Identification of DNA Methyltransferase/Demethylase Genes and 5-Azacytidine’s Impact on β-Elemene and Methylation in Curcuma wenyujin
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Sequence Analysis of CwC5-MTases and CwdMTases
2.2. Phylogenetic Tree Construction and Conserved Motifs Analysis
2.3. Plant Material and Stress Treatment
2.4. RNA Isolation, RT-qPCR Analysis and Determination of β-Elemene Content
2.4.1. RNA Extraction and Gene Expression Analysis
2.4.2. β-Elemene Content Determination
2.5. Methylation-Sensitive Amplified Polymorphism (MSAP) Analysis
2.5.1. DNA Isolation
2.5.2. MSAP Assay
2.5.3. Band Scoring
3. Results
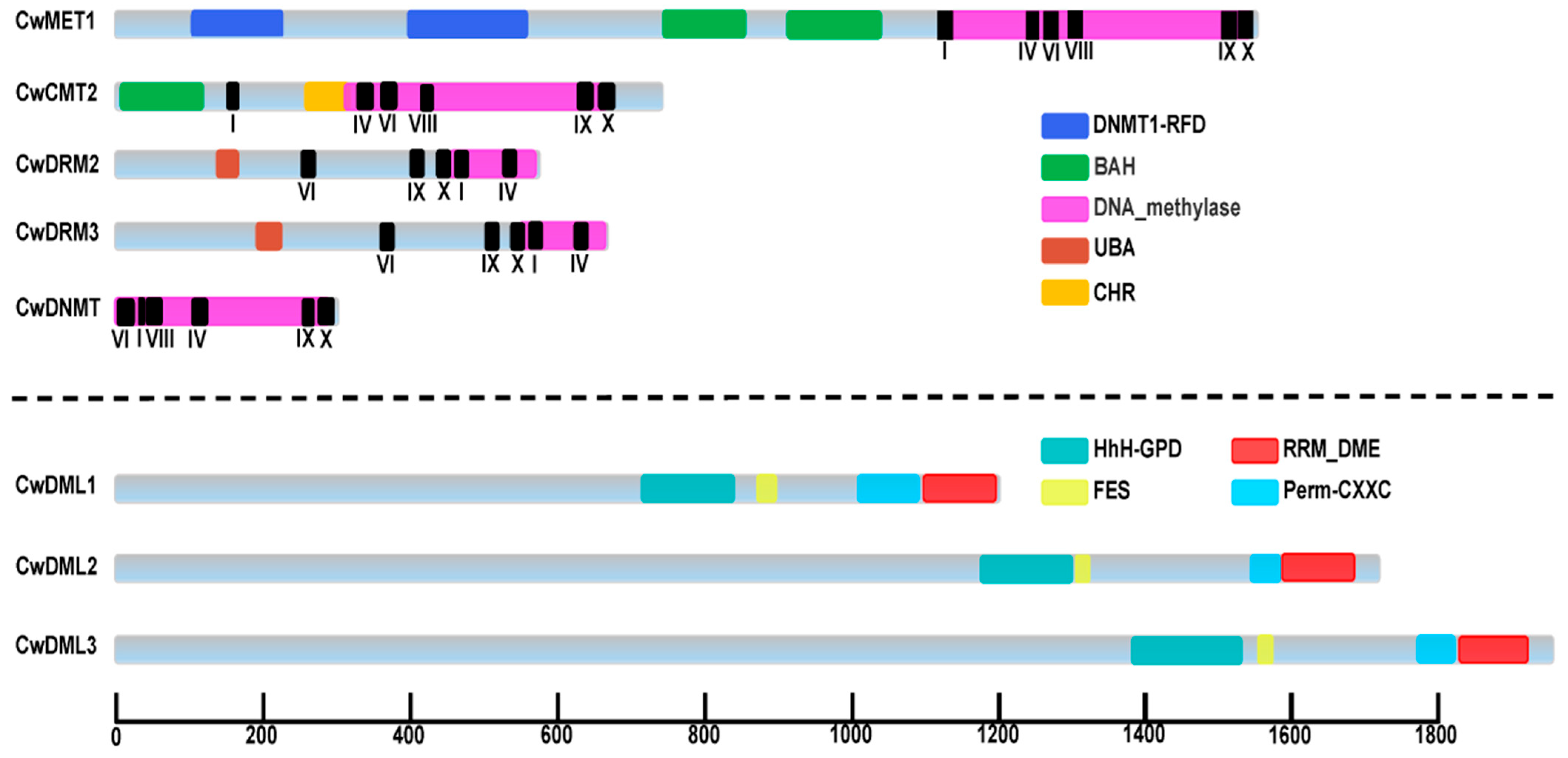
3.1. Identification and Classification of CwC5-MTase and CwdMTase Genes in C. wenyujin
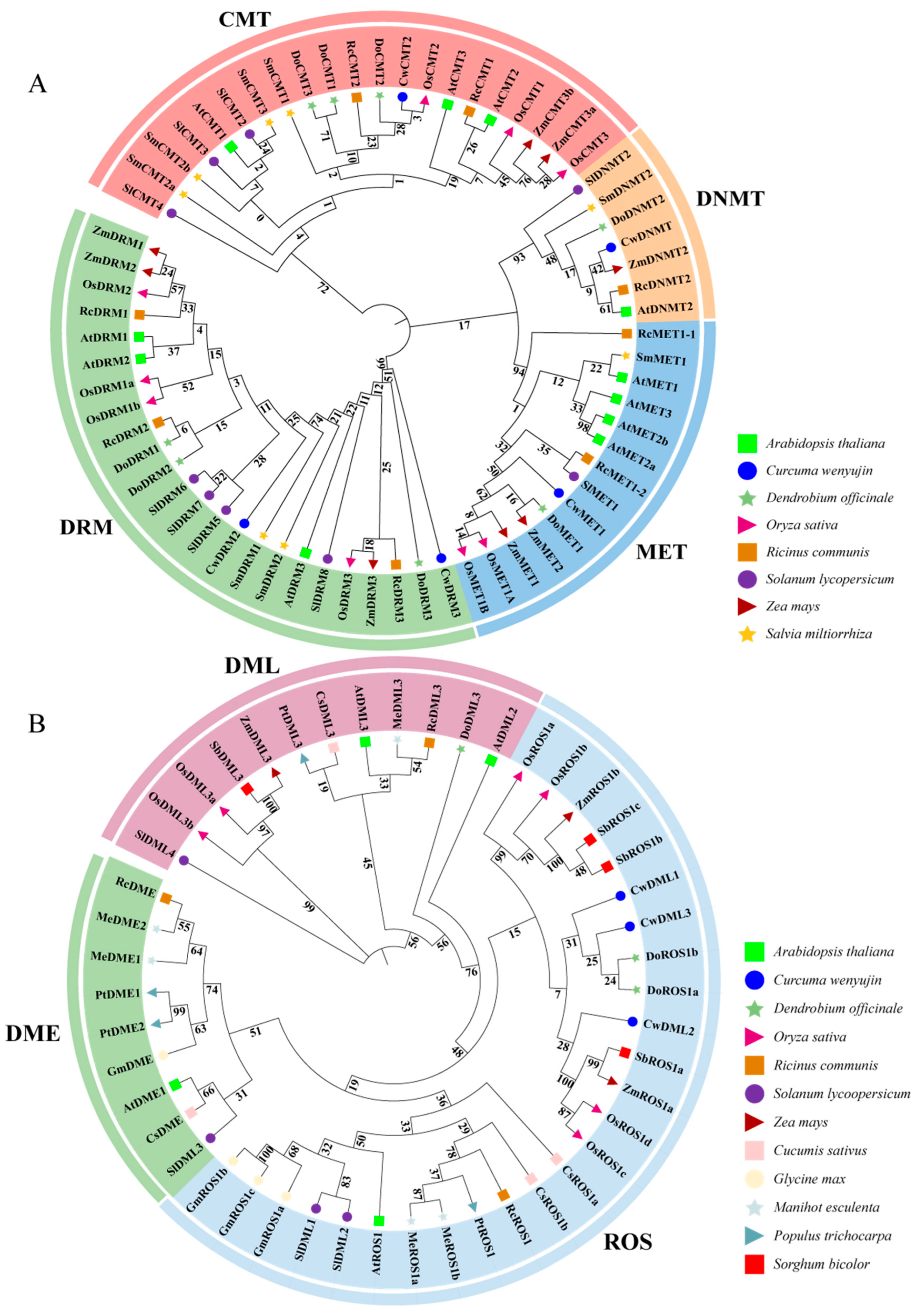
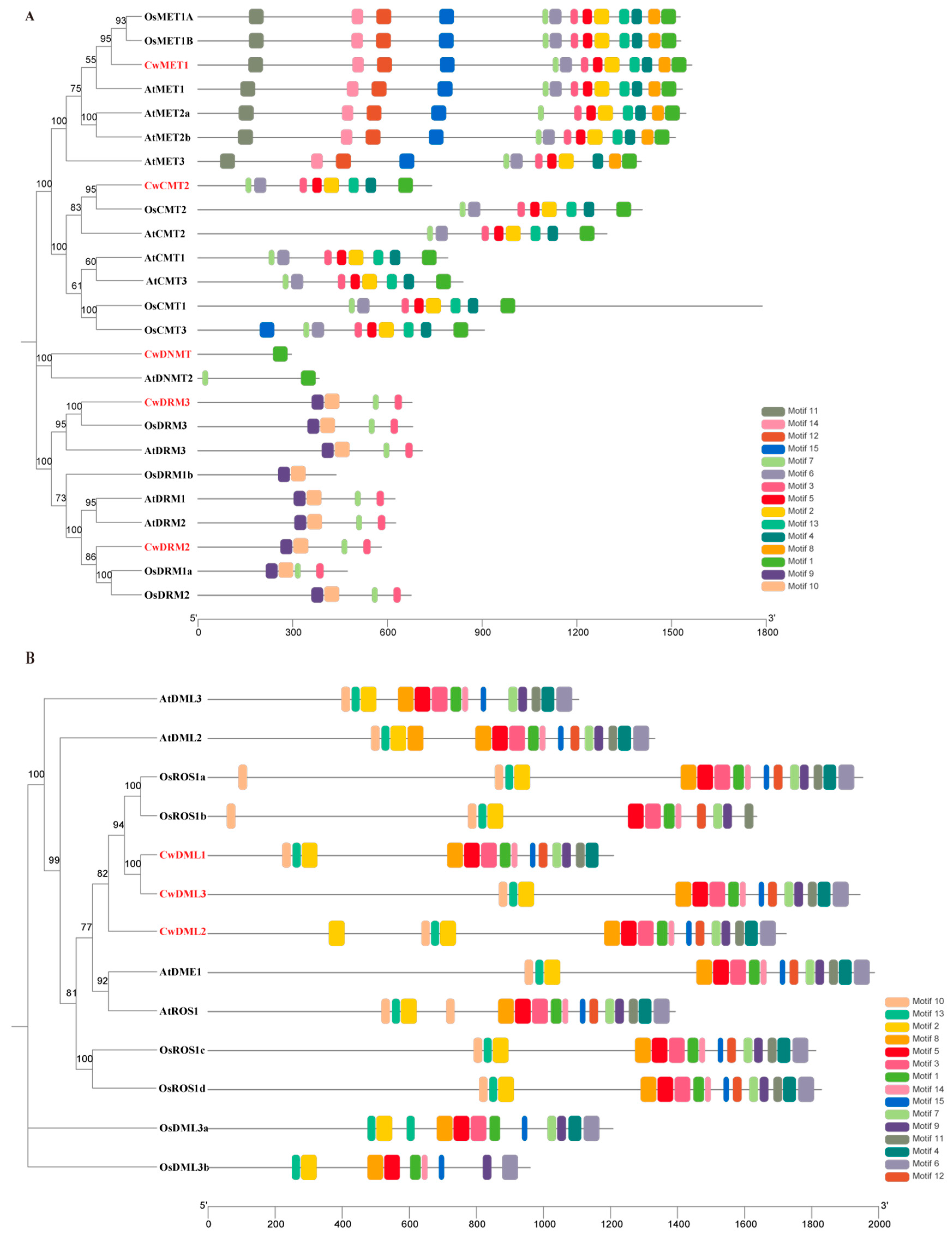
3.2. Phylogenetic and Conserved Motifs Analysis of C5-MTases and dMTases in C. wenyujin and Other Plant Species
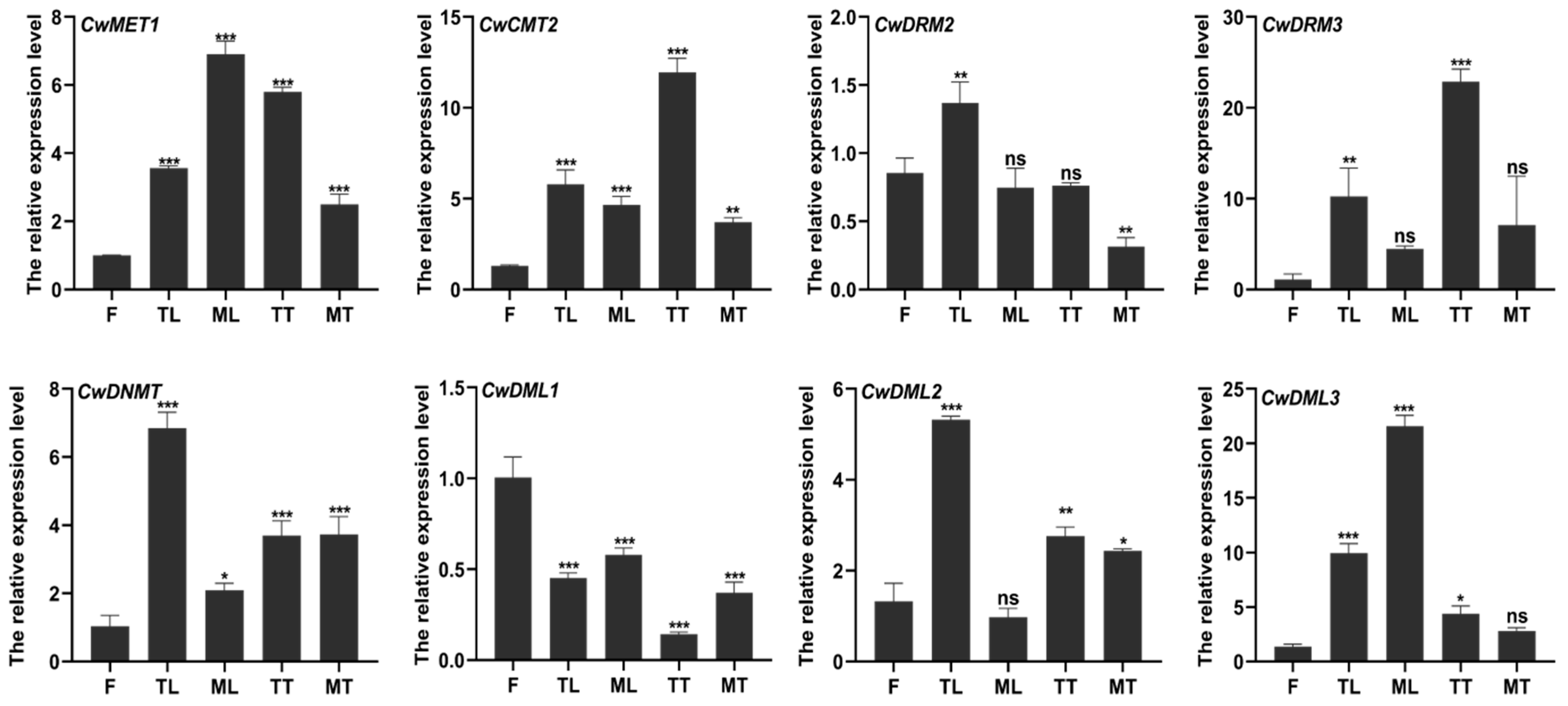
3.3. Transcript Abundance Analysis of CwC5-MTase and CwdMTase Genes in C. wenyujin
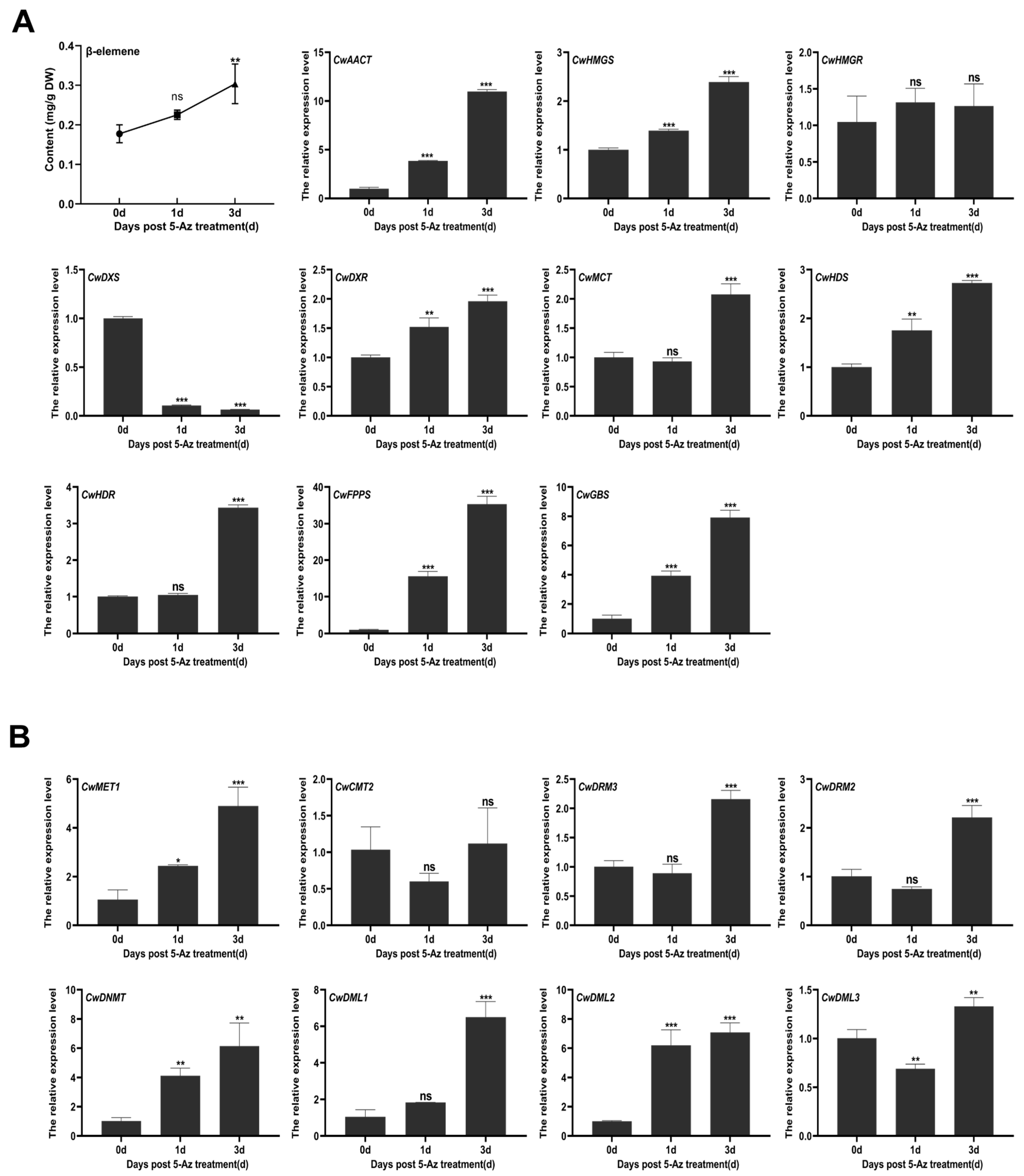
3.4. The Effects of 5-Az on the Expression of Key Genes in the Terpenoid Biosynthesis Pathway and β-Elemene Accumulation
3.5. The Effects of 5-Az on the DNA Methylation Pattern in C. wenyujin
3.6. Sequencing Analysis of Differentially Methylated DNA Fragments
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204–220. [Google Scholar] [CrossRef]
- He, X.J.; Chen, T.; Zhu, J.K. Regulation and function of DNA methylation in plants and animals. Cell Res. 2011, 21, 442–465. [Google Scholar] [CrossRef]
- Zhang, H.; Lang, Z.; Zhu, J.K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef]
- Lucibelli, F.; Valoroso, M.C.; Aceto, S. Plant DNA Methylation: An Epigenetic Mark in Development, Environmental Interactions, and Evolution. Int. J. Mol. Sci. 2022, 23, 8299. [Google Scholar] [CrossRef] [PubMed]
- Kankel, M.W.; Ramsey, D.E.; Stokes, T.L.; Flowers, S.K.; Haag, J.R.; Jeddeloh, J.A.; Riddle, N.C.; Verbsky, M.L.; Richards, E.J. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 2003, 163, 1109–1122. [Google Scholar] [CrossRef] [PubMed]
- Finnegan, E.J.; Kovac, K.A. Plant DNA methyltransferases. Plant Mol. Biol. 2000, 43, 189–201. [Google Scholar] [CrossRef]
- Lindroth, A.M.; Cao, X.; Jackson, J.P.; Zilberman, D.; McCallum, C.M.; Henikoff, S.; Jacobsen, S.E. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 2001, 292, 2077–2080. [Google Scholar] [CrossRef]
- Bartee, L.; Malagnac, F.; Bender, J. Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev. 2001, 15, 1753–1758. [Google Scholar] [CrossRef] [PubMed]
- Stroud, H.; Do, T.; Du, J.; Zhong, X.; Feng, S.; Johnson, L.; Patel, D.J.; Jacobsen, S.E. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 2014, 21, 64–72. [Google Scholar] [CrossRef]
- Cao, X.; Jacobsen, S.E. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc. Natl. Acad. Sci. USA 2002, 99 (Suppl. 4), 16491–16498. [Google Scholar] [CrossRef]
- Bewick, A.J.; Niederhuth, C.E.; Ji, L.; Rohr, N.A.; Griffin, P.T.; Leebens-Mack, J.; Schmitz, R.J. The evolution of CHROMOMETHYLASES and gene body DNA methylation in plants. Genome Biol. 2017, 18, 65. [Google Scholar] [CrossRef] [PubMed]
- Zemach, A.; Kim, M.Y.; Hsieh, P.H.; Coleman-Derr, D.; Eshed-Williams, L.; Thao, K.; Harmer, S.L.; Zilberman, D. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 2013, 153, 193–205. [Google Scholar] [CrossRef]
- Jeddeloh, J.A.; Stokes, T.L.; Richards, E.J. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 1999, 22, 94–97. [Google Scholar] [CrossRef]
- Goll, M.G.; Bestor, T.H. Eukaryotic Cytosine Methyltransferases. Annu. Rev. Biochem. 2005, 74, 481–514. [Google Scholar] [CrossRef]
- Jeltsch, A.; Ehrenhofer-Murray, A.; Jurkowski, T.P.; Lyko, F.; Reuter, G.; Ankri, S.; Nellen, W.; Schaefer, M.; Helm, M. Mechanism and biological role of Dnmt2 in Nucleic Acid Methylation. RNA Biol. 2016, 14, 1108–1123. [Google Scholar] [CrossRef]
- Zhang, H.; Gong, Z.; Zhu, J.K. Active DNA demethylation in plants: 20 years of discovery and beyond. J. Integr. Plant Biol. 2022, 64, 2217–2239. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, Q.; Kollie, L.; Dong, J.; Liang, Z. Molecular networks of secondary metabolism accumulation in plants: Current understanding and future challenges. Ind. Crops Prod. 2023, 201, 116901. [Google Scholar] [CrossRef]
- Bharti, P.; Mahajan, M.; Vishwakarma, A.K.; Bhardwaj, J.; Yadav, S.K. AtROS1 overexpression provides evidence for epigenetic regulation of genes encoding enzymes of flavonoid biosynthesis and antioxidant pathways during salt stress in transgenic tobacco. J. Exp. Bot. 2015, 66, 5959–5969. [Google Scholar] [CrossRef]
- Yu, L.; Sun, Y.; Zhang, X.; Chen, M.; Wu, T.; Zhang, J.; Xing, Y.; Tian, J.; Yao, Y. ROS1 promotes low temperature-induced anthocyanin accumulation in apple by demethylating the promoter of anthocyanin-associated genes. Hortic. Res. 2022, 9, uhac007. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Wang, N.; Chen, M.; Zhang, R.; Sun, Q.; Xu, H.; Zhang, Z.; Wang, Y.; Sui, X.; Wang, S.; et al. Methylation of MdMYB1 locus mediated by RdDM pathway regulates anthocyanin biosynthesis in apple. Plant Biotechnol. J. 2020, 18, 1736–1748. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Feng, S.; Jiao, Q.; Liu, C.; Zhang, W.; Chen, W.; Chen, X. Comparison of MdMYB1 sequences and expression of anthocyanin biosynthetic and regulatory genes between Malus domestica Borkh. cultivar ‘Ralls’ and its blushed sport. Euphytica 2011, 185, 157–170. [Google Scholar] [CrossRef]
- Han, M.; Lin, S.; Zhu, B.; Tong, W.; Xia, E.; Wang, Y.; Yang, T.; Zhang, S.; Wan, X.; Liu, J.; et al. Dynamic DNA Methylation Regulates Season-Dependent Secondary Metabolism in the New Shoots of Tea Plants. J. Agric. Food Chem. 2024, 72, 3984–3997. [Google Scholar] [CrossRef]
- Yuan, Y.; Wei, Y.; Yu, J.; Huang, L.Q. Relationship of epigenetic and Dao-di herbs. Zhongguo Zhong Yao Za Zhi 2015, 40, 2679–2683. [Google Scholar] [PubMed]
- Yang, D.; Huang, Z.; Jin, W.; Xia, P.; Jia, Q.; Yang, Z.; Hou, Z.; Zhang, H.; Ji, W.; Han, R. DNA methylation: A new regulator of phenolic acids biosynthesis in Salvia miltiorrhiza. Ind. Crops Prod. 2018, 124, 402–411. [Google Scholar] [CrossRef]
- Yang, B.C.; Lee, M.S.; Lin, M.K.; Chang, W.T. 5-Azacytidine increases tanshinone production in Salvia miltiorrhiza hairy roots through epigenetic modulation. Sci. Rep. 2022, 12, 9349. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Chen, Y.; Xia, Y.; Hong, X.; You, H.; Zhang, R.; Liang, Z.; Cui, Q.; Zhang, S.; Zhou, M.; et al. DNA methylation regulates biosynthesis of tanshinones and phenolic acids during growth of Salvia miltiorrhiza. Plant Physiol. 2024, 194, 2086–2100. [Google Scholar] [CrossRef]
- Yu, Z.; Zhang, G.; Teixeira da Silva, J.A.; Li, M.; Zhao, C.; He, C.; Si, C.; Zhang, M.; Duan, J. Genome-wide identification and analysis of DNA methyltransferase and demethylase gene families in Dendrobium officinale reveal their potential functions in polysaccharide accumulation. BMC Plant Biol. 2021, 21, 21. [Google Scholar] [CrossRef]
- Zha, L.; Liu, S.; Liu, J.; Jiang, C.; Yu, S.; Yuan, Y.; Yang, J.; Wang, Y.; Huang, L. DNA Methylation Influences Chlorogenic Acid Biosynthesis in Lonicera japonica by Mediating LjbZIP8 to Regulate Phenylalanine Ammonia-Lyase 2 Expression. Front. Plant Sci. 2017, 8, 1178. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, J.; Dong, J.; Kou, X.; Cui, Y.; Wang, S.; Long, Y.; Xing, Z. Identification and functional analysis of DNA methylation-related enzyme gene family in Eleutherococcus senticosus. Ind. Crops Prod. 2024, 213, 118412. [Google Scholar] [CrossRef]
- Dong, T.; Song, S.; Wang, Y.; Yang, R.; Chen, P.; Su, J.; Ding, X.; Liu, Y.; Duan, H. Effects of 5-azaC on Iridoid Glycoside Accumulation and DNA Methylation in Rehmannia glutinosa. Front. Plant Sci. 2022, 13, 913717. [Google Scholar] [CrossRef]
- Pandey, N.; Goswami, N.; Tripathi, D.; Rai, K.K.; Rai, S.K.; Singh, S.; Pandey-Rai, S. Epigenetic control of UV-B-induced flavonoid accumulation in Artemisia annua L. Planta 2018, 249, 497–514. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, C.; Lu, S. Identification and characterization of the cytosine-5 DNA methyltransferase gene family in Salvia miltiorrhiza. PeerJ 2018, 6, e4461. [Google Scholar] [CrossRef]
- Wang, Y.; Ai, P.; Li, S.; Kang, D.; Li, Z.; Wang, Z. Identification and Expression Analysis of Genes Related to DNA Methylation in Chrysanthemum * morifolium and C.nankingense. Acta Hortic. Sin. 2022, 49, 827–840. [Google Scholar]
- Zhang, X.; Zhu, L.; Xu, Z.; Wang, Z.; Dai, L. Characterization and differential expression of DNA methyltransferase and demethylase genes in response to abiotic stress in Isodon rubescens. BMC Plant Biol. 2025, 25, 755. [Google Scholar] [CrossRef]
- Li, Y.; Wu, Y.; Li, Y.; Guo, F. Review of the traditional uses, phytochemistry, and pharmacology of Curcuma wenyujin Y. H. Chen et C. Ling. J. Ethnopharmacol. 2021, 269, 113689. [Google Scholar] [CrossRef]
- Jiang, C.; Fei, X.; Pan, X.; Huang, H.; Qi, Y.; Wang, X.; Zhao, Q.; Li, F.; Zhang, L.; Shao, Q.; et al. Tissue-specific transcriptome and metabolome analyses reveal a gene module regulating the terpenoid biosynthesis in Curcuma wenyujin. Ind. Crops Prod. 2021, 170, 113758. [Google Scholar] [CrossRef]
- Chen, R.; Wei, Q.; Liu, Y.; Wei, X.; Chen, X.; Yin, X.; Xie, T. Transcriptome sequencing and functional characterization of new sesquiterpene synthases from Curcuma wenyujin. Arch. Biochem. Biophys. 2021, 709, 108986. [Google Scholar] [CrossRef]
- Wei, Q.; Lan, K.; Liu, Y.; Chen, R.; Hu, T.; Zhao, S.; Yin, X.; Xie, T. Transcriptome analysis reveals regulation mechanism of methyl jasmonate-induced terpenes biosynthesis in Curcuma wenyujin. PLoS ONE 2022, 17, e0270309. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, S.; Lan, K.; Wang, Q.; Ye, T.; Jin, H.; Hu, T.; Xie, T.; Wei, Q.; Yin, X. An Investigation of the JAZ Family and the CwMYC2-like Protein to Reveal Their Regulation Roles in the MeJA-Induced Biosynthesis of β-Elemene in Curcuma wenyujin. Int. J. Mol. Sci. 2023, 24, 15004. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Lan, K.; Wang, Q.; Su, Y.; Li, D.; Ma, J.; Hu, T.; Yin, X.; Wei, Q. Comprehensive characterization of the bHLH transcription factor family in Curcuma wenyujin and functional elucidation of CwbHLH27 in jasmonate-regulated sesquiterpenoid biosynthesis. Plant Physiol. Biochem. 2025, 220, 109527. [Google Scholar] [CrossRef] [PubMed]
- Hao, D.-C.; Xiao, P.-G. Deep in shadows: Epigenetic and epigenomic regulations of medicinal plants. Chin. Herb. Med. 2018, 10, 239–248. [Google Scholar] [CrossRef]
- Guo, W.; Ma, H.; Wang, C.-Z.; Wan, J.-Y.; Yao, H.; Yuan, C.-S. Epigenetic Studies of Chinese Herbal Medicine: Pleiotropic Role of DNA Methylation. Front. Pharmacol. 2021, 12, 790321. [Google Scholar] [CrossRef]
- Ma, X.; Tang, Y.; Feng, Z.; Yin, X.; Meng, Y.; Yin, X.; Xie, T. An organ-specific transcriptome atlas of Curcuma wenyujin: MicroRNAs, phasiRNAs, and metabolic pathways. Plant Genome 2025, 18, e20564. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhong, L.; Wu, X.; Fang, X.; Wang, J. Rapid alterations of gene expression and cytosine methylation in newly synthesized Brassica napus allopolyploids. Planta 2009, 229, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Figueroa, A. msap: A tool for the statistical analysis of methylation-sensitive amplified polymorphism data. Mol. Ecol. Resour. 2013, 13, 522–527. [Google Scholar] [CrossRef]
- Zeng, F.; Li, X.; Qie, R.; Li, L.; Ma, M.; Zhan, Y. Triterpenoid content and expression of triterpenoid biosynthetic genes in birch (Betula platyphylla Suk) treated with 5-azacytidine. J. For. Res. 2019, 31, 1843–1850. [Google Scholar] [CrossRef]
- Zhang, G.Q.; Xu, Q.; Bian, C.; Tsai, W.C.; Yeh, C.M.; Liu, K.W.; Yoshida, K.; Zhang, L.S.; Chang, S.B.; Chen, F.; et al. The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci. Rep. 2016, 6, 19029. [Google Scholar] [CrossRef]
- Kumar, S.; Cheng, X.; Klimasauskas, S.; Mi, S.; Posfai, J.; Roberts, R.J.; Wilson, G.G. The DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 1994, 22, 1–10. [Google Scholar] [CrossRef]
- Pósfai, J.; Bhagwat, A.S.; Pósfai, G.; Roberts, R.J. Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 1989, 17, 2421–2435. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Springer, N.M.; Muszynski, M.G.; Phillips, R.L.; Kaeppler, S.; Jacobsen, S.E. Conserved plant genes with similarity to mammalian de novo DNA methyltransferases. Proc. Natl. Acad. Sci. USA 2000, 97, 4979–4984. [Google Scholar] [CrossRef] [PubMed]
- Jeltsch, A. Circular permutations in the molecular evolution of DNA methyltransferases. J. Mol. Evol. 1999, 49, 161–164. [Google Scholar] [CrossRef] [PubMed]
- Bujnicki, J.M. Sequence permutations in the molecular evolution of DNA methyltransferases. BMC Evol. Biol. 2002, 2, 3. [Google Scholar] [CrossRef][Green Version]
- Pavlopoulou, A.; Kossida, S. Plant cytosine-5 DNA methyltransferases: Structure, function, and molecular evolution. Genomics 2007, 90, 530–541. [Google Scholar] [CrossRef]
- Gehring, M.; Huh, J.H.; Hsieh, T.F.; Penterman, J.; Choi, Y.; Harada, J.J.; Goldberg, R.B.; Fischer, R.L. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 2006, 124, 495–506. [Google Scholar] [CrossRef]
- Mok, Y.G.; Uzawa, R.; Lee, J.; Weiner, G.M.; Eichman, B.F.; Fischer, R.L.; Huh, J.H. Domain structure of the DEMETER 5-methylcytosine DNA glycosylase. Proc. Natl. Acad. Sci. USA 2010, 107, 19225–19230. [Google Scholar] [CrossRef]
- Ponferrada-Marín, M.I.; Parrilla-Doblas, J.T.; Roldán-Arjona, T.; Ariza, R.R. A discontinuous DNA glycosylase domain in a family of enzymes that excise 5-methylcytosine. Nucleic Acids Res. 2011, 39, 1473–1484. [Google Scholar] [CrossRef]
- Pei, L.; Zhang, L.; Li, J.; Shen, C.; Qiu, P.; Tu, L.; Zhang, X.; Wang, M. Tracing the origin and evolution history of methylation-related genes in plants. BMC Plant Biol. 2019, 19, 307. [Google Scholar] [CrossRef]
- Wang, S.; Zhao, X.; Li, C.; Dong, J.; Ma, J.; Long, Y.; Xing, Z. DNA methylation regulates the secondary metabolism of saponins to improve the adaptability of Eleutherococcus senticosus during drought stress. BMC Genom. 2024, 25, 330. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, G.; Yang, F.; Liang, Y.; Gao, Q.; Xiang, C.; Li, X.; Yang, R.; Zhang, G.; Jiang, H.; et al. Multilayered regulation of secondary metabolism in medicinal plants. Mol. Hortic. 2023, 3, 11. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Zhao, X.; Song, X.; Wang, S.; Zhao, X.; Liang, B.; Long, Y.; Xing, Z. Identification of Eleutherococcus senticosus NAC transcription factors and their mechanisms in mediating DNA methylation of EsFPS, EsSS, and EsSE promoters to regulate saponin synthesis. BMC Genom. 2024, 25, 536. [Google Scholar] [CrossRef]
- Mathieu, O.; Reinders, J.; Caikovski, M.; Smathajitt, C.; Paszkowski, J. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell 2007, 130, 851–862. [Google Scholar] [CrossRef]
- Williams, B.P.; Pignatta, D.; Henikoff, S.; Gehring, M. Methylation- sensitive expression of a DNA demethylase gene serves as an epigenetic rheostat. PLoS Genet. 2015, 11, e1005142. [Google Scholar] [CrossRef]
- Hu, L.; Li, N.; Xu, C.; Zhong, S.; Lin, X.; Yang, J.; Zhou, T.; Yuliang, A.; Wu, Y.; Chen, Y. Mutation of a major CG methylase in rice causes genome- wide hypomethylation, dysregulated genome expression, and seedling lethality. Proc. Natl. Acad. Sci. USA 2014, 111, 10642–10647. [Google Scholar] [CrossRef]
- Yu, Y.; Zhao, F.; Yue, Y.; Zhao, Y.; Zhou, D. Lysine acetylation of the histone acetyltransferase adaptor protein ADA2 is a mechanism of metabolic control of chromatin modification in plants. Nat. Plants 2024, 10, 439–452. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Dou, Y.; Zhao, S.; Zhao, C.; Chen, Y.; Jiang, M.; Shen, Z.; Chen, C. Rhizosphere microbes enhance plant resistance to cadmium through a root ROS-microbial IAA-root DNA methylation interkingdom signaling pathway. Cell Rep. 2025, 44, 116491. [Google Scholar] [CrossRef]
- Li, Y.; Chen, X.; Wang, J.; Zou, G.; Wang, L.; Li, X. Two responses to MeJA induction of R2R3-MYB transcription factors regulate flavonoid accumulation in Glycyrrhiza uralensis Fisch. PLoS ONE 2020, 15, e0236565. [Google Scholar] [CrossRef]
- Liu, L.; Yang, D.; Xing, B.; Zhang, C.; Lang, Z. SmMYB98b positive regulation to tanshinones in Salvia miltiorrhiza Bunge hairy roots. Plant Cell Tiss. Organ Cult. 2020, 140, 459–467. [Google Scholar] [CrossRef]
- Wu, Z.; Li, L.; Liu, H.; Yan, X.; Ma, Y.; Li, Y.; Chen, T.; Wang, C.; Xie, L.; Hao, X.; et al. AaMYB15, an R2R3-MYB TF in Artemisia annua, acts as a negative regulator of artemisinin biosynthesis. Plant Sci. 2021, 308, 110920. [Google Scholar] [CrossRef]
- Wang, X.; Zhou, X.; Li, Y.; Zhang, Z.; Dong, S.; Jin, Z.; Zhan, X.; Yang, S.; Wu, L.; Liu, L. Difference in DNA methylation pattern and expression of active ingredients between garden ginseng and ginseng under forest. Ind. Crops Prod. 2024, 219, 119120. [Google Scholar] [CrossRef]
- Hao, M.; Zhou, Y.; Zhou, J.; Zhang, M.; Yan, K.; Jiang, S.; Wang, W.; Peng, X.; Zhou, S. Cold-induced ginsenosides accumulation is associated with the alteration in DNA methylation and relative gene expression in perennial American ginseng (Panax quinquefolius L.) along with its plant growth and development process. J. Ginseng Res. 2020, 44, 747–755. [Google Scholar] [CrossRef]
- Huang, W.; Xiong, L.; Zhang, L.; Zhang, F.; Han, X.; Zhang, Y.; Zhang, L.; Yang, H. Study on content variation of flavonoids in different germplasm during development of Lonicerae Japonicae Flos. Chin. Tradit. Herb. Drugs 2022, 53, 3156–3164. [Google Scholar]
- Yu, X.; Yu, H.; Lu, Y.; Zhang, C.; Wang, H. Genetic and epigenetic variations underlying flavonoid divergence in Beihua and Sijihua honeysuckles. Epigenet. Insights 2024, 17, e002. [Google Scholar] [CrossRef]
- Chen, X.; Huang, C.; Li, K.; Liu, J.; Zheng, Y.; Feng, Y.; Kai, G.-Y. Recent advances in biosynthesis and pharmacology of β-elemene. Phytochem. Rev. 2022, 22, 169–186. [Google Scholar] [CrossRef]
- Schönbach, C.; Lu, L.; Liu, P.; Yang, Y.; Zhang, Y.; Wang, C.; Feng, J.; Wei, J. Transcriptome analysis of Curcuma wenyujin from Haikou and Wenzhou, and a comparison of the main constituents and related genes of Rhizoma Curcumae. PLoS ONE 2020, 15, e0242776. [Google Scholar] [CrossRef]
| Gene Name | ORF a (bp) | AA b (aa) | MW c (kDa) | GRAVY d Value | pI e | Predicted Subcellular Localization |
|---|---|---|---|---|---|---|
| CwMET1 | 4695 | 1564 | 176.37 | −0.469 | 5.69 | Cell membrane, Chloroplast, Cytoplasm, Nucleus. |
| CwCMT2 | 2223 | 740 | 84.08 | −0.397 | 6.34 | Nucleus |
| CwDRM2 | 1746 | 581 | 65.13 | −0.501 | 4.98 | Nucleus |
| CwDRM3 | 2037 | 678 | 76.76 | −0.499 | 4.86 | Chloroplast |
| CwDNMT | 891 | 296 | 33.98 | −0.231 | 5.43 | Chloroplast |
| CwDML1 | 3630 | 1209 | 136.49 | −0.564 | 5.3 | Nucleus |
| CwDML2 | 5175 | 1724 | 193.27 | −0.57 | 8.16 | Nucleus |
| CwDML3 | 5835 | 1944 | 217.41 | −0.58 | 5.92 | Nucleus |
| Type | No. of Bands and Ratio | |||
| HpaII | MspI | CK | 5-Az (3 d) | |
| 1 | 1 | I—Non-methylated | 704 | 700 |
| 1 | 0 | II—Hemi-methylated | 40 | 32 |
| 0 | 1 | III—Fully methylated | 33 | 35 |
| 0 | 0 | IV—Hypermethylated | 26 | 18 |
| Total amplified bands | 803 | 785 | ||
| Total methylated bands | 99 | 85 | ||
| Fully methylated bands | 59 | 53 | ||
| Hemi-methylated bands | 40 | 32 | ||
| Total methylated ratio (%) a | 12.33 | 10.83 | ||
| Fully methylated ratio (%) b | 7.35 | 6.75 | ||
| Hemi-methylated ratio (%) c | 4.98 | 4.08 | ||
| Non-methylated ratio (%) d | 87.67 | 89.17 | ||
| Description of Pattern | Classes | Banding Patterns | No. of Bands | Ratio | Methylation Pattern Alterations | |||
|---|---|---|---|---|---|---|---|---|
| Control | 5-Az (3d) | |||||||
| HspII | MspI | HspII | MspI | |||||
| No change | A1 | 1 | 1 | 1 | 1 | 312 | 87.29% | I → I |
| A2 | 1 | 0 | 1 | 0 | 24 | II → II | ||
| A3 | 0 | 1 | 0 | 1 | 21 | III → III | ||
| Demethylation | B1 | 1 | 0 | 1 | 1 | 15 | 9.05% | II → I |
| B2 | 0 | 1 | 1 | 1 | 4 | III → I | ||
| B3 | 0 | 0 | 1 | 1 | 5 | IV → I | ||
| B4 | 0 | 0 | 1 | 0 | 9 | IV → II | ||
| B5 | 0 | 0 | 0 | 1 | 4 | IV → III | ||
| Methylation | C1 | 1 | 1 | 1 | 0 | 4 | 3.67% | I → II |
| C2 | 1 | 1 | 0 | 1 | 6 | I → III | ||
| C3 | 1 | 1 | 0 | 0 | 0 | I → IV | ||
| C4 | 1 | 0 | 0 | 1 | 0 | II → III | ||
| C5 | 1 | 0 | 0 | 0 | 4 | II → IV | ||
| C6 | 0 | 1 | 0 | 0 | 1 | III → IV | ||
| Primer Combinations | Methylation Status | Accession | Description | E-Value |
|---|---|---|---|---|
| H14E11-1 | Methylated | LOC121986379 | Auxin-responsive protein IAA17-like | 2.8 × 10−29 |
| H14E11-2 | Methylated | LOC121976864 | Polygalacturonase At1g48100-like | 2 × 10−26 |
| H14E11-3 | Methylated | LOC121988566 | (S)-2-hydroxy-acidoxidase GLO1-like | 1.8 × 10−21 |
| H14E12 | Methylated | LOC122012221 | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase 2-like | 8.8 × 10−77 |
| H14E14-2 | Methylated | NC_048505.1 | Genome sequence, function unknown | 9 × 10−101 |
| H17E12 | Methylated | LOC121977767 | Kinesin-like protein KIN-7C | 2.7 × 10−51 |
| H18E13 | Methylated | OQ909706.1 | MYB protein | 3 × 10−9 |
| H14E13-1 | Demethylated | LOC122015080 | Metal tolerance protein 2-like | 3.5 × 10−50 |
| H14E13-2 | Demethylated | AP015041.1 | Genome sequence, function unknown | 2.3 × 10−3 |
| H16E20 | Demethylated | LOC122045127 | Protein SPIRAL1-like3 | 3.6 × 10−6 |
| H17E13 | Demethylated | LOC122046078 | Flocculation protein FLO11-like | 4.1 × 10−7 |
| H20E13 | Demethylated | LOC121995052 | Two-component response regulator ARR10-like | 3.2 × 10−24 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, X.; Ma, J.; Shen, Z.; Wang, Q.; Xu, M.; Hu, T.; Wei, Q.; Yin, X.; Ma, X. Identification of DNA Methyltransferase/Demethylase Genes and 5-Azacytidine’s Impact on β-Elemene and Methylation in Curcuma wenyujin. Biology 2025, 14, 1739. https://doi.org/10.3390/biology14121739
Yin X, Ma J, Shen Z, Wang Q, Xu M, Hu T, Wei Q, Yin X, Ma X. Identification of DNA Methyltransferase/Demethylase Genes and 5-Azacytidine’s Impact on β-Elemene and Methylation in Curcuma wenyujin. Biology. 2025; 14(12):1739. https://doi.org/10.3390/biology14121739
Chicago/Turabian StyleYin, Xiu, Jiawei Ma, Zhenlu Shen, Qian Wang, Mengdie Xu, Tianyuan Hu, Qiuhui Wei, Xiaopu Yin, and Xiaoxia Ma. 2025. "Identification of DNA Methyltransferase/Demethylase Genes and 5-Azacytidine’s Impact on β-Elemene and Methylation in Curcuma wenyujin" Biology 14, no. 12: 1739. https://doi.org/10.3390/biology14121739
APA StyleYin, X., Ma, J., Shen, Z., Wang, Q., Xu, M., Hu, T., Wei, Q., Yin, X., & Ma, X. (2025). Identification of DNA Methyltransferase/Demethylase Genes and 5-Azacytidine’s Impact on β-Elemene and Methylation in Curcuma wenyujin. Biology, 14(12), 1739. https://doi.org/10.3390/biology14121739






