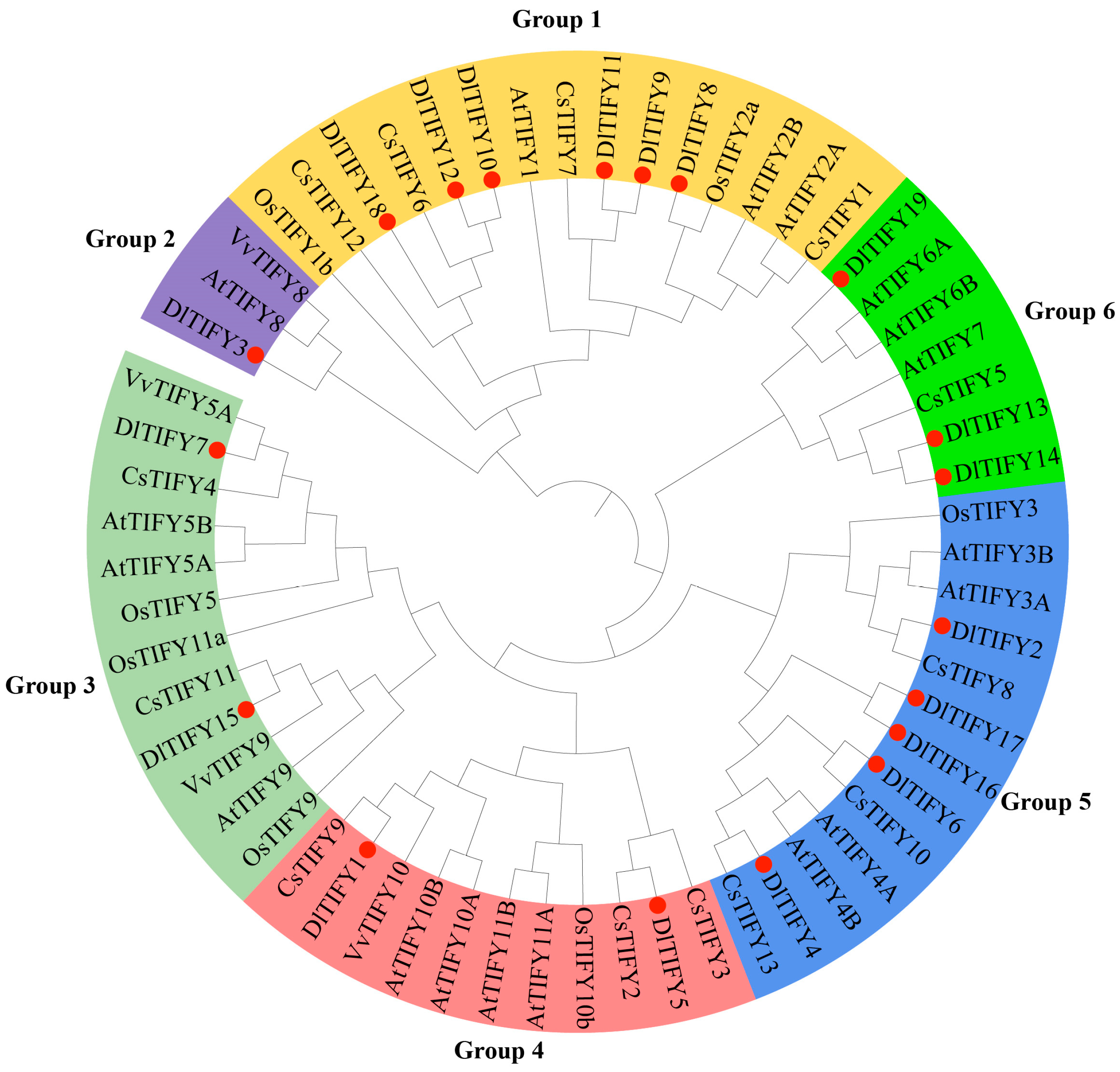
3.2. Conserved Domain and Phylogenetic Analysis of the DlTIFY Proteins
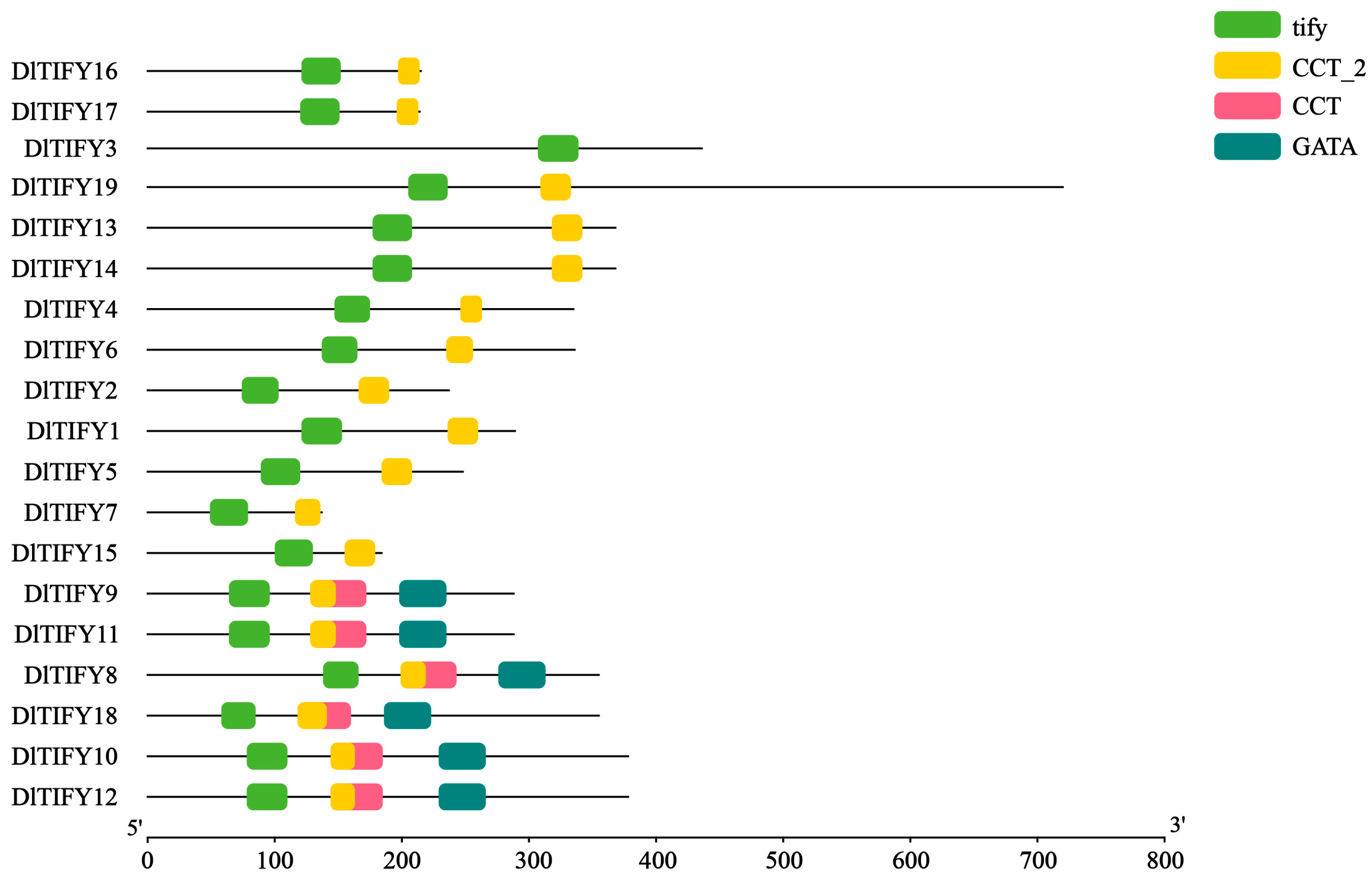
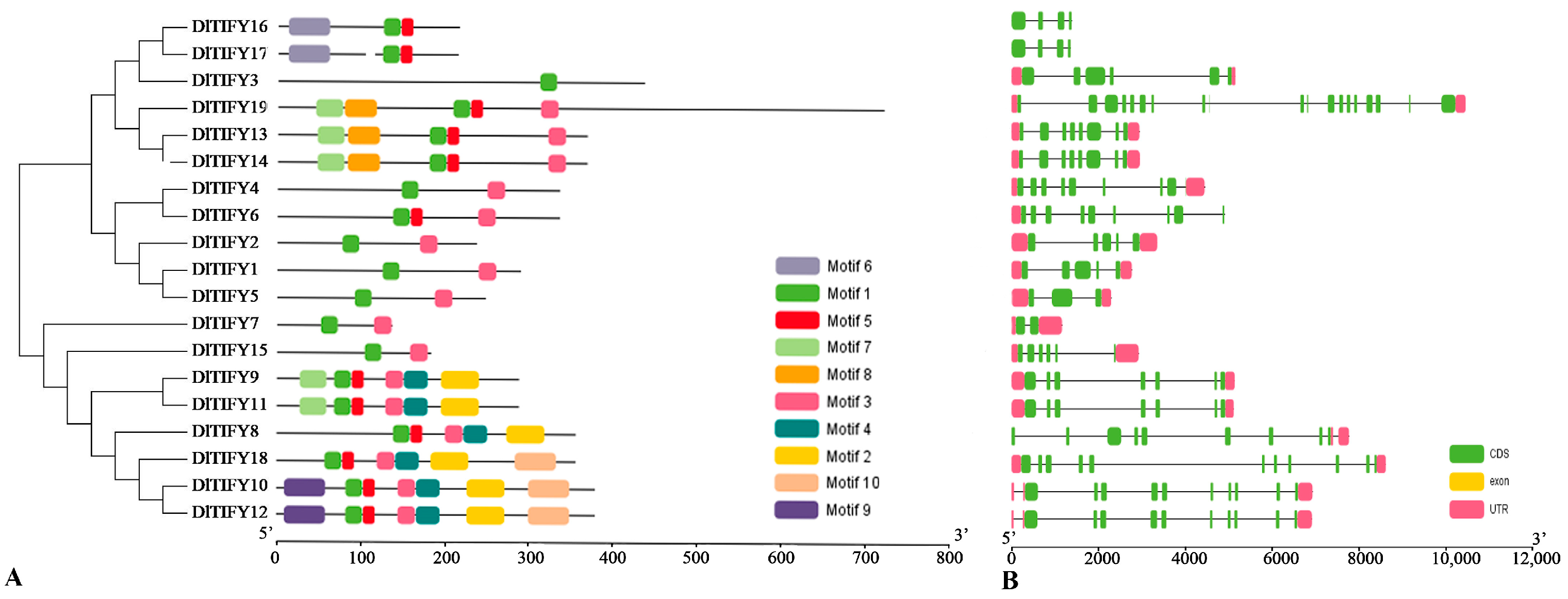
3.3. Conserved Motif and Gene Structure Analysis
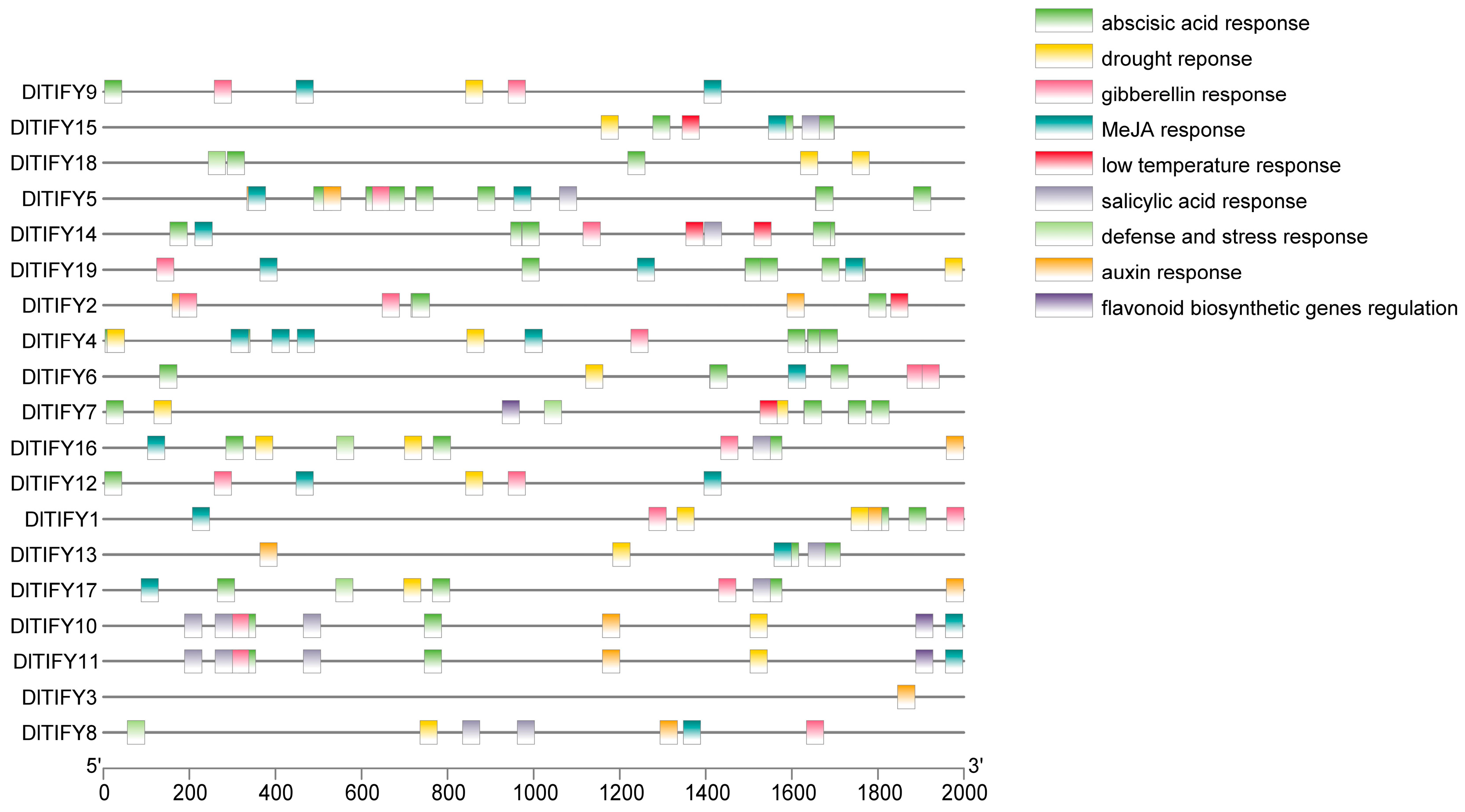
3.4. Analysis of Cis-Acting Elements of DlTIFY Genes
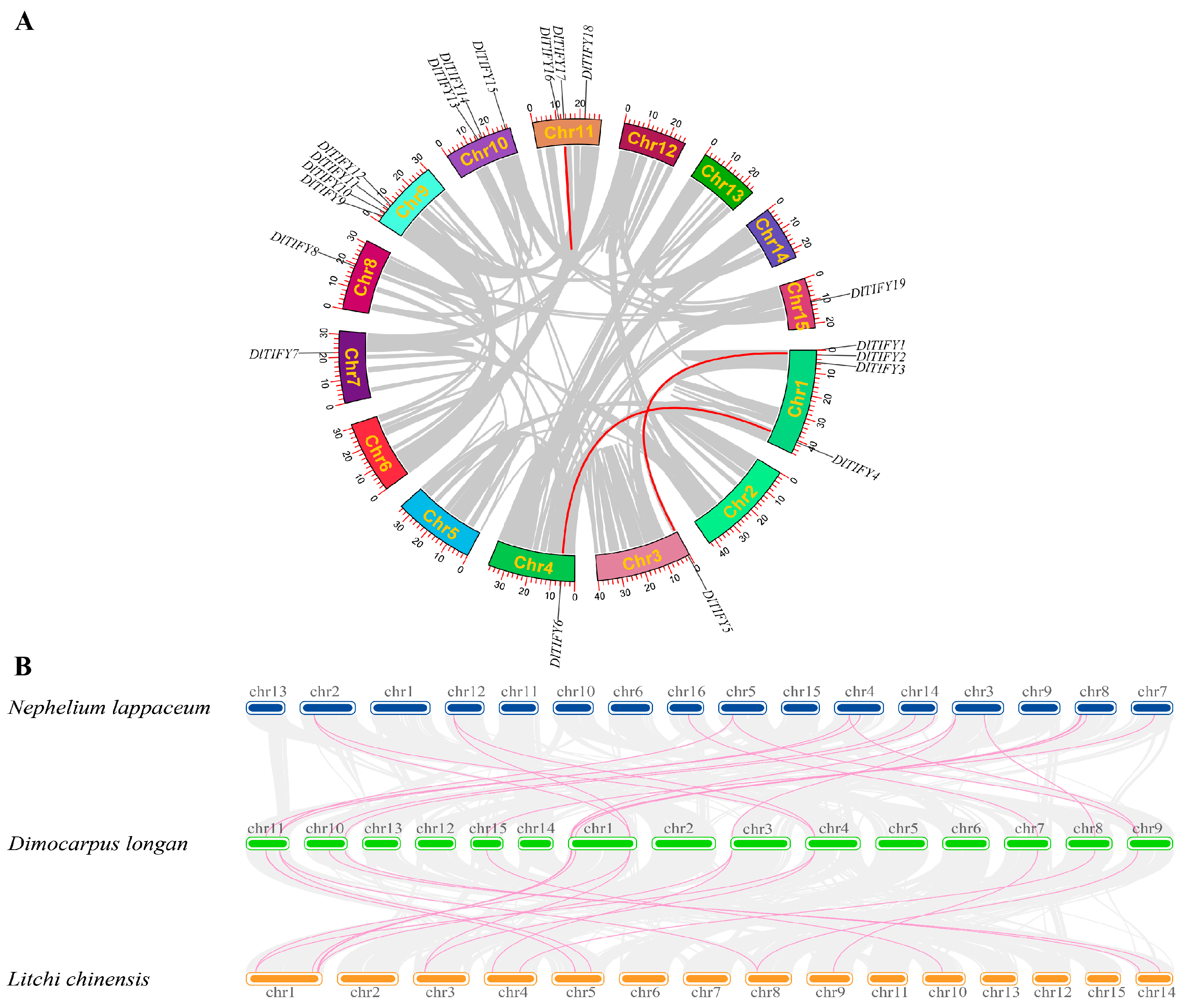
3.5. Chromosome Distribution and Gene Duplication Analysis of DlTIFY Genes
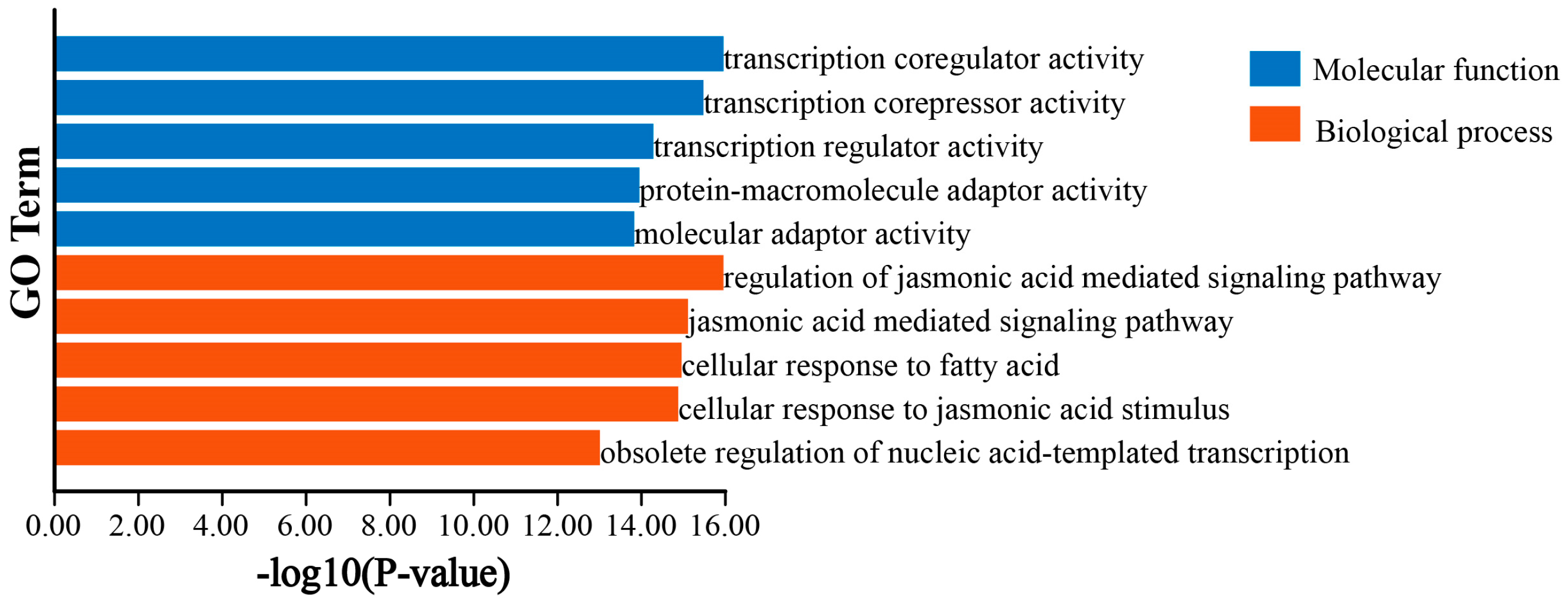
3.6. GO and KEGG Enrichment Analysis of DlTIFY Genes
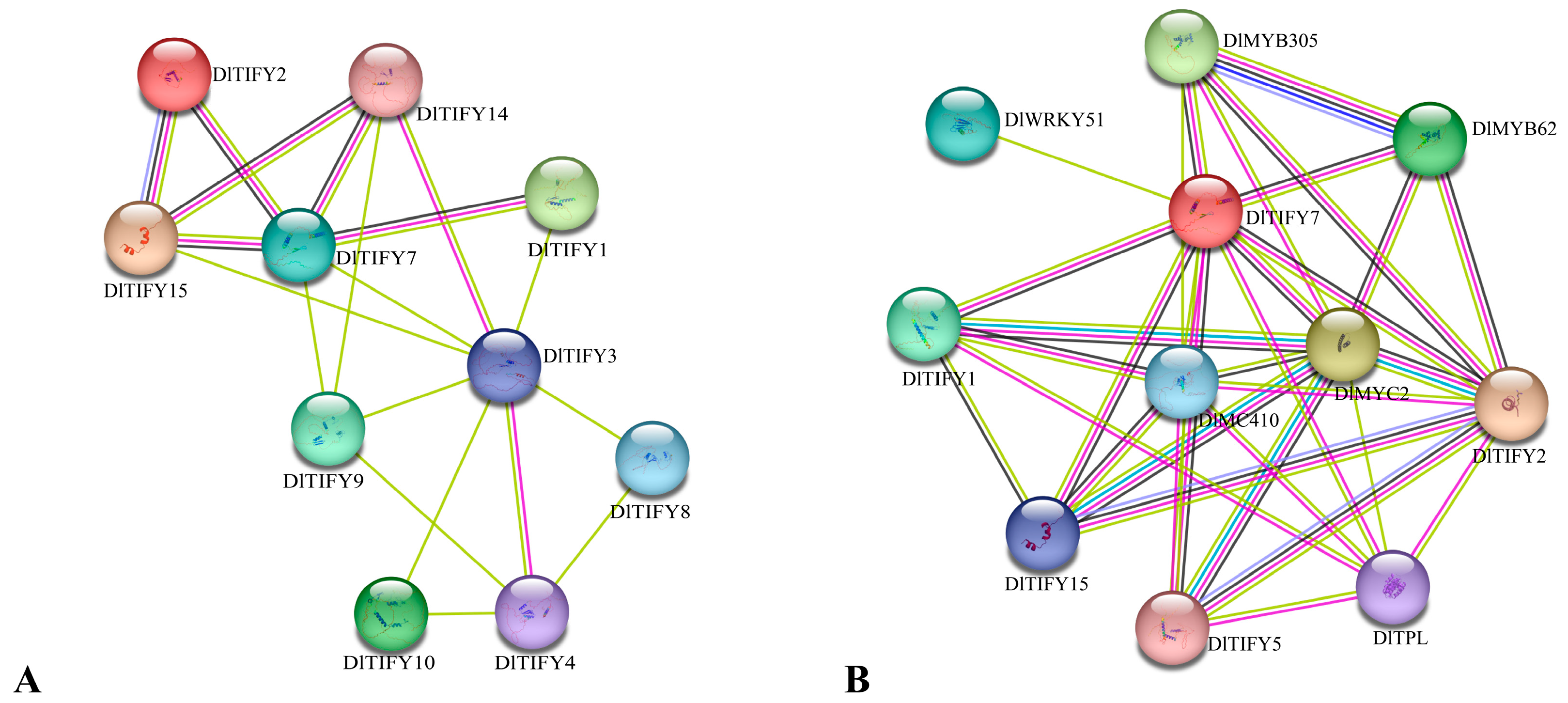
3.7. The Protein–Protein Interaction Network Analysis of the TIFY Family in Longan
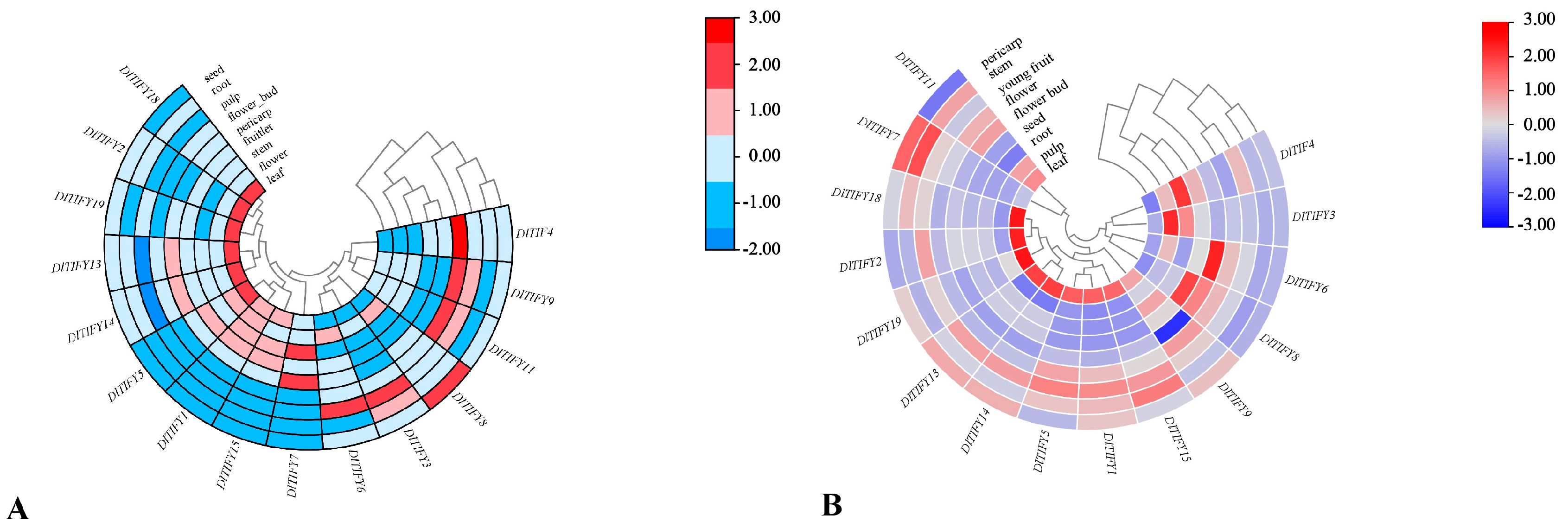
3.8. Expression Pattern of DlTIFY Genes in Different Tissues
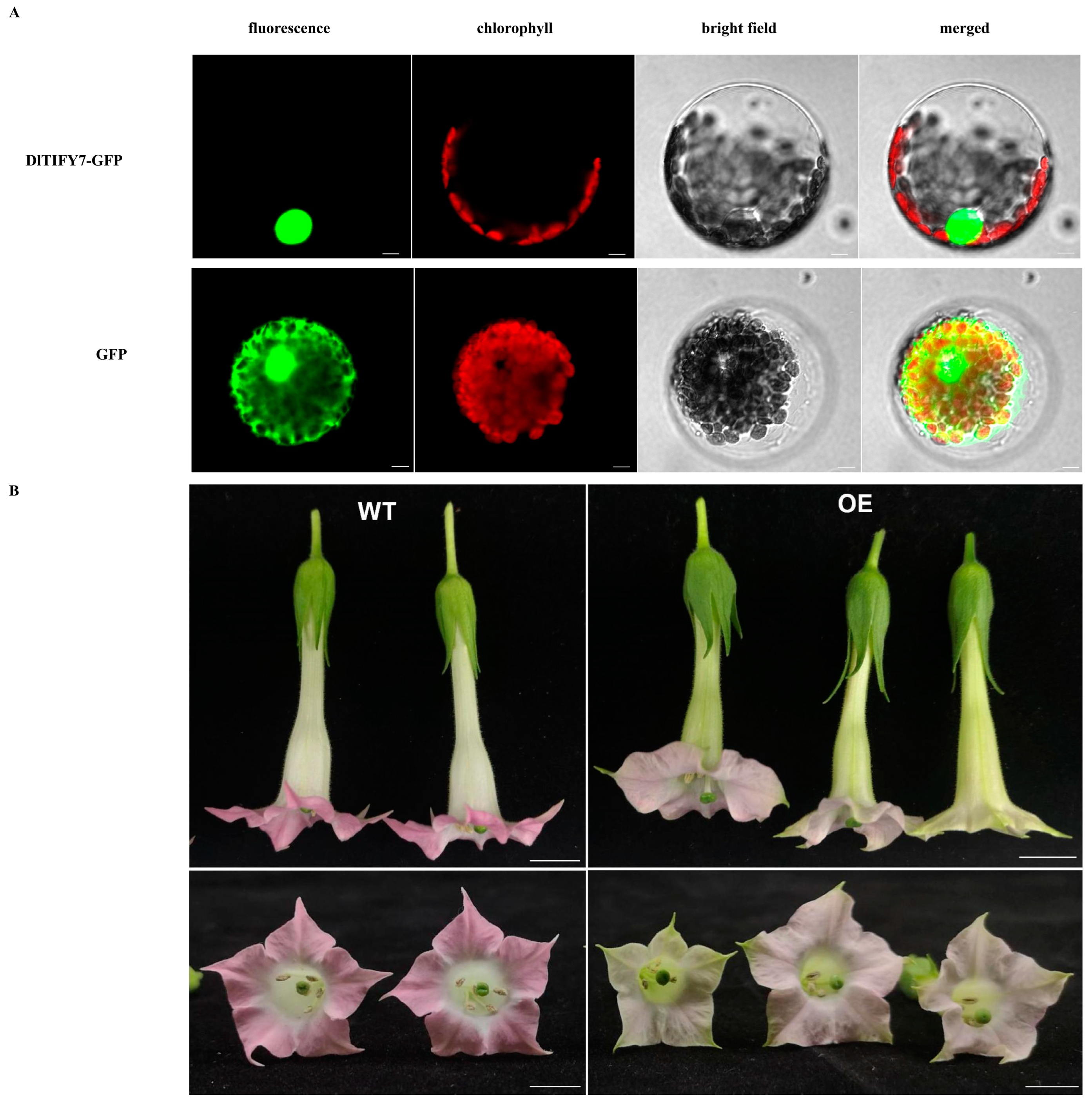
3.9. DlTIFY7 Is Localized to the Nucleus and Overexpression of DlTIFY7 Inhibits Anthocyanin Biosynthesis in Transgenic Tobacco
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nishii, A.; Takemura, M.; Fujita, H.; Shikata, M.; Yokota, A.; Kohchi, T. Characterization of a novel gene encoding a putative single zinc-finger protein, ZIM, expressed during the reproductive phase in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2000, 64, 1402–1409. [Google Scholar] [PubMed]
- Qi, T.; Song, S.; Ren, Q.; Wu, D.; Huang, H.; Chen, Y.; Fan, M.; Peng, W.; Ren, C.; Xie, D. The Jasmonate-ZIM-Domain Proteins Interact with the WD-Repeat/bHLH/MYB Complexes to Regulate Jasmonate-Mediated Anthocyanin Accumulation and Trichome Initiation in Arabidopsis thaliana. Plant Cell 2011, 23, 1795–1814. [Google Scholar] [PubMed]
- Shikata, M.; Matsuda, Y.; Ando, K.; Nishii, A.; Takemura, M.; Yokota, A.; Kohchi, T. Characterization of Arabidopsis ZIM, a member of a novel plant-specific GATA factor gene family. J. Exp. Bot. 2004, 55, 631–639. [Google Scholar] [PubMed]
- White, D.W. PEAPOD regulates lamina size and curvature in Arabidopsis. Proc. Nat. Acad. Sci. USA 2006, 103, 13238–13243. [Google Scholar]
- Hakata, M.; Kuroda, M.; Ohsumi, A.; Hirose, T.; Nakamura, H.; Muramatsu, M.; Ichikawa, H.; Yamakawa, H. Overexpression of a rice TIFY gene increases grain size through enhanced accumulation of carbohydrates in the stem. Biosci. Biotechnol. Biochem. 2012, 76, 2129–2134. [Google Scholar]
- Cai, Q.; Yuan, Z.; Chen, M.J.; Yin, C.S.; Luo, Z.J.; Zhao, X.X.; Liang, W.Q.; Hu, J.P.; Zhang, D.B. Jasmonic acid regulates spikelet development in rice. Nat. Commun. 2014, 5, 3476. [Google Scholar]
- Ye, H.; Du, H.; Tang, N.; Li, X.; Xiong, L. Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol. Biol. 2009, 71, 291–305. [Google Scholar]
- Ma, Y.; Shu, S.; Bai, S.; Tao, R.; Qian, M. Genome-wide survey and analysis of the TIFY gene family and its potential role in anthocyanin synthesis in Chinese sand pear (Pyrus pyrifolia). Tree Genet. Genomes 2018, 14, 25. [Google Scholar]
- Sheng, Y.; Yu, H.; Pan, H.F.; Qiu, K.L.; Xie, Q.M.; Chen, H.L.; Fu, S.L.; Zhang, J.Y.; Zhou, H. Genome-wide analysis of the gene structure, expression and protein interactions of the peach (Prunus persica) TIFY gene family. Front. Plant Sci. 2022, 13, 792802. [Google Scholar]
- Chen, K.Q.; Zhao, X.Y.; An, X.H.; Tian, Y.; Liu, D.D.; You, C.X.; Hao, Y.J. MdHIR proteins repress anthocyanin accumulation by interacting with the MdJAZ2 protein to inhibit its degradation in apples. Sci. Rep. 2017, 7, 44484. [Google Scholar]
- Wang, J.; Li, J.G.; Li, Z.Y.; Liu, B.; Zhang, L.L.; Guo, D.L.; Huang, S.L.; Qian, W.Q.; Guo, L. Genomic insights into longan evolution from a chromosome-level genome assembly and population genomics of longan accessions. Hortic. Res. 2022, 9, uhac021. [Google Scholar] [PubMed]
- Chen, C.J.; Wu, Y.; Li, J.W.; Wang, X.; Zeng, Z.H.; Xu, J.; Liu, Y.L.; Feng, J.T.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [PubMed]
- Li, J.W.; Chen, C.J.; Zeng, Z.H.; Wu, F.Q.; Feng, J.T.; Liu, B.; Mai, Y.X.; Chu, X.Y.; Wei, W.C.; Li, X.; et al. SapBase:A central portal for functional and comparative genomics of Sapindaceae species. J. Integr. Plant Biol. 2024, 66, 1561–1570. [Google Scholar]
- Yoo, S.-D.; Cho, Y.-H. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar]
- Bai, Y.H.; Meng, Y.J.; Huang, D.L.; Qi, Y.H.; Chen, M. Origin and evolutionary analysis of the plant-specific TIFY transcription factor family. Genomics 2011, 98, 128–136. [Google Scholar] [PubMed]
- Yucheng, Z.; Min, G.; Singer, S.D.; Fei, Z.J.; Wang, H.; Wang, X.P. Genome-Wide Identification and Analysis of the TIFY Gene Family in Grape. PLoS ONE 2012, 7, e44465. [Google Scholar]
- Tao, J.J.; Jia, H.M.; Wu, M.T.; Zhong, W.Q.; Jia, D.F.; Wang, Z.P.; Huang, C.H. Genome-wide identification and characterization of the TIFY gene family in kiwifruit. BMC Genom. 2022, 23, 179. [Google Scholar]
- Mei, C.; Zhang, X.Y.; Yan, P.; Aisajan, M.; Feng, B.B.; Ma, K.; Han, L.Q.; Dong, L.X.; Wang, J.X. Identification of TIFY family in apple and their expression analysis under insect stress. Acta Hortic. Sin. 2021, 48, 233–242. [Google Scholar]
- Fedorova, L.; Fedorov, A. Introns in gene evolution. Genetica 2003, 118, 123–131. [Google Scholar]
- Freeling, M. Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 2009, 60, 433–453. [Google Scholar]
- Wang, N.; Xiang, Y.; Fang, L.; Wang, Y.J.; Xin, H.P.; Li, S.H. Patterns of gene duplication and their contribution to expansion of gene families in grapevine. Plant Mol. Biol. Rep. 2013, 31, 852–861. [Google Scholar]
- Xu, L.; Liu, A.; Wang, T.; Wang, Y.; Li, L.; Wu, P. Characterization and coexpression analysis of the TIFY family genes in Euryale ferox related to leaf development. Plants 2023, 12, 2323. [Google Scholar] [CrossRef] [PubMed]
- Gabriela, A.G.A.; Jasmin, D.; Corina, S.S.; Marieluise, W.; Von, R.E.; Laurens, P.; Alain, G.; Justine, B.; Ulrike, Z. The Non-JAZ TIFY Protein TIFY8 of Arabidopsis thaliana Interacts with the HD-ZIP III Transcription Factor REVOLUTA and Regulates Leaf Senescence. Int. J. Mol. Sci. 2023, 24, 3079. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, S.; Chico, J.M.; Solano, R. The jasmonate pathway: The ligand, the receptor and the core signalling module. Curr. Opin. Plant Biol. 2009, 12, 539–547. [Google Scholar]
- Gao, Q.M.; Venugopal, S.; Navarre, D.; Kachroo, A. Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol. 2011, 155, 464–476. [Google Scholar]
- Song, S.; Qi, T.; Huang, H.; Ren, Q.; Wu, D.; Chang, C.; Peng, W.; Liu, Y.; Peng, J.; Xie, D. The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 2011, 23, 1000–1013. [Google Scholar] [PubMed]
- Goossens, J.; Swinnen, G.; Vanden, R.B.; Pauwels, L.; Goossens, A. Change of a conserved amino acid in the MYC 2 and MYC 3 transcription factors leads to release of JAZ repression and increased activity. New Phytol. 2015, 206, 1229–1237. [Google Scholar]
- Heidari, P.; Faraji, S.; Ahmadizadeh, M.; Ahmar, S.; Mora-Poblete, F. New insights into structure and function of TIFY genes in Zea mays and Solanum lycopersicum: A genome-wide comprehensive analysis. Front. Genet. 2021, 12, 657970. [Google Scholar]
- Liu, L.M.; Bai, N.; Zheng, Y.Q.; Chen, L.J.; Zong, Y.; Ye, L.Y.; Li, Y.Q.; Liao, F.L.; Lu, M.; Yang, L.; et al. Genome-wide identification and analysis of TIFY family in highbush blueberry and their responses to exogenous jasmonic acid. Sci. Hortic. 2022, 305, 111391. [Google Scholar]
- Lian, C.L.; Zhang, B.; Li, J.J.; Yang, H.; Liu, X.Y.; Ma, R.; Zhang, F.; Liu, J.; Yang, J.F.; Lan, J.X.; et al. Genome-wide identification, characterization and expression pattern analysis of TIFY family members in Artemisia argyi. BMC Genom. 2024, 25, 925. [Google Scholar]
- Wu, H.; Ye, H.Y.; Yao, R.F.; Zhang, T.; Xiong, L.Z. OsJAZ9 acts as a transcriptional regulator in jasmonate signaling and modulates salt stress tolerance in rice. Plant Sci. 2015, 232, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Cai, H.; Luo, X.; Bai, X.; Deyholos, M.K.; Chen, Q.; Chen, C.; Ji, W.; Zhu, Y. Over-expression of a novel JAZ family gene from Glycine soja, increases salt and alkali stress tolerance. Biochem. Biophys. Res. Commun. 2012, 426, 273–279. [Google Scholar] [CrossRef]
- You, X.M.; Zhu, S.S.; Zhang, W.W.; Zhang, J.; Wang, C.M.; Jing, R.N.; Chen, W.W.; Wu, H.M.; Cai, Y.; Feng, Z.M.; et al. OsPEX5 regulates rice spikelet development through modulating jasmonic acid biosynthesis. New Phytol. 2019, 224, 712–724. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.C.; Tian, J.Q.; Liu, Y.L.; Chen, X.F.; Li, S.Q.; Persson, S.; Lu, D.; Chen, M.J.; Luo, Z.J.; Zhang, D.B.; et al. Ectopic expression of OsJAZ6, which interacts with OsJAZ1, alters JA signaling and spikelet development in rice. Plant J. 2021, 108, 1083–1096. [Google Scholar] [CrossRef]
- Zhu, D.; Li, R.T.; Liu, X.; Sun, M.Z.; Wu, J.; Zhang, N.; Zhu, Y.M. The positive regulatory roles of the TIFY10 proteins in plant responses to alkaline stress. PLoS ONE 2014, 9, e111984. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Yang, R.J.; Zhang, T.T.; Zheng, D.Y.; Li, X.L.; Zhang, Z.B.; Li, L.G.; Wu, Z.Y. ZmTIFY16, a novel maize TIFY transcription factor gene, promotes root growth and development and enhances drought and salt tolerance in Arabidopsis and Zea mays. Plant Growth Regul. 2023, 100, 149–160. [Google Scholar] [CrossRef]
- Falcone Ferreyra, M.L.; Rius, S.P.; Casati, P. Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 2012, 3, 222. [Google Scholar] [CrossRef]
- Schenke, D.; Utami, H.P.; Zhou, Z.; Gallegos, M.T.; Cai, D.G. Suppression of UV-B stress induced flavonoids by biotic stress: Is there reciprocal crosstalk? Plant Physiol. Biochem. 2019, 134, 53–63. [Google Scholar] [CrossRef]
- Ma, G.; Zhang, L.; Yamamoto, R.; Kojima, N.; Yahata, M.; Kato, M. Molecular characterization of a flavanone 3-hydroxylase gene from citrus fruit reveals its crucial roles in anthocyanin accumulation. BMC Plant Biol. 2023, 23, 233. [Google Scholar] [CrossRef]
- Postiglione, A.E.; Delange, A.M.; Ali, M.F.; Wang, E.Y.; Houben, M.; Hahn, S.L.; Khoury, M.G.; Roark, C.M.; Davis, M.; Reid, R.W.; et al. Flavonols improve tomato pollen thermotolerance during germination and tube elongation by maintaining reactive oxygen species homeostasis. Plant Cell 2024, 36, 4511–4534. [Google Scholar] [CrossRef]
- Pei, D.; Ren, Y.H.; Yu, W.B.; Zhang, P.A.; Dong, T.Y.; Jia, H.F.; Fang, J.G. The roles of brassinosteroids and methyl jasmonate on postharvest grape by regulating the interaction between VvDWF4 and VvTIFY5A. Plant Sci. 2023, 336, 111830. [Google Scholar] [CrossRef] [PubMed]
- Hao, Q.; Li, T.; Lu, G.; Wang, S.; Li, Z.; Gu, C.; Kong, F.; Shu, Q.; Li, Y. Chlorophyllase (PsCLH) and light-harvesting chlorophyll a/b binding protein 1 (PsLhcb1) and PsLhcb5 maintain petal greenness in Paeonia suffruticosa ‘Lv Mu Yin Yu’. J. Adv. Res. 2024, in press.
- Dombrecht, B.; Xue, G.P.; Sprague, S.J.; Kirkegaard, J.A.; Ross, J.J.; Reid, J.B.; Fitt, G.P.; Sewelam, N.; Schenk, P.M.; Manners, J.M.; et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 2007, 19, 2225–2245. [Google Scholar] [PubMed]
- Hong, G.J.; Xue, X.Y.; Mao, Y.B.; Wang, L.J.; Chen, X.Y. Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 2012, 24, 2635–2648. [Google Scholar] [CrossRef]
- Shoji, T.; Hashimoto, T. Tobacco MYC2 regulates jasmonate-inducible nicotine biosynthesis genes directly and by way of the NIC2-locus ERF genes. Plant Cell Physiol. 2011, 52, 1117–1130. [Google Scholar]
| Gene Name | Gene ID | Chromosome Location | CDS/bp | Size/aa | pI | Molecular Mass/ku | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| DlTIFY1 | Dil.01g002090 | Chr1: 1595544~1598311 | 867 | 288 | 8.94 | 30,911.09 | Nucleus |
| DlTIFY2 | Dil.01g002730 | Chr1: 2125173~2128525 | 711 | 236 | 5.18 | 24,954.09 | Nucleus |
| DlTIFY3 | Dil.01g005880 | Chr1: 4817410~4822560 | 1308 | 435 | 8.98 | 46,050.99 | Nucleus |
| DlTIFY4 | Dil.01g031450 | Chr1: 40844418~40848860 | 1005 | 334 | 8.68 | 35,903.26 | Nucleus |
| DlTIFY5 | Dil.03g000830 | Chr3: 675367~677656 | 744 | 247 | 9.08 | 26,945.32 | Nucleus |
| DlTIFY6 | Dil.04g007360 | Chr4: 5796350~5801241 | 1008 | 335 | 8.64 | 36,650.41 | Nucleus |
| DlTIFY7 | Dil.07g012790 | Chr7: 22023102~22024262 | 411 | 136 | 9.60 | 15,454.49 | Nucleus |
| DlTIFY8 | Dil.08g009850 | Chr8: 19156311~19164074 | 1065 | 354 | 8.41 | 38,771.16 | Nucleus |
| DlTIFY9 | Dil.09g007890 | Chr9: 6380164~6385297 | 864 | 287 | 6.15 | 31,263.42 | Nucleus |
| DlTIFY10 | Dil.09g007900 | Chr9: 6387450~6394375 | 1134 | 377 | 4.69 | 41,380.79 | Nucleus |
| DlTIFY11 | Dil.09g008150 | Chr9: 6682786~6687897 | 864 | 287 | 6.15 | 31,263.42 | Nucleus |
| DlTIFY12 | Dil.09g008160 | Chr9: 6690088~6697001 | 1134 | 377 | 4.69 | 41,380.79 | Nucleus |
| DlTIFY13 | Dil.10g008370 | Chr10: 16658489~16661430 | 1104 | 367 | 8.86 | 38,651.59 | Nucleus |
| DlTIFY14 | Dil.10g008390 | Chr10: 16722598~16725548 | 1104 | 367 | 8.86 | 38,574.55 | Nucleus |
| DlTIFY15 | Dil.10g020310 | Chr10: 27282532~27285457 | 552 | 183 | 9.30 | 20,614.34 | Nucleus |
| DlTIFY16 | Dil.11g007770 | Chr11: 12937525~12938905 | 645 | 214 | 9.15 | 24,279.82 | Nucleus |
| DlTIFY17 | Dil.11g007950 | Chr11: 13210964~13212321 | 642 | 213 | 9.30 | 24,185.7 | Nucleus |
| DlTIFY18 | Dil.11g015490 | Chr11: 22161800~22170409 | 1065 | 354 | 5.03 | 39,204.58 | Nucleus |
| DlTIFY19 | Dil.15g013250 | Chr15: 10560028~10570474 | 2160 | 719 | 7.21 | 77,478.53 | Cytoplasm, Nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qing, H.; Wu, Z.; Mo, X.; Wei, J.; Shi, Y.; Guo, H.; Xu, J.; Ding, F.; Zhang, S. Genome-Wide Identification of the TIFY Family in Longan and Their Potential Functional Analysis in Anthocyanin Synthesis. Biology 2025, 14, 364. https://doi.org/10.3390/biology14040364
Qing H, Wu Z, Mo X, Wei J, Shi Y, Guo H, Xu J, Ding F, Zhang S. Genome-Wide Identification of the TIFY Family in Longan and Their Potential Functional Analysis in Anthocyanin Synthesis. Biology. 2025; 14(4):364. https://doi.org/10.3390/biology14040364
Chicago/Turabian StyleQing, Haowei, Ziang Wu, Xiao Mo, Jinjv Wei, Yuyu Shi, Huiqin Guo, Jiongzhi Xu, Feng Ding, and Shuwei Zhang. 2025. "Genome-Wide Identification of the TIFY Family in Longan and Their Potential Functional Analysis in Anthocyanin Synthesis" Biology 14, no. 4: 364. https://doi.org/10.3390/biology14040364
APA StyleQing, H., Wu, Z., Mo, X., Wei, J., Shi, Y., Guo, H., Xu, J., Ding, F., & Zhang, S. (2025). Genome-Wide Identification of the TIFY Family in Longan and Their Potential Functional Analysis in Anthocyanin Synthesis. Biology, 14(4), 364. https://doi.org/10.3390/biology14040364





