The Gut Microbiota and Colorectal Cancer: Understanding the Link and Exploring Therapeutic Interventions
Simple Summary
Abstract
1. Introduction
2. Colorectal Cancer Pathophysiology
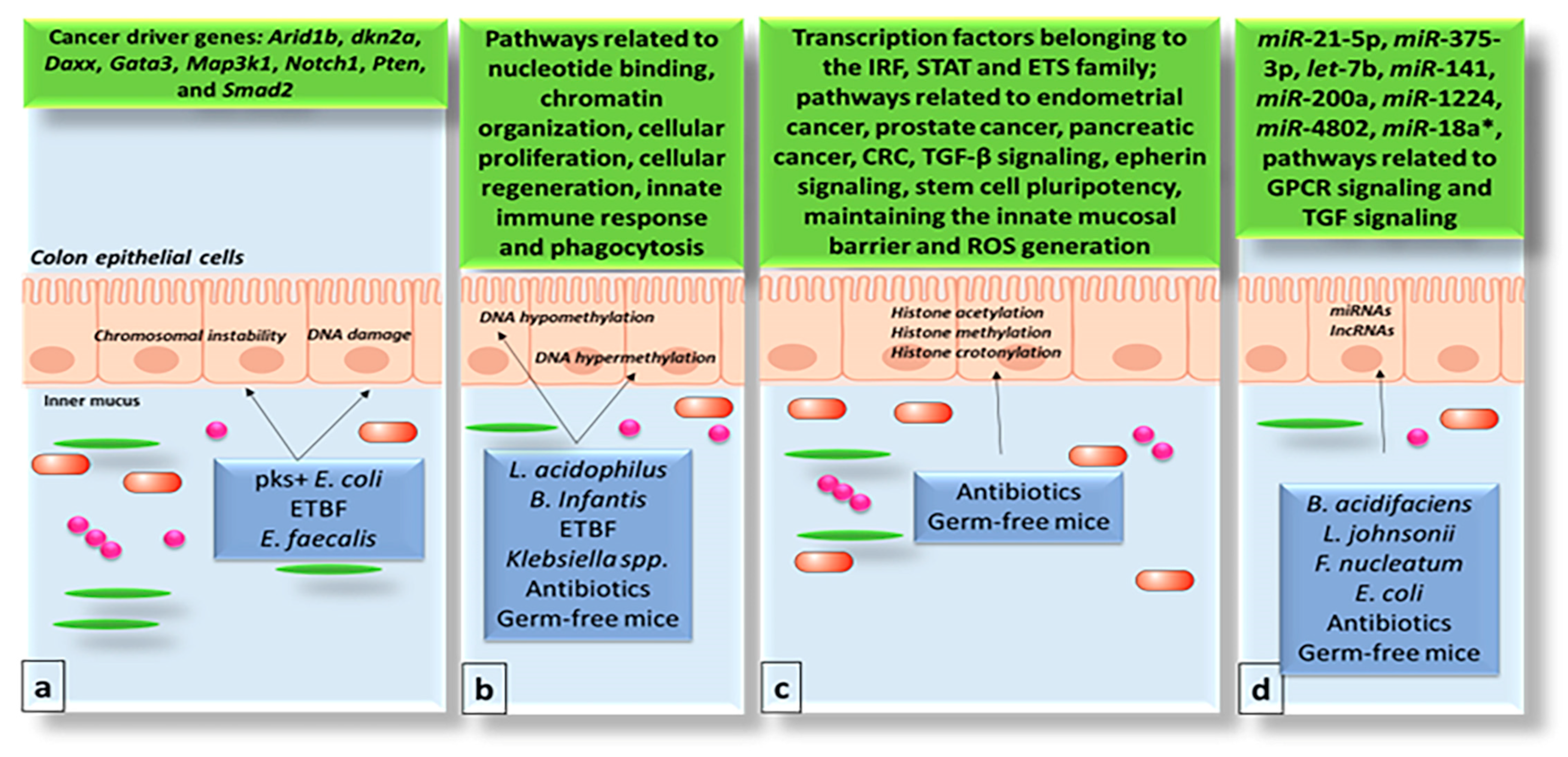
3. Impact of the Gut Microbiome on the Genome and Epigenome of Colon Epithelial Cells
4. The Gut Microbiome and Chromatin Structure
5. The Gut Microbiome and Non-Coding RNAs
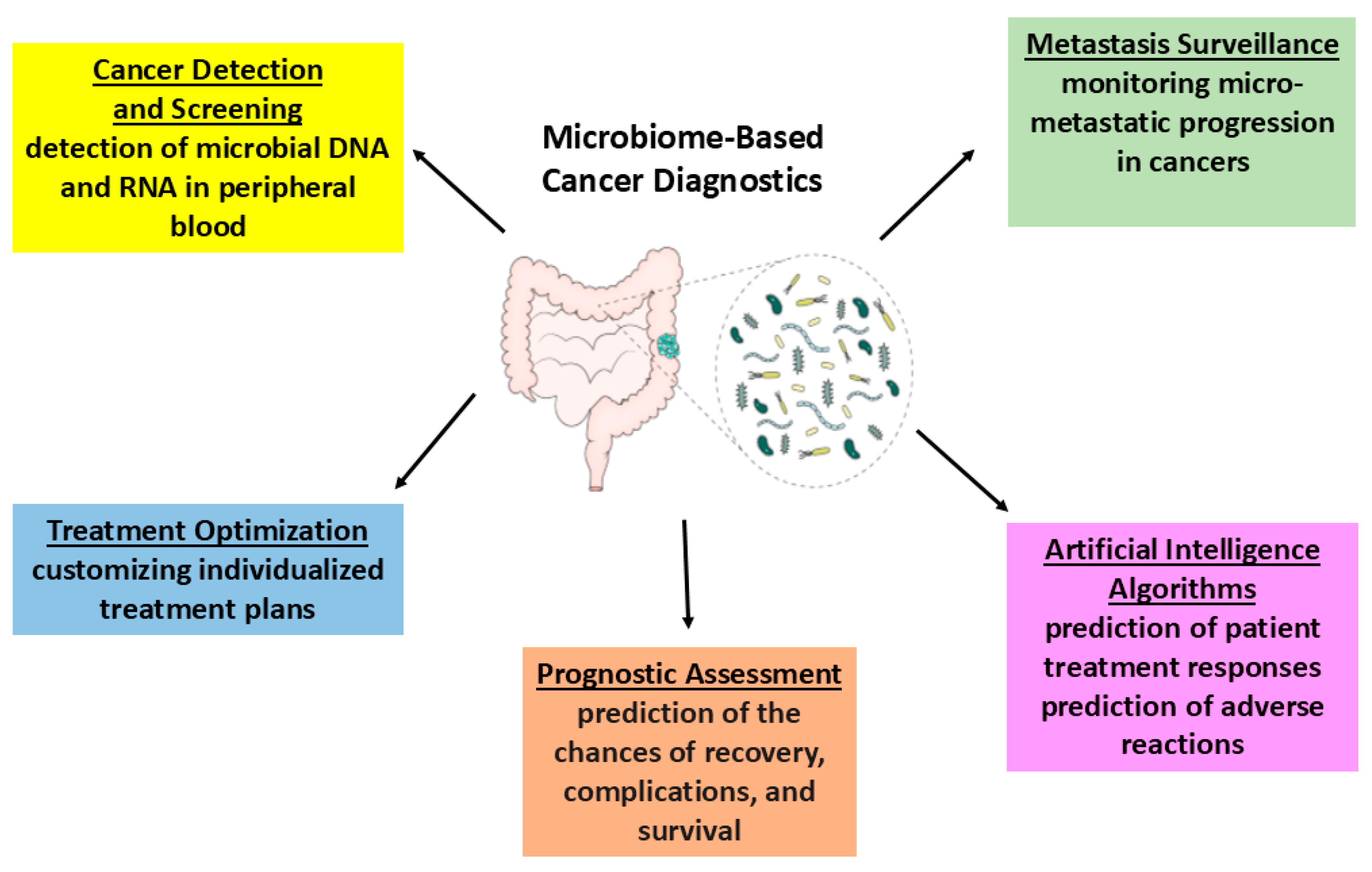
6. Recent and Future Diagnostic Strategies
6.1. Urine-Derived Extracellular Vesicles (EVs)
6.2. Histopathology Imaging Using Artificial Intelligence
6.3. Engineered Bacteria to Detect Cancer in the Gut
7. Treatment Implications
8. Potential Applications of the Gut Microbiome in CRC Prevention and Treatment
8.1. Probiotics
8.2. Prebiotics
8.3. Postbiotics
8.4. Faecal Microbiota Transplantation
8.5. Bacteriophage Therapy
8.6. Fiber Diet
9. Gut Microbiota Impact on Gastrointestinal Therapy
10. Conclusions
11. Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Pineros, M.; Znaor, A.; Bray, F. Cancer statistics for the year 2020: An overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef] [PubMed]
- Global Burden of Disease 2019 Cancer Collaboration; Kocarnik, J.M.; Compton, K.; Dean, F.E.; Fu, W.; Gaw, B.L.; Harvey, J.D.; Henrikson, H.J.; Lu, D.; Pennini, A.; et al. Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life Years for 29 Cancer Groups From 2010 to 2019: A Systematic Analysis for the Global Burden of Disease Study 2019. JAMA Oncol. 2022, 8, 420–444. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Sharma, P.; Sarma, D.K.; Kumawat, M.; Tiwari, R.; Verma, V.; Nagpal, R.; Kumar, M. Implication of Obesity and Gut Microbiome Dysbiosis in the Etiology of Colorectal Cancer. Cancers 2023, 15, 1913. [Google Scholar] [CrossRef] [PubMed]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef]
- Fusco, W.; Lorenzo, M.B.; Cintoni, M.; Porcari, S.; Rinninella, E.; Kaitsas, F.; Lener, E.; Mele, M.C.; Gasbarrini, A.; Collado, M.C.; et al. Short-Chain Fatty-Acid-Producing Bacteria: Key Components of the Human Gut Microbiota. Nutrients 2023, 15, 2211. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, Q.; Zhang, H.; Yi, Z.; Ma, H.; Wang, X.; Wang, J.; Liu, Y.; Zheng, Y.; Fang, W.; et al. Colorectal cancer microbiome programs DNA methylation of host cells by affecting methyl donor metabolism. Genome Med. 2024, 16, 77. [Google Scholar] [CrossRef]
- Zhou, H.; Beltran, J.F.; Brito, I.L. Host-microbiome protein-protein interactions capture disease-relevant pathways. Genome Biol. 2022, 23, 72. [Google Scholar] [CrossRef]
- Kim, D.; Hofstaedter, C.E.; Zhao, C.; Mattei, L.; Tanes, C.; Clarke, E.; Lauder, A.; Sherrill-Mix, S.; Chehoud, C.; Kelsen, J.; et al. Optimizing methods and dodging pitfalls in microbiome research. Microbiome 2017, 5, 52. [Google Scholar] [CrossRef]
- Sun, S.; Wang, H.; Tsilimigras, M.C.; Howard, A.G.; Sha, W.; Zhang, J.; Su, C.; Wang, Z.; Du, S.; Sioda, M.; et al. Does geographical variation confound the relationship between host factors and the human gut microbiota: A population-based study in China. BMJ Open 2020, 10, e038163. [Google Scholar] [CrossRef]
- Zhao, M.; Chu, J.; Feng, S.; Guo, C.; Xue, B.; He, K.; Li, L. Immunological mechanisms of inflammatory diseases caused by gut microbiota dysbiosis: A review. Biomed. Pharmacother. 2023, 164, 114985. [Google Scholar] [CrossRef]
- Ge, Y.; Wang, X.; Guo, Y.; Yan, J.; Abuduwaili, A.; Aximujiang, K.; Yan, J.; Wu, M. Correction to: Gut microbiota influence tumor development and Alter interactions with the human immune system. J. Exp. Clin. Cancer Res. 2021, 40, 334. [Google Scholar] [CrossRef] [PubMed]
- Van Dingenen, L.; Segers, C.; Wouters, S.; Mysara, M.; Leys, N.; Kumar-Singh, S.; Malhotra-Kumar, S.; Van Houdt, R. Dissecting the role of the gut microbiome and fecal microbiota transplantation in radio- and immunotherapy treatment of colorectal cancer. Front. Cell Infect. Microbiol. 2023, 13, 1298264. [Google Scholar] [CrossRef] [PubMed]
- Pandey, H.; Tang, D.W.T.; Wong, S.H.; Lal, D. Gut Microbiota in Colorectal Cancer: Biological Role and Therapeutic Opportunities. Cancers 2023, 15, 866. [Google Scholar] [CrossRef] [PubMed]
- Veziant, J.; Villeger, R.; Barnich, N.; Bonnet, M. Gut Microbiota as Potential Biomarker and/or Therapeutic Target to Improve the Management of Cancer: Focus on Colibactin-Producing Escherichia coli in Colorectal Cancer. Cancers 2021, 13, 2215. [Google Scholar] [CrossRef]
- Zhao, L.; Cho, W.C.; Nicolls, M.R. Colorectal Cancer-Associated Microbiome Patterns and Signatures. Front. Genet. 2021, 12, 787176. [Google Scholar] [CrossRef]
- Dougherty, M.W.; Jobin, C. Intestinal bacteria and colorectal cancer: Etiology and treatment. Gut Microbes 2023, 15, 2185028. [Google Scholar] [CrossRef]
- Wielandt, A.M.; Hurtado, C.; Moreno, C.M.; Villarroel, C.; Castro, M.; Estay, M.; Simian, D.; Martinez, M.; Vial, M.T.; Kronberg, U.; et al. Characterization of Chilean patients with sporadic colorectal cancer according to the three main carcinogenic pathways: Microsatellite instability, CpG island methylator phenotype and Chromosomal instability. Tumour Biol. 2020, 42, 1010428320938492. [Google Scholar] [CrossRef]
- Ternet, C.; Kiel, C. Signaling pathways in intestinal homeostasis and colorectal cancer: KRAS at centre stage. Cell Commun. Signal 2021, 19, 31. [Google Scholar] [CrossRef]
- Zella, D.; Gallo, R.C. Viruses and Bacteria Associated with Cancer: An Overview. Viruses 2021, 13, 1039. [Google Scholar] [CrossRef]
- Agagunduz, D.; Cocozza, E.; Cemali, O.; Bayazit, A.D.; Nani, M.F.; Cerqua, I.; Morgillo, F.; Saygili, S.K.; Berni Canani, R.; Amero, P.; et al. Understanding the role of the gut microbiome in gastrointestinal cancer: A review. Front. Pharmacol. 2023, 14, 1130562. [Google Scholar] [CrossRef]
- Ibeanu, G.C.; Rowaiye, A.B.; Okoli, J.C.; Eze, D.U. Microbiome Differences in Colorectal Cancer Patients and Healthy Individuals: Implications for Vaccine Antigen Discovery. Immunotargets Ther. 2024, 13, 749–774. [Google Scholar] [CrossRef] [PubMed]
- Esfandiari, F.; Bakhshi, B.; Shahbazi, T.; Derakhshan-Nezhad, E.; Bahroudi, M.; Minaeeian, S.; Boustanshenas, M.; Alborzi, F.; Behboudi, B.; Fazeli, M.S. Significant difference in gut microbiota Bifidobacterium species but not Lactobacillus species in colorectal cancer patients in comparison with healthy volunteers using quantitative real-time PCR. PLoS ONE 2024, 19, e0294053. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Yang, M.; Dong, W.; Liu, T.; Song, X.; Gu, Y.; Wang, S.; Liu, Y.; Abla, Z.; Qiao, X.; et al. Gut Dysbiosis and Abnormal Bile Acid Metabolism in Colitis-Associated Cancer. Gastroenterol. Res. Pract. 2021, 2021, 6645970. [Google Scholar] [CrossRef]
- Liu, X.; Jin, G.; Tang, Q.; Huang, S.; Zhang, Y.; Sun, Y.; Liu, T.; Guo, Z.; Yang, C.; Wang, B.; et al. Early life Lactobacillus rhamnosus GG colonisation inhibits intestinal tumour formation. Br. J. Cancer 2022, 126, 1421–1431. [Google Scholar] [CrossRef]
- Liu, Y.; Lau, H.C.; Cheng, W.Y.; Yu, J. Gut Microbiome in Colorectal Cancer: Clinical Diagnosis and Treatment. Genom. Proteom. Bioinform. 2023, 21, 84–96. [Google Scholar] [CrossRef]
- Sawicki, T.; Ruszkowska, M.; Danielewicz, A.; Niedzwiedzka, E.; Arlukowicz, T.; Przybylowicz, K.E. A Review of Colorectal Cancer in Terms of Epidemiology, Risk Factors, Development, Symptoms and Diagnosis. Cancers 2021, 13, 2025. [Google Scholar] [CrossRef]
- Das, C.; Tyler, J.K. Histone exchange and histone modifications during transcription and aging. Biochim. Biophys. Acta 2013, 1819, 332–342. [Google Scholar] [CrossRef]
- Jung, G.; Hernandez-Illan, E.; Moreira, L.; Balaguer, F.; Goel, A. Epigenetics of colorectal cancer: Biomarker and therapeutic potential. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 111–130. [Google Scholar] [CrossRef]
- Allen, J.; Sears, C.L. Impact of the gut microbiome on the genome and epigenome of colon epithelial cells: Contributions to colorectal cancer development. Genome Med. 2019, 11, 11. [Google Scholar] [CrossRef]
- Fellows, R.; Denizot, J.; Stellato, C.; Cuomo, A.; Jain, P.; Stoyanova, E.; Balazsi, S.; Hajnady, Z.; Liebert, A.; Kazakevych, J.; et al. Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat. Commun. 2018, 9, 105. [Google Scholar] [CrossRef]
- Donohoe, D.R.; Holley, D.; Collins, L.B.; Montgomery, S.A.; Whitmore, A.C.; Hillhouse, A.; Curry, K.P.; Renner, S.W.; Greenwalt, A.; Ryan, E.P.; et al. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov. 2014, 4, 1387–1397. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Chen, S.; Zang, D.; Sun, H.; Sun, Y.; Chen, J. Butyrate as a promising therapeutic target in cancer: From pathogenesis to clinic (Review). Int. J. Oncol. 2024, 64, 44. [Google Scholar] [CrossRef] [PubMed]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Gerstein, M.B.; Rozowsky, J.; Yan, K.K.; Wang, D.; Cheng, C.; Brown, J.B.; Davis, C.A.; Hillier, L.; Sisu, C.; Li, J.J.; et al. Comparative analysis of the transcriptome across distant species. Nature 2014, 512, 445–448. [Google Scholar] [CrossRef]
- Harrow, J.; Frankish, A.; Gonzalez, J.M.; Tapanari, E.; Diekhans, M.; Kokocinski, F.; Aken, B.L.; Barrell, D.; Zadissa, A.; Searle, S.; et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012, 22, 1760–1774. [Google Scholar] [CrossRef]
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermuller, J.; Hofacker, I.L.; et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007, 316, 1484–1488. [Google Scholar] [CrossRef]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef]
- Poller, W.; Dimmeler, S.; Heymans, S.; Zeller, T.; Haas, J.; Karakas, M.; Leistner, D.M.; Jakob, P.; Nakagawa, S.; Blankenberg, S.; et al. Non-coding RNAs in cardiovascular diseases: Diagnostic and therapeutic perspectives. Eur. Heart J. 2018, 39, 2704–2716. [Google Scholar] [CrossRef]
- Zhou, L.; Zhu, Y.; Sun, D.; Zhang, Q. Emerging Roles of Long non-coding RNAs in The Tumor Microenvironment. Int. J. Biol. Sci. 2020, 16, 2094–2103. [Google Scholar] [CrossRef]
- Viennois, E.; Chassaing, B.; Tahsin, A.; Pujada, A.; Wang, L.; Gewirtz, A.T.; Merlin, D. Host-derived fecal microRNAs can indicate gut microbiota healthiness and ability to induce inflammation. Theranostics 2019, 9, 4542–4557. [Google Scholar] [CrossRef]
- Xing, J.; Liao, Y.; Zhang, H.; Zhang, W.; Zhang, Z.; Zhang, J.; Wang, D.; Tang, D. Impacts of MicroRNAs Induced by the Gut Microbiome on Regulating the Development of Colorectal Cancer. Front. Cell Infect. Microbiol. 2022, 12, 804689. [Google Scholar] [CrossRef] [PubMed]
- Nakata, K.; Sugi, Y.; Narabayashi, H.; Kobayakawa, T.; Nakanishi, Y.; Tsuda, M.; Hosono, A.; Kaminogawa, S.; Hanazawa, S.; Takahashi, K. Commensal microbiota-induced microRNA modulates intestinal epithelial permeability through the small GTPase ARF4. J. Biol. Chem. 2017, 292, 15426–15433. [Google Scholar] [CrossRef] [PubMed]
- Bullman, S.; Pedamallu, C.S.; Sicinska, E.; Clancy, T.E.; Zhang, X.; Cai, D.; Neuberg, D.; Huang, K.; Guevara, F.; Nelson, T.; et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017, 358, 1443–1448. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.; Kim, N.E.; Park, J.; Shin, C.M.; Kim, N.; Lee, D.H.; Park, J.Y.; Choi, C.H.; Kim, J.G.; Park, Y.S. Analysis of the gut microbiome using extracellular vesicles in the urine of patients with colorectal cancer. Korean J. Intern. Med. 2023, 38, 27–38. [Google Scholar] [CrossRef]
- Thomer, L.; Schneewind, O.; Missiakas, D. Pathogenesis of Staphylococcus aureus Bloodstream Infections. Annu. Rev. Pathol. 2016, 11, 343–364. [Google Scholar] [CrossRef]
- Ren, X.; Palmer, L.D. Acinetobacter Metabolism in Infection and Antimicrobial Resistance. Infect. Immun. 2023, 91, e0043322. [Google Scholar] [CrossRef]
- Shakhpazyan, N.K.; Mikhaleva, L.M.; Bedzhanyan, A.L.; Gioeva, Z.V.; Mikhalev, A.I.; Midiber, K.Y.; Pechnikova, V.V.; Biryukov, A.E. Exploring the Role of the Gut Microbiota in Modulating Colorectal Cancer Immunity. Cells 2024, 13, 1437. [Google Scholar] [CrossRef]
- Wang, K.S.; Yu, G.; Xu, C.; Meng, X.H.; Zhou, J.; Zheng, C.; Deng, Z.; Shang, L.; Liu, R.; Su, S.; et al. Accurate diagnosis of colorectal cancer based on histopathology images using artificial intelligence. BMC Med. 2021, 19, 76. [Google Scholar] [CrossRef]
- Hildebrand, L.A.; Pierce, C.J.; Dennis, M.; Paracha, M.; Maoz, A. Artificial Intelligence for Histology-Based Detection of Microsatellite Instability and Prediction of Response to Immunotherapy in Colorectal Cancer. Cancers 2021, 13, 391. [Google Scholar] [CrossRef]
- Takamatsu, M.; Yamamoto, N.; Kawachi, H.; Nakano, K.; Saito, S.; Fukunaga, Y.; Takeuchi, K. Prediction of lymph node metastasis in early colorectal cancer based on histologic images by artificial intelligence. Sci. Rep. 2022, 12, 2963. [Google Scholar] [CrossRef]
- Shimada, Y.; Okuda, S.; Watanabe, Y.; Tajima, Y.; Nagahashi, M.; Ichikawa, H.; Nakano, M.; Sakata, J.; Takii, Y.; Kawasaki, T.; et al. Histopathological characteristics and artificial intelligence for predicting tumor mutational burden-high colorectal cancer. J. Gastroenterol. 2021, 56, 547–559. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Mo, Z.; Zhu, W.; Liao, B.; Yang, Y.; Wu, F.X. Prediction of Target-Drug Therapy by Identifying Gene Mutations in Lung Cancer With Histopathological Stained Image and Deep Learning Techniques. Front. Oncol. 2021, 11, 642945. [Google Scholar] [CrossRef] [PubMed]
- Kather, J.N.; Pearson, A.T.; Halama, N.; Jager, D.; Krause, J.; Loosen, S.H.; Marx, A.; Boor, P.; Tacke, F.; Neumann, U.P.; et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 2019, 25, 1054–1056. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.H.; Gilkes, D.M.; Phillip, J.M.; Narkar, A.; Cheng, T.W.; Marchand, J.; Lee, M.H.; Li, R.; Wirtz, D. Single-cell morphology encodes metastatic potential. Sci. Adv. 2020, 6, eaaw6938. [Google Scholar] [CrossRef]
- D’Urso, F.; Broccolo, F. Applications of Artificial Intelligence in Microbiome Analysis and Probiotic Interventions—An Overview and Perspective Based on the Current State of the Art. Appl. Sci. 2024, 14, 8627. [Google Scholar] [CrossRef]
- Hernandez Medina, R.; Kutuzova, S.; Nielsen, K.N.; Johansen, J.; Hansen, L.H.; Nielsen, M.; Rasmussen, S. Machine learning and deep learning applications in microbiome research. ISME Commun. 2022, 2, 98. [Google Scholar] [CrossRef]
- Aryal, S.; Alimadadi, A.; Manandhar, I.; Joe, B.; Cheng, X. Machine Learning Strategy for Gut Microbiome-Based Diagnostic Screening of Cardiovascular Disease. Hypertension 2020, 76, 1555–1562. [Google Scholar] [CrossRef]
- Chehelgerdi, M.; Chehelgerdi, M.; Allela, O.Q.B.; Pecho, R.D.C.; Jayasankar, N.; Rao, D.P.; Thamaraikani, T.; Vasanthan, M.; Viktor, P.; Lakshmaiya, N.; et al. Progressing nanotechnology to improve targeted cancer treatment: Overcoming hurdles in its clinical implementation. Mol. Cancer 2023, 22, 169. [Google Scholar] [CrossRef]
- Cooper, R.M.; Wright, J.A.; Ng, J.Q.; Goyne, J.M.; Suzuki, N.; Lee, Y.K.; Ichinose, M.; Radford, G.; Ryan, F.J.; Kumar, S.; et al. Engineered bacteria detect tumor DNA. Science 2023, 381, 682–686. [Google Scholar] [CrossRef]
- Evangelou, K.; Belogiannis, K.; Papaspyropoulos, A.; Petty, R.; Gorgoulis, V.G. Escape from senescence: Molecular basis and therapeutic ramifications. J. Pathol. 2023, 260, 649–665. [Google Scholar] [CrossRef]
- Sedlak, J.C.; Yilmaz, O.H.; Roper, J. Metabolism and Colorectal Cancer. Annu. Rev. Pathol. 2023, 18, 467–492. [Google Scholar] [CrossRef] [PubMed]
- Agagunduz, D.; Celik, E.; Cemali, O.; Bingol, F.G.; Ozenir, C.; Ozogul, F.; Capasso, R. Probiotics, Live Biotherapeutic Products (LBPs), and Gut-Brain Axis Related Psychological Conditions: Implications for Research and Dietetics. Probiotics Antimicrob. Proteins 2023, 15, 1014–1031. [Google Scholar] [CrossRef] [PubMed]
- Gamallat, Y.; Meyiah, A.; Kuugbee, E.D.; Hago, A.M.; Chiwala, G.; Awadasseid, A.; Bamba, D.; Zhang, X.; Shang, X.; Luo, F.; et al. Lactobacillus rhamnosus induced epithelial cell apoptosis, ameliorates inflammation and prevents colon cancer development in an animal model. Biomed. Pharmacother. 2016, 83, 536–541. [Google Scholar] [CrossRef] [PubMed]
- Li, S.C.; Lin, H.P.; Chang, J.S.; Shih, C.K. Lactobacillus acidophilus-Fermented Germinated Brown Rice Suppresses Preneoplastic Lesions of the Colon in Rats. Nutrients 2019, 11, 2718. [Google Scholar] [CrossRef]
- Tegopoulos, K.; Stergiou, O.S.; Kiousi, D.E.; Tsifintaris, M.; Koletsou, E.; Papageorgiou, A.C.; Argyri, A.A.; Chorianopoulos, N.; Galanis, A.; Kolovos, P. Genomic and Phylogenetic Analysis of Lactiplantibacillus plantarum L125, and Evaluation of Its Anti-Proliferative and Cytotoxic Activity in Cancer Cells. Biomedicines 2021, 9, 1718. [Google Scholar] [CrossRef]
- Dehghani, N.; Tafvizi, F.; Jafari, P. Cell cycle arrest and anti-cancer potential of probiotic Lactobacillus rhamnosus against HT-29 cancer cells. Bioimpacts 2021, 11, 245–252. [Google Scholar] [CrossRef]
- Salemi, R.; Vivarelli, S.; Ricci, D.; Scillato, M.; Santagati, M.; Gattuso, G.; Falzone, L.; Libra, M. Lactobacillus rhamnosus GG cell-free supernatant as a novel anti-cancer adjuvant. J. Transl. Med. 2023, 21, 195. [Google Scholar] [CrossRef]
- Tiptiri-Kourpeti, A.; Spyridopoulou, K.; Santarmaki, V.; Aindelis, G.; Tompoulidou, E.; Lamprianidou, E.E.; Saxami, G.; Ypsilantis, P.; Lampri, E.S.; Simopoulos, C.; et al. Lactobacillus casei Exerts Anti-Proliferative Effects Accompanied by Apoptotic Cell Death and Up-Regulation of TRAIL in Colon Carcinoma Cells. PLoS ONE 2016, 11, e0147960. [Google Scholar] [CrossRef]
- Faghfoori, Z.; Faghfoori, M.H.; Saber, A.; Izadi, A.; Yari Khosroushahi, A. Anticancer effects of bifidobacteria on colon cancer cell lines. Cancer Cell Int. 2021, 21, 258. [Google Scholar] [CrossRef]
- Kim, S.W.; Kim, H.M.; Yang, K.M.; Kim, S.A.; Kim, S.K.; An, M.J.; Park, J.J.; Lee, S.K.; Kim, T.I.; Kim, W.H.; et al. Bifidobacterium lactis inhibits NF-kappaB in intestinal epithelial cells and prevents acute colitis and colitis-associated colon cancer in mice. Inflamm. Bowel Dis. 2010, 16, 1514–1525. [Google Scholar] [CrossRef]
- Li, Q.; Hu, W.; Liu, W.X.; Zhao, L.Y.; Huang, D.; Liu, X.D.; Chan, H.; Zhang, Y.; Zeng, J.D.; Coker, O.O.; et al. Streptococcus thermophilus Inhibits Colorectal Tumorigenesis Through Secreting beta-Galactosidase. Gastroenterology 2021, 160, 1179–1193e1114. [Google Scholar] [CrossRef] [PubMed]
- Quevrain, E.; Maubert, M.A.; Michon, C.; Chain, F.; Marquant, R.; Tailhades, J.; Miquel, S.; Carlier, L.; Bermudez-Humaran, L.G.; Pigneur, B.; et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut 2016, 65, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Pandey, K.R.; Naik, S.R.; Vakil, B.V. Probiotics, prebiotics and synbiotics—A review. J. Food Sci. Technol. 2015, 52, 7577–7587. [Google Scholar] [CrossRef]
- de Vrese, M.; Schrezenmeir, J. Probiotics, prebiotics, and synbiotics. Adv. Biochem. Eng. Biotechnol. 2008, 111, 1–66. [Google Scholar] [CrossRef]
- Halmos, E.P.; Christophersen, C.T.; Bird, A.R.; Shepherd, S.J.; Muir, J.G.; Gibson, P.R. Consistent Prebiotic Effect on Gut Microbiota With Altered FODMAP Intake in Patients with Crohn’s Disease: A Randomised, Controlled Cross-Over Trial of Well-Defined Diets. Clin. Transl. Gastroenterol. 2016, 7, e164. [Google Scholar] [CrossRef]
- Raisch, J.; Buc, E.; Bonnet, M.; Sauvanet, P.; Vazeille, E.; de Vallee, A.; Dechelotte, P.; Darcha, C.; Pezet, D.; Bonnet, R.; et al. Colon cancer-associated B2 Escherichia coli colonize gut mucosa and promote cell proliferation. World J. Gastroenterol. 2014, 20, 6560–6572. [Google Scholar] [CrossRef]
- Chen, P.W.; Liu, Z.S.; Kuo, T.C.; Hsieh, M.C.; Li, Z.W. Prebiotic effects of bovine lactoferrin on specific probiotic bacteria. Biometals 2017, 30, 237–248. [Google Scholar] [CrossRef]
- Mahdavi, M.; Laforest-Lapointe, I.; Masse, E. Preventing Colorectal Cancer through Prebiotics. Microorganisms 2021, 9, 1325. [Google Scholar] [CrossRef]
- Hawcroft, G.; Volpato, M.; Marston, G.; Ingram, N.; Perry, S.L.; Cockbain, A.J.; Race, A.D.; Munarini, A.; Belluzzi, A.; Loadman, P.M.; et al. The omega-3 polyunsaturated fatty acid eicosapentaenoic acid inhibits mouse MC-26 colorectal cancer cell liver metastasis via inhibition of PGE2-dependent cell motility. Br. J. Pharmacol. 2012, 166, 1724–1737. [Google Scholar] [CrossRef]
- Wang, W.; Yang, J.; Nimiya, Y.; Lee, K.S.S.; Sanidad, K.; Qi, W.; Sukamtoh, E.; Park, Y.; Liu, Z.; Zhang, G. omega-3 Polyunsaturated fatty acids and their cytochrome P450-derived metabolites suppress colorectal tumor development in mice. J. Nutr. Biochem. 2017, 48, 29–35. [Google Scholar] [CrossRef]
- Fidelis, M.; Santos, J.S.; Escher, G.B.; Rocha, R.S.; Cruz, A.G.; Cruz, T.M.; Marques, M.B.; Nunes, J.B.; do Carmo, M.A.V.; de Almeida, L.A.; et al. Polyphenols of jabuticaba [Myrciaria jaboticaba (Vell.) O.Berg] seeds incorporated in a yogurt model exert antioxidant activity and modulate gut microbiota of 1,2-dimethylhydrazine-induced colon cancer in rats. Food Chem. 2021, 334, 127565. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, J.; Ledesma, E.; Monte, J.; Millan, E.; Costa, P.; de la Fuente, V.G.; Garcia, M.T.F.; Martinez-Camblor, P.; Villar, C.J.; Lombo, F. Traditional Processed Meat Products Re-designed Towards Inulin-rich Functional Foods Reduce Polyps in Two Colorectal Cancer Animal Models. Sci. Rep. 2019, 9, 14783. [Google Scholar] [CrossRef] [PubMed]
- Salminen, S.; Collado, M.C.; Endo, A.; Hill, C.; Lebeer, S.; Quigley, E.M.M.; Sanders, M.E.; Shamir, R.; Swann, J.R.; Szajewska, H.; et al. Publisher Correction: Publisher Correction: The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 671. [Google Scholar] [CrossRef]
- Liang, B.; Xing, D. The Current and Future Perspectives of Postbiotics. Probiotics Antimicrob. Proteins 2023, 15, 1626–1643. [Google Scholar] [CrossRef]
- Song, D.; Wang, X.; Ma, Y.; Liu, N.N.; Wang, H. Beneficial insights into postbiotics against colorectal cancer. Front. Nutr. 2023, 10, 1111872. [Google Scholar] [CrossRef]
- Li, N.; Russell, W.M.; Douglas-Escobar, M.; Hauser, N.; Lopez, M.; Neu, J. Live and heat-killed Lactobacillus rhamnosus GG: Effects on proinflammatory and anti-inflammatory cytokines/chemokines in gastrostomy-fed infant rats. Pediatr. Res. 2009, 66, 203–207. [Google Scholar] [CrossRef]
- Wang, L.; Tang, L.; Feng, Y.; Zhao, S.; Han, M.; Zhang, C.; Yuan, G.; Zhu, J.; Cao, S.; Wu, Q.; et al. A purified membrane protein from Akkermansia muciniphila or the pasteurised bacterium blunts colitis associated tumourigenesis by modulation of CD8(+) T cells in mice. Gut 2020, 69, 1988–1997. [Google Scholar] [CrossRef]
- Wang, S.; Ahmadi, S.; Nagpal, R.; Jain, S.; Mishra, S.P.; Kavanagh, K.; Zhu, X.; Wang, Z.; McClain, D.A.; Kritchevsky, S.B.; et al. Lipoteichoic acid from the cell wall of a heat killed Lactobacillus paracasei D3-5 ameliorates aging-related leaky gut, inflammation and improves physical and cognitive functions: From C. elegans to mice. Geroscience 2020, 42, 333–352. [Google Scholar] [CrossRef]
- Li, Q.; Ding, C.; Meng, T.; Lu, W.; Liu, W.; Hao, H.; Cao, L. Butyrate suppresses motility of colorectal cancer cells via deactivating Akt/ERK signaling in histone deacetylase dependent manner. J. Pharmacol. Sci. 2017, 135, 148–155. [Google Scholar] [CrossRef]
- Mowat, C.; Dhatt, J.; Bhatti, I.; Hamie, A.; Baker, K. Short chain fatty acids prime colorectal cancer cells to activate antitumor immunity. Front. Immunol. 2023, 14, 1190810. [Google Scholar] [CrossRef]
- Biazzo, M.; Deidda, G. Fecal Microbiota Transplantation as New Therapeutic Avenue for Human Diseases. J. Clin. Med. 2022, 11, 4119. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Allen-Vercoe, E.; Petrof, E.O. Fecal microbiota transplantation: In perspective. Ther. Adv. Gastroenterol. 2016, 9, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Chen, Z.; Cai, J. Fecal microbiota transplantation: Emerging applications in autoimmune diseases. J. Autoimmun. 2023, 141, 103038. [Google Scholar] [CrossRef]
- Wu, K.; Luo, Q.; Liu, Y.; Li, A.; Xia, D.; Sun, X. Causal relationship between gut microbiota and gastrointestinal diseases: A mendelian randomization study. J. Transl. Med. 2024, 22, 92. [Google Scholar] [CrossRef]
- Rosshart, S.P.; Vassallo, B.G.; Angeletti, D.; Hutchinson, D.S.; Morgan, A.P.; Takeda, K.; Hickman, H.D.; McCulloch, J.A.; Badger, J.H.; Ajami, N.J.; et al. Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell 2017, 171, 1015–1028e1013. [Google Scholar] [CrossRef]
- Wong, S.H.; Zhao, L.; Zhang, X.; Nakatsu, G.; Han, J.; Xu, W.; Xiao, X.; Kwong, T.N.Y.; Tsoi, H.; Wu, W.K.K.; et al. Gavage of Fecal Samples From Patients With Colorectal Cancer Promotes Intestinal Carcinogenesis in Germ-Free and Conventional Mice. Gastroenterology 2017, 153, 1621–1633.e6. [Google Scholar] [CrossRef]
- Huang, J.; Zheng, X.; Kang, W.; Hao, H.; Mao, Y.; Zhang, H.; Chen, Y.; Tan, Y.; He, Y.; Zhao, W.; et al. Metagenomic and metabolomic analyses reveal synergistic effects of fecal microbiota transplantation and anti-PD-1 therapy on treating colorectal cancer. Front. Immunol. 2022, 13, 874922. [Google Scholar] [CrossRef]
- El Haddad, L.; Mendoza, J.F.; Jobin, C. Bacteriophage-mediated manipulations of microbiota in gastrointestinal diseases. Front. Microbiol. 2022, 13, 1055427. [Google Scholar] [CrossRef]
- Dong, X.; Pan, P.; Zheng, D.W.; Bao, P.; Zeng, X.; Zhang, X.Z. Bioinorganic hybrid bacteriophage for modulation of intestinal microbiota to remodel tumor-immune microenvironment against colorectal cancer. Sci. Adv. 2020, 6, eaba1590. [Google Scholar] [CrossRef]
- De Almeida, C.V.; de Camargo, M.R.; Russo, E.; Amedei, A. Role of diet and gut microbiota on colorectal cancer immunomodulation. World J. Gastroenterol. 2019, 25, 151–162. [Google Scholar] [CrossRef]
- Oh, N.S.; Lee, J.Y.; Kim, Y. The growth kinetics and metabolic and antioxidant activities of the functional synbiotic combination of Lactobacillus gasseri 505 and Cudrania tricuspidata leaf extract. Appl. Microbiol. Biotechnol. 2016, 100, 10095–10106. [Google Scholar] [CrossRef] [PubMed]
- Oh, N.S.; Lee, J.Y.; Kim, Y.T.; Kim, S.H.; Lee, J.H. Cancer-protective effect of a synbiotic combination between Lactobacillus gasseri 505 and a Cudrania tricuspidata leaf extract on colitis-associated colorectal cancer. Gut Microbes 2020, 12, 1785803. [Google Scholar] [CrossRef] [PubMed]
- Dikeocha, I.J.; Al-Kabsi, A.M.; Hussin, S.; Alshawsh, M.A. Role of probiotics in patients with colorectal cancer: A systematic review protocol of randomised controlled trial studies. BMJ Open 2020, 10, e038128. [Google Scholar] [CrossRef]
- Shrifteylik, A.; Maiolini, M.; Dufault, M.; Austin, D.L.; Subhadra, B.; Lamichhane, P.; Deshmukh, R.R. A Current Review on the Role of Prebiotics in Colorectal Cancer. Biologics 2023, 3, 209–231. [Google Scholar] [CrossRef]
- Xie, W.; Zhong, Y.S.; Li, X.J.; Kang, Y.K.; Peng, Q.Y.; Ying, H.Z. Postbiotics in colorectal cancer: Intervention mechanisms and perspectives. Front. Microbiol. 2024, 15, 1360225. [Google Scholar] [CrossRef]
- Miller, P.L.; Carson, T.L. Mechanisms and microbial influences on CTLA-4 and PD-1-based immunotherapy in the treatment of cancer: A narrative review. Gut Pathog. 2020, 12, 43. [Google Scholar] [CrossRef]
- Serna, G.; Ruiz-Pace, F.; Hernando, J.; Alonso, L.; Fasani, R.; Landolfi, S.; Comas, R.; Jimenez, J.; Elez, E.; Bullman, S.; et al. Fusobacterium nucleatum persistence and risk of recurrence after preoperative treatment in locally advanced rectal cancer. Ann. Oncol. 2020, 31, 1366–1375. [Google Scholar] [CrossRef]
- Kim, K.; Castro, E.J.T.; Shim, H.; Advincula, J.V.G.; Kim, Y.W. Differences Regarding the Molecular Features and Gut Microbiota Between Right and Left Colon Cancer. Ann. Coloproctol. 2018, 34, 280–285. [Google Scholar] [CrossRef]
- Sillo, T.O.; Beggs, A.D.; Middleton, G.; Akingboye, A. The Gut Microbiome, Microsatellite Status and the Response to Immunotherapy in Colorectal Cancer. Int. J. Mol. Sci. 2023, 24, 5767. [Google Scholar] [CrossRef]
- Wong, C.C.; Yu, J. Gut microbiota in colorectal cancer development and therapy. Nat. Rev. Clin. Oncol. 2023, 20, 429–452. [Google Scholar] [CrossRef]
- Qi, X.; Liu, Y.; Hussein, S.; Choi, G.; Kimchi, E.T.; Staveley-O’Carroll, K.F.; Li, G. The Species of Gut Bacteria Associated with Antitumor Immunity in Cancer Therapy. Cells 2022, 11, 3684. [Google Scholar] [CrossRef] [PubMed]
- Gazzaniga, F.S.; Kasper, D.L. The gut microbiome and cancer response to immune checkpoint inhibitors. J. Clin. Investig. 2025, 135. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudian, F.; Gheshlagh, S.R.; Hemati, M.; Farhadi, S.; Eslami, M. The influence of microbiota on the efficacy and toxicity of immunotherapy in cancer treatment. Mol. Biol. Rep. 2024, 52, 86. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.W.; Wang, T.; Liu, Y.Y. Artificial Intelligence for Microbiology and Microbiome Research. arXiv 2024, arXiv:2411.01098. [Google Scholar]
- Wu, H. Charting the microbial frontier: A comprehensive guidebook for advancing microbiome research. FEMS Microbiol. Rev. 2025, 49, fuae033. [Google Scholar] [CrossRef]
- Haraoui, L.P.; Blaser, M.J. The Microbiome and Infectious Diseases. Clin. Infect. Dis. 2023, 77, S441–S446. [Google Scholar] [CrossRef]
- Mohsin Khurshid, M.S.H.A. Human MicrobiomeTechniques, Strategies, and Therapeutic Potential; Springer: Singapore, 2024. [Google Scholar]
- Yadav, M.; Chauhan, N.S. Microbiome therapeutics: Exploring the present scenario and challenges. Gastroenterol. Rep. 2022, 10, goab046. [Google Scholar] [CrossRef]


| General | Species | Effect | Mechanism | Reference |
|---|---|---|---|---|
| Lactobacillus | L. acidophilus | Anti-inflammatory activity |
Can modulate the MAPK signaling pathway and TLR-2-mediated NF-κB activation in inflammatory intestinal epithelial cells (IECs), thereby regulating inflammatory responses.
Reduce the secretion of interleukin-8 (IL-8) and the expression of phosphorylated NF-κB (p-p65), p-p38 MAPK, VCAM-1, and COX-2 in inflammatory conditions, indicating decreased inflammation.
Combining specific probiotics, such as Bifidobacterium lactis, enhances TLR-2 expression in intestinal epithelial cells subjected to LPS and TNF-α, improving immune response against inflammation. | [64] |
| L. plantarum (L. plantarum L125) | Anti-proliferative, anti-clonogenic, and anti-migration activity using cell-free culture supernatant |
Exhibit anti-proliferative effects through cell surface molecules and excreted metabolites, including exopolysaccharides (EPS), peptidoglycans, conjugated linolenic acids (CLA), and S-layer proteins, which have been implicated in inducing cell death. | [65] | |
| L. rhamnosus | Arrest cancer cell growth Apoptosis induction Synergistic action with 5-Fluorouracil and Irinotecan Anti-proliferative activity |
Secretes bioproducts that trigger a reduction in cancer cell viability.
Induces mitotic arrest in the G2/M phase of the cell cycle, preventing cell division.
Exhibits a positive synergistic effect with both 5-Fluorouracil and Irinotecan by sensitizing cancer cells and increasing pro-apoptotic gene expression.
The activation of Bax leads to the intrinsic mitochondrial pathway, promoting apoptosis.
Apoptosis is induced via the activation of caspase-3 and caspase-9, alongside the release of cytochrome c.
Results in a decrease in the expression of anti-apoptotic genes, such as Bcl-2, and genes involved in cell cycle progression, including cyclin D1 and cyclin E, as well as ERBB2, which is associated with cell proliferation. | [66,67] | |
| L. casei ATCC393 | Cell viability reduction Apoptosis induction |
Probiotic treatment modulates the expression of genes associated with cell cycle progression and tumor growth.
Downregulation:Survivin (BIRC5): the downregulation of survivin expression reduces anti-apoptotic activity, thereby promoting apoptosis in cancer cells.
Upregulation:
| [68] | |
| Bifidobacterium | B. lactis | Apoptosis induction |
The combination of specific Bifidobacterium species induces both intrinsic and extrinsic apoptosis pathways, enhancing the apoptotic response in tumor cells.
The presence of certain Bifidobacterium species leads to the inhibition of Nuclear Factor kappa-light-chain-enhancer of activated B cells (NF-κB) activity, which is crucial for regulating inflammatory responses and tumor survival. | [69] |
| B. longum | Anti-inflammatory and cancer prevention effects |
By suppressing the degradation of IκB-α, there is a subsequent decline in its translocation into the nucleus and its DNA binding activity. This results in decreased concentrations of NF-κB and interleukin-6 (IL-6), thereby reducing inflammation and tumor progression.
The inhibition of cyclooxygenase-2 (COX-2), an NF-κB-dependent mediator, leads to the suppression of proliferation and metastasis in colitis-related cancer, thus contributing to a decrease in oncogenic signaling pathways. | [70] | |
| Streptococcus | S. thermophilus | Anti-proliferative protective. CRC cell viability reduction |
The production of β-galactosidases inhibits the Warburg metabolic phenotype and disrupts the Hippo signaling pathway in colorectal cancer (CRC) cells, leading to decreased tumor cell proliferation and survival.
The treatment increases the abundance of beneficial probiotic species, such as Bifidobacterium and Lactobacillus, while simultaneously reducing pathogenic bacteria associated with CRC, thereby promoting a healthier gut microbiota composition. | [71] |
| Faecalibacterium | F. prausnitsii | Anti-inflammatory effect |
The probiotic produces a protein known as the Microbial Anti-inflammatory Molecule (MAM), which exerts inhibitory effects on the NF-κB pathway in various intestinal epithelial cells. This action results in a reduction in colitis in mouse models, highlighting its potential in mitigating intestinal inflammation. | [72] |
| Prebiotic Substance | Source | Effect | Reference |
|---|---|---|---|
| Polyunsaturated fatty acids (PUFAs) | Abundant in fish and fish oil supplements |
Omega-3 polyunsaturated fatty acids (PUFAs) modulate eicosanoid profiles in both circulation and colorectal tumor tissues. This modulation leads to a reduction in ω-6-series metabolites while increasing the levels of ω-3-series metabolites, such as docosahexaenoic acid (DHA, 22:6ω-3) and eicosapentaenoic acid (EPA, 20:5ω-3), which play significant roles in mediating anti-cancer and anti-angiogenic effects.
Eicosapentaenoic acid (EPA) is thought to exert its antineoplastic effects by inhibiting cyclooxygenase-2 (COX-2)-dependent synthesis of prostaglandin E2 (PGE2) and reducing the de novo production of PGE3, thereby contributing to the suppression of tumor growth. | [79,80] |
| Polyphenols | Jaboticaba (Myrciaria jaboticaba) seed extract (LJE) |
Phenolic compounds, including castalagin, vescalagin, procyanidin A2, and the ellagic acid found in LJE, exhibit pro-oxidant and cytotoxic effects against cancer cells, contributing to the inhibition of tumor cell proliferation.
These phenolic compounds also demonstrate antioxidant properties by reducing the generation of reactive oxygen species (ROS) in normal human cell lines, such as IMR90, thereby protecting healthy cells from oxidative stress. | [81] |
| Inulin | Garlic, onion, artichoke, asparagus, and chicory |
There is an increase in well-known propionate-producing bacterial species from the
Bacteroidaceae and Prevotellaceae
families, which are beneficial for gut health and linked to anti-inflammatory effects.
A reduction in Firmicutes, particularly Ruminococcus gnavus, has been observed, which corresponds to a decrease in inflammatory conditions associated with gut dysbiosis.
An increase in the Faecalibacterium genus, recognized for its butyrate-producing capabilities, is noted. This genus exhibits anti-inflammatory effects primarily through the inhibition of NF-κB signaling, contributing to improved gut homeostasis. | [82] |
| Substance | Example | Source | Effect | Reference |
|---|---|---|---|---|
| Inactivated microbial cells | Heat-killed LGG | Lactobacillus rhamnosus GG |
Substances lead to an increase in anti-inflammatory mediators, promoting a more balanced immune response and reducing overall inflammation.
These substances also decrease the inflammatory response induced by
E. coli
lipopolysaccharides (LPS), thereby mitigating the effects of pathogenic stimuli and contributing to intestinal health. | [86] |
| Cell components | Amuc_1100 | A protein from the outer membrane of Akkermansia muciniphila |
These substances induce an anti-tumor immune response, enhancing the body’s ability to combat cancer cell growth.
They also improve colitis by modulating cytotoxic T lymphocyte (CTL) activity, resulting in increased CTL concentrations in both the colon and mesenteric lymph nodes. This modulation upregulates CTL activity and tumor necrosis factor-alpha (TNF-α), leading to the apoptosis of tumor cells. | [87] |
| Lipoteichoic acid (LTA) | Cell wall component of Lactobacillus paracasei D3-5 |
These substances reduce age-related leaky gut and associated inflammation by modulating the TLR-2/p38-MAPK/NF-κB signaling pathway, leading to the increased expression of mucin (Muc2), which enhances the intestinal barrier function. | [88] | |
| Metabolites secreted by gut microbiota. | SCFAs including acetate, propionate and butyrate; enzymes; bacteriocins; reuterin; acetoin; organic acids | Members of gut microbiota example: Faecalibacterium prausnitzii |
Butyrate blocks the motility-dependent activation of histone deacetylase 3 (HDAC3)-dependent signaling pathways, resulting in the inhibition of migration and the invasion of colorectal cancer (CRC) cells.
Both butyrate and propionate trigger an antitumor immune response by activating a positive feedback loop that upregulates genes involved in immune activation. This process promotes the activation of cytotoxic T lymphocytes (CD8+) and enhances the secretion of interferon-gamma (IFN-γ). | [89,90] |
| Cell-free supernatants (CFS) | Exopolysaccharides (EPS), peptidoglycans and conjugated linolenic acids (CLA) | L. plantarum (L. plantarum L125) |
Cell death is induced through interactions with cell surface molecules and/or the action of excreted metabolites, including exopolysaccharides (EPS), peptidoglycans, conjugated linolenic acids (CLA), and S-layer proteins, which collectively promote apoptotic pathways in target cells. | [65] |
| Category | Details | Effect | Reference |
|---|---|---|---|
| Microbial Impacts | Short-Chain Fatty Acids (SCFAs) | Produced by fermentation of dietary fibers; anti-inflammatory and anti-carcinogenic properties. | [55] |
| Secondary Bile Acids | Produced by gut bacteria from primary bile acids; can induce DNA damage and promote carcinogenesis. | [114] | |
| Therapeutic Targets | Bacteroides fragilis | Enhances efficacy of immune checkpoint inhibitors (ICIs) by promoting T-cell activation. | [115] |
| Akkermansia muciniphila | Associated with increased infiltration of CD8+ T cells into tumors. | [116] | |
| Bacteroides thetaiotaomicron | Can inhibit effectiveness of ICIs by inducing immunosuppressive environments. | [116] | |
| AI Applications | DeepMicro | Utilizes high-throughput sequencing data to detect patterns, classify microbes, and predict disease associations. | [117] |
| Meta-Spec | Integrates host and microbial information to map microbiome patterns related to diseases. | [118] | |
| MicroPheno | Predicts phenotypic traits from microbiome data, aiding in the identification of disease-associated microbial features. | [118] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zalila-Kolsi, I.; Dhieb, D.; Osman, H.A.; Mekideche, H. The Gut Microbiota and Colorectal Cancer: Understanding the Link and Exploring Therapeutic Interventions. Biology 2025, 14, 251. https://doi.org/10.3390/biology14030251
Zalila-Kolsi I, Dhieb D, Osman HA, Mekideche H. The Gut Microbiota and Colorectal Cancer: Understanding the Link and Exploring Therapeutic Interventions. Biology. 2025; 14(3):251. https://doi.org/10.3390/biology14030251
Chicago/Turabian StyleZalila-Kolsi, Imen, Dhoha Dhieb, Hussam A. Osman, and Hadjer Mekideche. 2025. "The Gut Microbiota and Colorectal Cancer: Understanding the Link and Exploring Therapeutic Interventions" Biology 14, no. 3: 251. https://doi.org/10.3390/biology14030251
APA StyleZalila-Kolsi, I., Dhieb, D., Osman, H. A., & Mekideche, H. (2025). The Gut Microbiota and Colorectal Cancer: Understanding the Link and Exploring Therapeutic Interventions. Biology, 14(3), 251. https://doi.org/10.3390/biology14030251







