Early Detection of the Pathogenetic Variants of Homologous Recombination Repair Genes in Prostate Cancer: Critical Analysis and Experimental Design
Simple Summary
Abstract
1. Introduction
2. DDR Analysis: Prognostic Role in Different Stages of PCa
2.1. Incidence
2.2. Role in Non-Metastatic PCa
2.3. Role in Metastatic PC
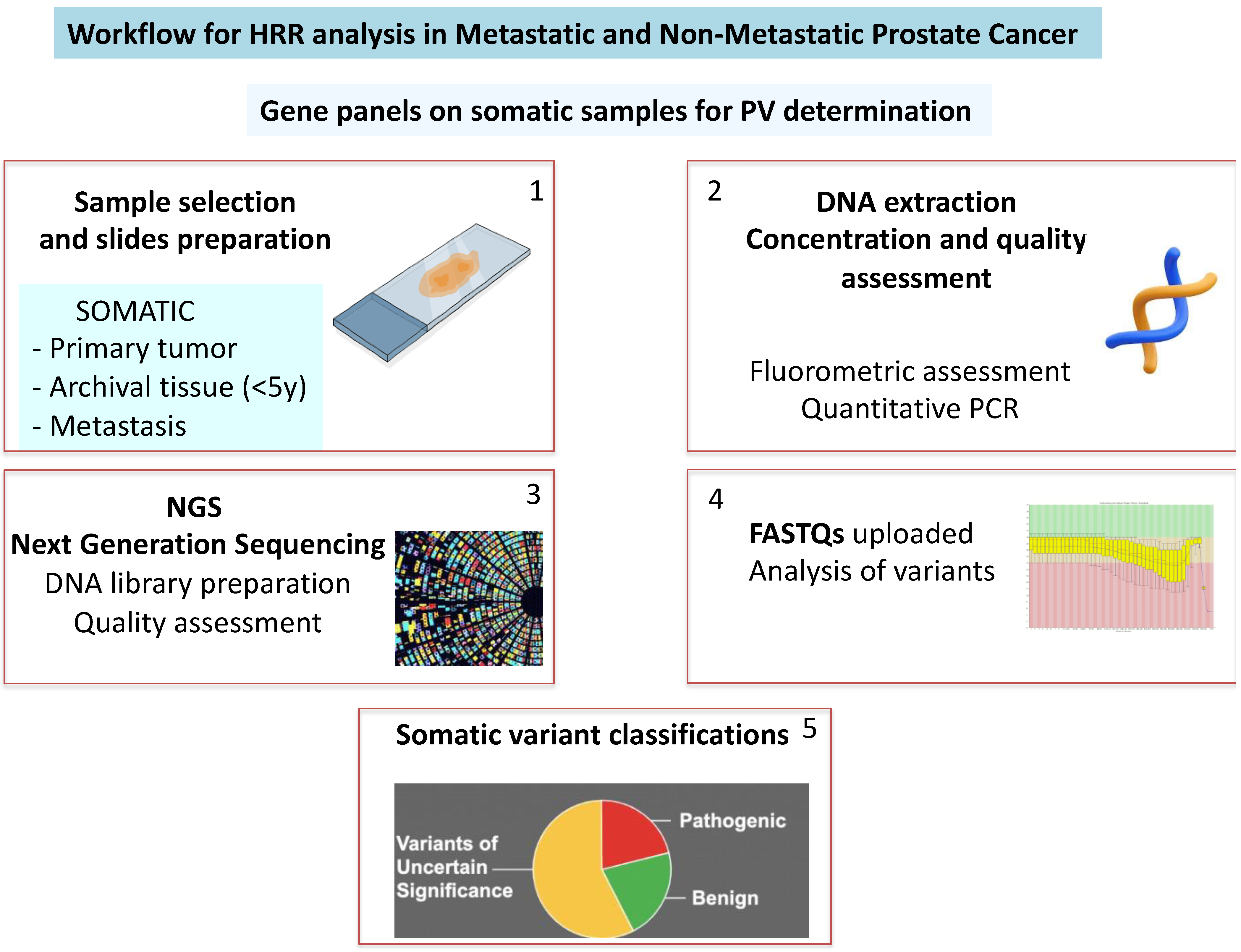
3. How to Detect
3.1. Somatic Samples: Recent vs. Archived
3.2. Germline Analysis in PC
- Benign variants (PP < 0.001);
- Likely benign or of limited clinical significance (PP: 0.001–0.049);
- Uncertain significance (PP: 0.05–0.949);
- Likely pathogenic (PP: 0.95–0.99);
- Pathogenic (PP > 0.99).
3.3. Circulating DNA (cDNA)
4. Purpose for Experimental Design: Personal Experience
4.1. Methods
4.1.1. Urologic Evaluation
4.1.2. Pathologic Preparation
4.1.3. Genetic Analysis
4.2. Findings
| Patient | Age (Years) | Familiarity | Aggressiveness | Early Progression | TNM Staging System | Years from Sample Collection | Slice | Cellularity | Concentration of the Extracted DNA (ng/uL) | Quality of the Extracted DNA | NGS Library Concentration (nMol) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| (12 Months) | |||||||||||
| 1 | 58 | no | ISUP 4 | yes | M1 | 1 | A | 60% | 3.9 | Low | Not performed |
| B | 60% | 6.4 | Low | Not performed | |||||||
| 7 | 69 | no | ISUP 3 | yes | M1 | 1 | A | 70% | 10.6 | Medium | Low |
| B | 60% | 11.9 | Medium | Low | |||||||
| 9 | 65 | no | ISUP 4 | yes | M1 | 0 | A | 40% | 11.5 | Medium | Low |
| 10 | 78 | no | ISUP 5 | yes | M1 | 0 | A | 80% | 6.2 | Medium | Low |
| 11 | 67 | no | ISUP 3 | yes | M0 | 1 | A | 40% | 7.8 | Medium | Low |
| B | 50% | 90.0 | Low | Not performed | |||||||
| C | 50% | 4.0 | Low | Not performed | |||||||
| 12 | 54 | no | ISUP 3 | no | M0 | 4 | A | 30% | 3.9 | Low | Not performed |
| B | 20% | 5.5 | Medium | Low | |||||||
| 15 | 66 | no | ISUP 3 | yes | M0 | 2 | A | 40% | 75.6 | Medium | Low |
| B | 60% | 6.9 | Medium | Low | |||||||
| 17 | 66 | yes (1 brother) | ISUP 3 | yes | M0 | 1 | A | 20% | 4.5 | Low | Not performed |
| B | 40% | 3.8 | Low | Not performed | |||||||
| 18 | 78 | no | ISUP 4 | yes | M1 | 0 | A | 80% | 4.2 | Low | Not performed |
| 22 | 76 | no | ISUP 4 | yes | M1 | 1 | A | 40% | 1.8 | Low | Not performed |
| B | 50% | 0.2 | Low | Not performed |
4.3. Discussion
5. Conclusions
- The incidence of pathogenic variants (PVs) in HRR genes among men with metastatic PCa ranges from 11% to 33%, which is notably higher than in non-metastatic prostate cancer (nmPC). Within the metastatic setting, BRCA2 mutations are more prevalent compared to other HRR gene mutations.
- Identifying somatic or germline HRR PVs, particularly BRCA2 mutations, plays a crucial role in personalizing treatment with PARP inhibitors in metastatic castration-resistant prostate cancer (mCRPC). This approach has shown significant improvements in radiographic progression-free survival (rPFS) and overall survival (OS). As a result, this strategy has been recommended by international guidelines and has received approval from both the FDA and EMA.
- -
- To better define clinical and pathological characteristics of newly diagnosed prostate cancer associated with DDR gene defects.
- -
- To offer a platform for approaching personalized medicine based on the genetic assessment of PVs in DDR genes in a non-metastatic stage.
- -
- To define whether the expression of PVs of DDR genes is also relevant in non-metastatic prostate cancer at high risk.
- -
- To define whether BRCA2 remains the main PV expressed also in non-metastatic prostate cancer cases or other PVs for different DDR genes are similarly expressed and useful.
- -
- To simplify the detection of PVs of DDR genes, exploring not only the somatic but also other approaches.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Castro, E.; Romero-Laorden, N.; Del Pozo, A.; Lozano, R.; Medina, A.; Puente, J.; Piulats, J.M.; Lorente, D.; Saez, M.I.; Morales-Barrera, R.; et al. PROREPAIR-B: A Prospective Cohort Study of the Impact of Germline DNA Repair Mutations on the Outcomes of Patients With Metastatic Castration-Resistant Prostate Cancer. J. Clin. Oncol. 2019, 37, 490–503. [Google Scholar] [CrossRef] [PubMed]
- Sciarra, A.; Frisenda, M.; Bevilacqua, G.; Gentilucci, A.; Cattarino, S.; Mariotti, G.; Del Giudice, F.; Di Pierro, G.B.; Viscuso, P.; Casale, P.; et al. How the Analysis of the Pathogenetic Variants of DDR Genes Will Change the Management of Prostate Cancer Patients. Int. J. Mol. Sci. 2023, 24, 674. [Google Scholar] [CrossRef]
- Sciarra, A.; Santarelli, V.; Santodirocco, L.; Frisenda, M.; Salciccia, S.; Casale, P.; Forte, F.; Mariotti, G.; Moriconi, M.; Cattarino, S.; et al. Is It Time to Anticipate the Use of PARP Inhibition in ProstateCancer Patients? Curr. Oncol. 2023, 30, 8054–8067. [Google Scholar] [CrossRef]
- Parsons, M.T.; Tudini, E.; Li, H.; Hahnen, E. Large scale multifactorial likelihood quantitative analysis of BRCA1 and BRCA2 variants: An ENIGMA resource to support clinical variant classification. Hum. Mutat. 2019, 40, 1557–1578. [Google Scholar] [CrossRef]
- Caldecott, K.W. Mammalian single-strand break repair: Mechanisms and links with chromatin. DNA Repair 2007, 6, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Rouleau, M.; Patel, A.; Hendzel, M. PARP inhibition: PARP1 and beyond. Nat. Rev. Cancer 2010, 10, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Castro, E.; Goh, C.; Olmos, D. Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J. Clin. Oncol. 2013, 31, 1748–1757. [Google Scholar] [CrossRef]
- European Urological Association (EAU) guidelines Prostate Cancer. 2024. Available online: https://uroweb.org/guidelines/prostate-cancer (accessed on 10 April 2024).
- Hussain, M.; Corcoran, C.; Sibilla, C.; Fizazi, K.; Saad, F.; Shore, N.; Sandhu, S.; Mateo, J.; Olmos, D.; Mehra, N.; et al. Tumor Genomic Testing for >4000 Men with Metastatic Castration-resistant Prostate Cancer in the Phase III Trial PROfound (Olaparib). Clin. Cancer Res. 2022, 28, 1518–1530. [Google Scholar] [CrossRef]
- Wei, Y.; Wu, J.; Gu, W.; Qin, X.; Dai, B.; Lin, G.; Gan, H.; Freedland, S.J.; Zhu, Y.; Ye, D. Germline DNA Repair Gene Mutation Landscape in Chinese Prostate Cancer Patients. Eur. Urol. 2019, 76, 280–283. [Google Scholar] [CrossRef]
- Pritchard, C.C.; Mateo, J.; Walsh, M.F.; De Sarkar, N.; Abida, W.; Beltran, H.; Garofalo, A.; Gulati, R.; Carreira, S.; Eeles, R.; et al. Inherited DNA-Repair Gene Mutations in Men with Metastatic Prostate Cancer. N. Engl. J. Med. 2016, 375, 443–453. [Google Scholar] [CrossRef]
- ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 2020, 578, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Page, E.C.; Bancroft, E.K.; Brook, M.N.; Assel, M.; Hassan Al Battat, M.; Thomas, S.; Taylor, N.; Chamberlain, A.; Pope, J.; Raghallaigh, H.N.; et al. Interim Results from the IMPACT Study: Evidence for Prostate-specific Antigen Screening in BRCA2 Mutation Carriers. Eur. Urol. 2019, 76, 831–842. [Google Scholar] [CrossRef]
- Chen, R.C.; Rumble, R.B.; Loblaw, D.A. Active surveillance for the management of localized prostate cancer (Cancer Care Ontario Guideline): American Society of Clinical Oncology Clinical Practice Guideline Endorsement. J. Clin. Oncol. 2016, 34, 2182–2190. [Google Scholar] [CrossRef] [PubMed]
- Carter, H.B.; Helfand, B.; Mamawala, M. Germline Mutations in ATM and BRCA1/2 Are Associated with Grade Reclassification in Men on Active Surveillance for Prostate Cancer. Eur. Urol. 2019, 75, 743–749. [Google Scholar] [CrossRef] [PubMed]
- Castro, E.; Goh, C.; Leongamornlert, D. Effect of BRCA Mutations on Metastatic Relapse and Cause-specific Survival After Radical Treatment for Localised Prostate Cancer. Eur. Urol. 2015, 68, 186–193. [Google Scholar] [CrossRef]
- Martinez Chanza, N.; Bernard, B.; Barthelemy, P.; Accarain, A.; Paesmans, M.; Desmyter, L.; T’Kint de Roodenbeke, D.; Gil, T.; Sideris, S.; Roumeguere, T.; et al. Prevalence and clinical impact of tumor BRCA1 and BRCA2 mutations in patients presenting with localized or metastatic hormone-sensitive prostate cancer. Prostate Cancer Prostatic Dis. 2022, 25, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Marshall, C.H.; Fu, W.; Wang, H.; Baras, A.S.; Lotan, T.L.; Antonarakis, E.S. Prevalence of DNA repair gene mutations in localized prostate cancer according to clinical and pathologic features: Association of Gleason score and tumor stage. Prostate Cancer Prostatic Dis. 2019, 22, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Lang, S.H.; Swift, S.L.; White, H.; Misso, K.; Kleijnen, J.; Quek, R.G.W. A systematic review of the prevalence of DNA damage response gene mutations in prostate cancer. Int. J. Oncol. 2019, 55, 597–616. [Google Scholar] [CrossRef]
- Kim, I.E., Jr.; Kim, S.; Srivastava, A. Similar incidence of DNA damage response pathway alterations between clinically localized and metastatic prostate cancer. BMC Urol. 2019, 19, 33. [Google Scholar] [CrossRef] [PubMed]
- Annala, M.; Struss, W.J.; Warner, E.W.; Beja, K.; Vandekerkhove, G.; Wong, A.; Khalaf, D.; Seppälä, I.L.; So, A.; Lo, G.; et al. Treatment Outcomes and Tumor Loss of Heterozygosity in Germline DNA Repair-deficient Prostate Cancer. Eur. Urol. 2017, 72, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Antonarakis, E.S.; Lu, C.; Luber, B.; Liang, C.; Wang, H.; Chen, Y.; Silberstein, J.L.; Piana, D.; Lai, Z.; Chen, Y.; et al. Germline DNA-repair Gene Mutations and Outcomes in Men with Metastatic Castration-resistant Prostate Cancer Receiving First-line Abiraterone and Enzalutamide. Eur. Urol. 2018, 74, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Carr, T.H.; Adelman, C.; Barnicle, A.; Kozarewa, I.; Luke, S.; Lai, Z.; Hollis, S.; Dougherty, B.; Harrington, E.A.; Kang, J.; et al. Homologous Recombination Repair Gene Mutation Characterization by Liquid Biopsy: A Phase II Trial of Olaparib and Abiraterone in Metastatic Castrate-Resistant Prostate Cancer. Cancers 2021, 13, 5830. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.R.; Scher, H.I.; Sandhu, S.; Efstathiou, E.; Lara, P.N., Jr.; Yu, E.Y.; George, D.J.; Chi, K.N.; Saad, F.; Ståhl, O.; et al. Niraparib in patients with metastatic castration-resistant prostate cancer and DNA repair gene defects (GALAHAD): A multicentre, open-label, phase 2 trial. Lancet Oncol. 2022, 23, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Torga, G.; Pienta, K.J. Patient-Paired Sample Congruence Between 2 Commercial Liquid Biopsy Tests. JAMA Oncol. 2018, 4, 868–870. [Google Scholar] [CrossRef]
- Li, M.M.; Datto, M.; Duncavage, E.J.; Kulkarni, S.; Lindeman, N.I.; Roy, S.; Tsimberidou, A.M.; Vnencak-Jones, C.L.; Wolff, D.J.; Younes, A.; et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 2017, 19, 4–23. [Google Scholar] [CrossRef] [PubMed]
- Horak, P.; Griffith, M.; Danos, A.M.; Pitel, B.A.; Madhavan, S.; Liu, X.; Chow, C.; Williams, H.; Carmody, L.; Barrow-Laing, L.; et al. Standards for the classification of pathogenicity of somatic variants in cancer (oncogenicity): Joint recommendations of Clinical Genome Resource (ClinGen), Cancer Genomics Consortium (CGC), and Variant Interpretation for Cancer Consortium (VICC). Genet Med. 2022, 24, 986–998. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Zhang, B.; Ahrenfeldt, J.; Joseph, J.V.; Riedel, M.; Gao, Z.; Thomsen, S.K.; Christensen, D.S.; Bak, R.O.; Hager, H.; et al. CRISPR/Cas9 model of prostate cancer identifies Kmt2c deficiency as a metastatic driver by Odam/Cabs1 gene cluster expression. Nat. Commun. 2024, 15, 2088. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Garcia-Bassets, I.; Benner, C.; Li, W.; Su, X.; Zhou, Y.; Qiu, J.; Liu, W.; Kaikkonen, M.U.; Ohgi, K.A.; et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 2011, 474, 390–394. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Park, S.H.; Zhao, J.C.; Fong, K.W.; Li, S.; Lee, Y.; Yang, Y.A.; Sridhar, S.; Lu, X.; Abdulkadir, S.A.; et al. Targeting FOXA1-mediated repression of TGF-β signaling suppresses castration-resistant prostate cancer progression. J. Clin. Investig. 2019, 129, 569–582. [Google Scholar] [CrossRef]
- Grasso, C.S.; Wu, Y.-M.; Robinson, D.R.; Cao, X.; Dhanasekaran, S.M.; Khan, A.P. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012, 487, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Burkhart, D.L.; Sage, J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat. Rev. Cancer 2008, 8, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Zhu, Y.; Xiao, C.; Li, R.; Cao, X.; Kang, R.; Wang, X.; Li, E. Novel insights into RB1 in prostate cancer lineage plasticity and drug resistance. Tumori J. 2024, 110, 252–263. [Google Scholar] [CrossRef]
- Zhang, J.; Ye, Y.; Xu, Z.; Luo, M.; Wu, C. Histone methyltransferase KMT2D promotes prostate cancer progression through paracrine IL-6 signaling. Biochem. Biophys. Res. Commun. 2023, 655, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Grochot, R.; Carreira, S.; Miranda, S.; Figueiredo, I.; Claudia, B. Germline ATM mutations detected by somatic DNA sequencing in Lethal Prostate cancer. Eur. Urol. Open Sci. 2023, 52, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Jamaspishvilli, T.; Berman, D.M.; Ross, A.E.; Sher, H.I.; De Marzo, A.M.; Squire, J.A.; Lotan, T.L. Clinical implications of PTEN loss in prostate cancer. Nat. Rev. Urol. 2018, 15, 222–234. [Google Scholar] [CrossRef]
| Patients | Age (Years) | Familiarity | Aggressiveness | Early Progression | TNM Staging System | Years from Sample Collection | Slice | Cellularity | Concentration of the Extracted DNA (ng/ul) | Quality of the Extracted DNA | NGS Libraries’ Concentration (nMol) | Genetic Results | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (12 Months) | Gene | Variant | Classification | Variant Allele Frequency (%) | Microsatellites Instability | |||||||||||
| 2 | 65 | no | ISUP 4 | yes | M1 | 1 | A | 80% | 12.0 | Medium | Low | NGS failed | ||||
| B | 70% | 93.6 | Medium | 46.5 | DDR2 | NM_006182.4:c.106A > C, p.Met36Leu | TIER III | 52.0% | no | |||||||
| 3 | 59 | no | ISUP 3 | no | M0 | 1 | A | 40% | 61.0 | Medium | 91.3 | ATM | NM_000051.4:c.3044_3045delinsCCTT, p.Gln1015Profs | TIER I | 47.0% | no |
| B | 50% | 52.4 | Medium | 128.5 | ATM | NM_000051.4:c.3044_3045delinsCCTT, p.Gln1015Profs | TIER I | 45.90% | no | |||||||
| 4 | 66 | no | ISUP 3 | yes | M0 | 2 | A | 30% | 58.8 | Medium | 81.3 | wild type | no | |||
| 5 | 78 | no | ISUP 3 | yes | M0 | 4 | A | 30% | 5.54 | Medium | Low | NGS failed | ||||
| B | 30% | 7.36 | Medium | 36.3 | wild type | no | ||||||||||
| 6 | 68 | yes (1 brother) | ISUP3 | yes | M0 | 1 | A | 30% | 27.4 | Low | n.p. | NGS n.p. | ||||
| B | 60% | 68.4 | Medium | 87.2 | RB1 BRCA1 KMT2D | NM_000321.3:c.1976A > G, p.Tyr659Cys NM_007294.4:c.5150del, p.Phe1717SerfsNM_003482.4:c.9979C > T, p.Gln3327Ter | TIER III TIER I TIER II | 50.1% 46.9% 2.8% | no | |||||||
| 8 | 68 | no | ISUP 5 | yes | M0 | 1 | A | 35% | 77.8 | Medium | 65.4 | wild type | no | |||
| B | 40% | 93.8 | Medium | 139.6 | ATM FOXA1 | NM_000051.4:c.2272_2301del, p.Glu758_Thr767del NM_004496.5:c.787del, p.Gln263ArgfsTer58 | TIER III TIER III | 6.4% 6.2% | no | |||||||
| C | 40% | 6.83 | Medium | Low | NGS failed | |||||||||||
| 13 | 66 | no | ISUP 3 | no | M0 | 2 | A | 30% | 23.0 | Medium | Low | NGS failed | ||||
| B | 40% | 27.8 | Medium | 29.7 | wild type | no | ||||||||||
| 14 | 57 | no | ISUP 5 | yes | M0 | 0 | A | 70% | 54.8 | Medium | 29.2 | ATM | NM_000051.4:c.2093C > G, p.Ser698Ter | TIER I | 55.0% | no |
| B | 80% | 118 | Medium | Low | NGS failed | |||||||||||
| C | 70% | 76.6 | Medium | Low | NGS failed | |||||||||||
| 16 | 57 | no | ISUP 4 | no | M0 | 2 | A | 80% | 38.4 | Medium | Low | NGS failed | ||||
| B | 80% | 93.6 | Medium | 36.4 | wild type | no | ||||||||||
| 19 | 64 | no | ISUP 4 | no | M0 | 0 | A | 75% | 7.98 | Medium | 46.9 | wild type | no | |||
| B | 60% | 3.92 | Medium | 28.6 | wild type | no | ||||||||||
| 20 | 65 | no | ISUP 5 | yes | M1 | 1 | A | 70% | 13.5 | Medium | 93.2 | CDH1 FANCA PLCB4 | NM_004360.5:c.1912dup, p.Trp638Leufs NM_000135.4:c.11C > G, p.Ser4Trp NM_000933.4:c.713C > T, p.Thr238Met | TIER II TIER III TIER III | 12.4% 46.0% 43.2% | no |
| B | 70% | 7.14 | Medium | 76.5 | CDH1 FANCA PLCB4 | NM_004360.5:c.1912dup, p.Trp638Leufs NM_000135.4:c.11C > G, p.Ser4Trp NM_000933.4:c.713C > T, p.Thr238Met | TIER II TIER III TIER III | 10.5% 44.6% 42.9% | no | |||||||
| 21 | 76 | yes (1 brother) | ISUP 3 | yes | M0 | 1 | A | 50% | 11.4 | Medium | 44.6 | FOXA1 PTCH1 | NM_004496.5:c.806A > T, p.Glu269Val NM_000264.5:c.2492A > G, p.Tyr831Cys | TIER III TIER III | 50.5% 53.5% | no |
| B | 40% | 11.9 | Medium | 63.7 | FOXA1 PTCH1 PTEN | NM_004496.5:c.806A > T, p.Glu269Val NM_000264.5:c.2492A > G, p.Tyr831Cys NM_000314.8:c.955_958del, p.Thr319Ter | TIER III TIER III TIER I | 45.4% 46.2% 4.8% | no | |||||||
| Pathogenetic Variants DDR Genes | Definition | Control Activity | Frequency in Prostate Cancer from the Literature |
|---|---|---|---|
| FOXA1 [31] | Forkhead box A1 gene | Partner of androgen and estrogen receptors | 3–5% |
| RB1 [32] | Retinoblastoma gene1 | Tumor suppressor gene encoding protein pRb | 5–20% |
| KMT2D [34] | Histone-lysine N-methyltransferase 2D gene | Oncogene influencing tumor cell proliferation. Possible association with Interleukine-6 and microenvironment activity | 5–8% |
| ATM [35] | Ataxia telengiectasia mutated gene | Tumor suppressor gene encoding PI3K-related serine/threonine protein kinase. Identification of DNA damage | 1.5–7% |
| PTEN [36] | Phosphatase and tensin homolog gene | Tumor suppressor gene related to the PI3K-AKT pathway | 20–50% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bottillo, I.; Sciarra, A.; Bevilacqua, G.; Gentilucci, A.; Sciarra, B.; Santarelli, V.; Salciccia, S.; Bacigalupo, F.; Pastacaldi, F.; Ciccone, M.P.; et al. Early Detection of the Pathogenetic Variants of Homologous Recombination Repair Genes in Prostate Cancer: Critical Analysis and Experimental Design. Biology 2025, 14, 117. https://doi.org/10.3390/biology14020117
Bottillo I, Sciarra A, Bevilacqua G, Gentilucci A, Sciarra B, Santarelli V, Salciccia S, Bacigalupo F, Pastacaldi F, Ciccone MP, et al. Early Detection of the Pathogenetic Variants of Homologous Recombination Repair Genes in Prostate Cancer: Critical Analysis and Experimental Design. Biology. 2025; 14(2):117. https://doi.org/10.3390/biology14020117
Chicago/Turabian StyleBottillo, Irene, Alessandro Sciarra, Giulio Bevilacqua, Alessandro Gentilucci, Beatrice Sciarra, Valerio Santarelli, Stefano Salciccia, Francesca Bacigalupo, Francesco Pastacaldi, Maria Pia Ciccone, and et al. 2025. "Early Detection of the Pathogenetic Variants of Homologous Recombination Repair Genes in Prostate Cancer: Critical Analysis and Experimental Design" Biology 14, no. 2: 117. https://doi.org/10.3390/biology14020117
APA StyleBottillo, I., Sciarra, A., Bevilacqua, G., Gentilucci, A., Sciarra, B., Santarelli, V., Salciccia, S., Bacigalupo, F., Pastacaldi, F., Ciccone, M. P., De Marchis, L., Santini, D., Magliocca, F. M., Merenda, E., Forte, F., & Grammatico, P. (2025). Early Detection of the Pathogenetic Variants of Homologous Recombination Repair Genes in Prostate Cancer: Critical Analysis and Experimental Design. Biology, 14(2), 117. https://doi.org/10.3390/biology14020117








