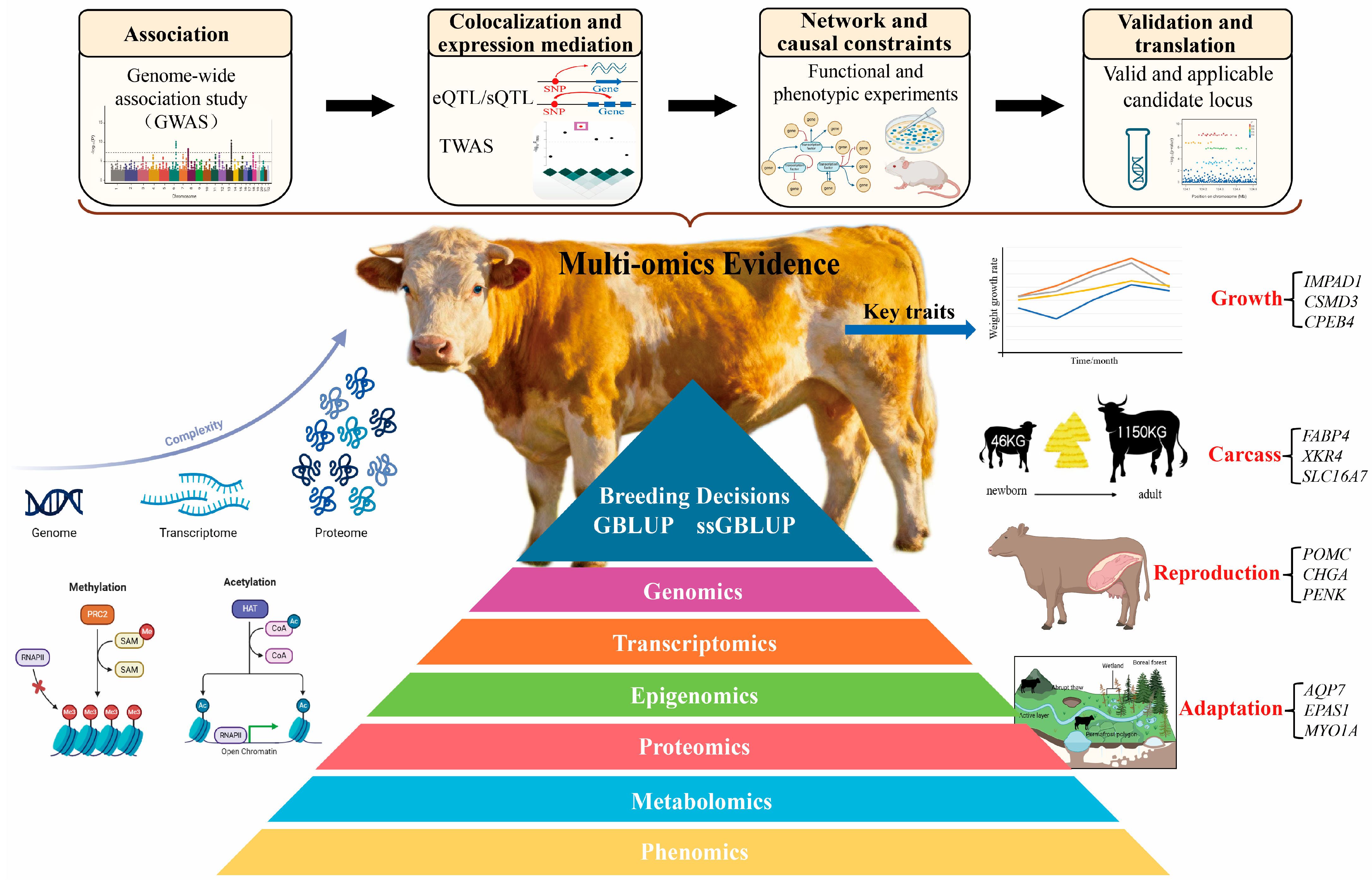
Omics Evidence Chains for Complex Traits in Beef Cattle: From Cross-Layer Colocalization to Genetic Evaluation and Application
Simple Summary
Abstract
1. Introduction
2. Single-Omics Evidence and Trait Dissection Progress from Association to Prioritized Candidates
2.1. Discovery-Stage Signals for Growth and Development
2.2. Discovery-Stage Signals for Carcass and Meat Quality
2.3. Discovery-Stage Signals for Reproductive Traits
2.4. Discovery-Stage Signals for Environmental Adaptation and Resilience
3. Cross-Omics Integration and Causal Localization Move Candidates Toward Translation
3.1. Integrated Multi-Omics Evidence for Growth and Development
| Module or Pathway | Representative Candidates | Evidence | Suggested Application | Practical Application (Breeding/Monitoring/Management) | References |
|---|---|---|---|---|---|
| Body-size and bone-growth hub | PLAG1–LCORL/NCAPG | Cross-ancestry GWAS and meta-GWAS followed by TWAS and colocalization (PP4 increased); tissue expression consistent | Stability anchor to benchmark other loci; track effect direction | Breeding evaluation: benchmark loci for growth index; Farm monitoring: track growth curve consistency | [22,30] |
| ECM and muscle-fiber formation | IMPAD1; PENK; STC2; CPEB4 | WGCNA or GRN centrality increased; mediation; colocalization with body-size traits | Add informative priors in indices for size and bone mass | Breeding: informative priors in ssGBLUP; Monitoring: bone-density/ECM markers | [31,32,33,34] |
| Cooperating gene clusters | CSMD3; LAP3; SYN3; FAM19A5; TIMP3 | Module explains variation with tissue eQTL support | Modular weighting for body-size and bone profile | Management: integrate cluster weighting into carcass evaluation pipeline | [32,33,34] |
| Methods and workflow | eQTL; TWAS; ATAC; methylation | From association, through regulation and network constraints, to validation; containerized; batch control, QC, and ID mapping | Reproducible pipeline; Minimum Information sheets | Batch management: ensure reproducible QC, ID mapping, and metadata tracking | [5,7,9] |
3.2. Integrated Multi-Omics Evidence for Carcass and Meat Quality
| Module or Pathway | Representative Candidates | Evidence | Suggested Application | Practical Application (Breeding/Monitoring/Management) | References |
|---|---|---|---|---|---|
| Lipid-droplet biogenesis, transport, and desaturation (three-segment) | FABP4; SCD; ADIRF | Joint enrichment in transcriptome, methylome, and metabolome; concordant with IMF percentage and fatty-acid profile | Use fatty-acid profile and desaturation index as mediating phenotypes for validation | Breeding: include desaturation index as genomic weight for meat-quality selection; Monitoring: track fatty-acid profile and IMF % via NIR or biochemical assays; Management: adjust feeding regime based on IMF trend | [35,36,37,38,78] |
| Fat distribution and marbling | PLIN1; SLCO4C1; SLC16A7; SLC22 family | Coordinated epigenetic and transcript coupling; covaries with marbling and tenderness | Field Warner–Bratzler shear force and near-infrared monitoring; link to indices | Breeding: integrate marbling score and tenderness into multi-trait selection; Monitoring: on-farm infrared sensors for carcass grading; Management: feedback loop between carcass data and finishing diets | [79] |
| Subcutaneous backfat thickness | XKR4 | Multi-breed association replicated | Add distribution weight in the index | Breeding: use as stability marker for fat deposition; Monitoring: ultrasound or digital imaging for back-fat tracking; Management: optimize energy balance in finishing phase | [44] |
| Implementation and evaluation | IMF percentage; MUFA to SFA ratio; C18:1; WBSF; NIR | Cross-layer colocalization leading to a monitorable-phenotype loop; G × E recorded | Finishing Minimum Information Sheet; model batch as a random effect with a diet by genotype interaction | Breeding: validate across herds to refine index weighting; Monitoring: collect standardized finishing data; Management: apply batch QC and diet-genotype recording via unified templates | [40,41,72,73] |
3.3. Integrated Multi-Omics Evidence for Reproductive Traits
| Module or Layer | Representative Candidates | Evidence | Suggested Application | Practical Application (Breeding/Monitoring/Management) | References |
|---|---|---|---|---|---|
| Neuroendocrine upstream | POMC; CHGA; PENK | Transcriptome with co-expression implicates puberty initiation; TWAS and colocalization support | Build a Puberty Program Score mapped to APU and AFC | Breeding: include puberty score as genomic predictor; Monitoring: measure puberty onset or cyclicity via hormonal assays; Management: schedule synchronization protocols by maturity stage | [46] |
| Ovarian and uterine microenvironment | ALKBH5-BMP15 (m6A) | m6A with splicing and noncoding RNA coupling; associated with puberty timing | Perturbation and rescue in cumulus–oocyte complexes and granulosa cells | Breeding: prioritize fertility alleles in index; Monitoring: track follicle growth, oocyte quality; Management: nutritional and hormonal adjustment to support maturation | [9,47] |
| Male gametogenesis | circRNA–target networks | Single-cell RNA sequencing and single-cell ATAC or CUT and Tag show stage-specific regulation | Validate with organoids and in vitro spermatogenesis systems | Breeding: select bulls based on spermatogenic stability markers; Monitoring: semen-quality scoring (motility, circRNA biomarkers); Management: manage temperature and stress conditions in AI centers | [57,81] |
| Recording and modeling | APU; AFC; HCR or HP; CCR; CI; SC | Low heritability (h2) requires large cohorts and standardization; control season, nutrition, and health status | Include stayability and open days in indices | Breeding: integrate stayability in lifetime-productivity index; Monitoring: record open days and conception rate; Management: implement reproductive-data logging and seasonal adjustment | [23] |
3.4. Integrated Multi-Omics Evidence for Environmental Adaptation and Resilience
| Module or Pathway | Representative Candidates | Evidence | Suggested Application | Practical Application (Breeding/Monitoring/Management) | References |
|---|---|---|---|---|---|
| Cold-adaptation track | PRDM16; AQP3; AQP7 | Population signals consistent with cold-tolerance phenotypes; single-cell and epigenomic support | Metabolomics informing browning and energy-substrate preferences | Breeding: include thermogenic and lipid-oxidation markers in adaptive index; Monitoring: measure body-temperature resilience and metabolite profile in cold season; Management: optimize feeding and housing for cold regions | [66,75,83] |
| Heat-adaptation track | MYO1A; TECPR2 | Repeated across heat-tolerance studies; aligns with THI, body temperature, and behavior | Joint modeling with THI and behavioral phenotypes | Breeding: incorporate heat-tolerance loci in tropical index; Monitoring: track THI, panting score, body temp; Management: implement shade/cooling/watering schedule by genotype | [68,69,70] |
| Structural variation | EPAS1; EGLN1 | Introgression with structural variation and distal regulation consistent with altitude and the partial pressure of oxygen; tissue and environment-specific expression | Selection scans, environmental association, and regulatory evidence; multi-site and multi-season reaction norms | Breeding: select for hypoxia-resistant genotypes; Monitoring: use hematologic and oxygen-saturation indicators; Management: plan herd movement or breeding by altitude | [3,43,82] |
| Regional translation | THI, altitude, and aridity–humidity by genotype | Environment-specific expression with metabolomics, G × E prediction, and reaction-norm validation | Design regional deployment and diet stratification schemes | Breeding: establish ecozone-specific sub-panels; Monitoring: link genotype with local THI records; Management: tailor diet and breeding schedule per region | [62,63] |
4. Multi-Omics Evidence Chains, G × E, and Functional Validation Connect Association to Causality and Translation for Breeding Applications
4.1. A Framework for Causal Inference and Localization Brings Correlation Closer to Causation
4.2. Multiscale Functional Validation Turns Statistical Signals into Biological Mechanisms
4.3. G × E and Reaction Norms Bring the Environment into the Causal Chain
4.4. Multi-Omics Mechanisms Are Translated into Breeding Decisions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Daniel, P.; Natalia, S.; Nicolazzi, E.L.; Machugh, D.E.; Park, S.D.E.; Licia, C.; Rodrigo, M.; Bruford, M.W.; Pablo, O.T. Domestication of cattle: Two or three events? Evol. Appl. 2018, 12, 123–136. [Google Scholar] [CrossRef]
- Decker, J.E.; McKay, S.D.; Rolf, M.M.; Kim, J.; Molina Alcala, A.; Sonstegard, T.S.; Hanotte, O.; Gotherstrom, A.; Seabury, C.M.; Praharani, L.; et al. Worldwide patterns of ancestry, divergence, and admixture in domesticated cattle. PLoS Genet. 2014, 10, e1004254. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Zhang, F.; Li, S.; Luo, X.; Peng, L.; Dong, Z.; Pausch, H.; Leonard, A.S.; Crysnanto, D.; Wang, S.; et al. Structural variation and introgression from wild populations in East Asian cattle genomes confer adaptation to local environment. Genome Biol. 2023, 24, 211. [Google Scholar] [CrossRef]
- FAO. Meat Market Review: Overview of Global Market Developments in 2023; FAO: Rome, Italy, 2024. [Google Scholar]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-omics approaches to disease. Genome Biol. 2017, 18, 83. [Google Scholar] [CrossRef]
- Stefan, G.; Kevin, C.; Washam, C.L.; Allen, G.; Jordan, B.; Robeson, M.S.; Byrum, S.D. Multi-omics data integration considerations and study design for biological systems and disease. Mol. Omics 2021, 17, 170–185. [Google Scholar] [CrossRef] [PubMed]
- Picard, M.; Scott-Boyer, M.P.; Bodein, A.; Olivier, P.; Droit, A. Integration strategies of multi-omics data for machine learning analysis. Comput. Struct. Biotechnol. J. 2021, 19, 3735–3746. [Google Scholar] [CrossRef]
- Oakley, A.E.; Clifton, D.K.; Steiner, R.A. Kisspeptin signaling in the brain. Endocr. Rev. 2009, 30, 713–743. [Google Scholar] [CrossRef]
- Wang, L.; Khunsriraksakul, C.; Markus, H.; Chen, D.; Zhang, F.; Chen, F.; Zhan, X.; Carrel, L.; Liu, D.J.; Jiang, B. Integrating single cell expression quantitative trait loci summary statistics to understand complex trait risk genes. Nat. Commun. 2024, 15, 4260. [Google Scholar] [CrossRef] [PubMed]
- Fontanesi, L. Metabolomics and livestock genomics: Insights into a phenotyping frontier and its applications in animal breeding. Anim. Front. 2016, 6, 73–79. [Google Scholar] [CrossRef]
- Wadood, A.A.; Bordbar, F.; Zhang, X. Integrating omics approaches in livestock biotechnology: Innovations in production and reproductive efficiency. Front. Anim. Sci. 2025, 15, 1946. [Google Scholar] [CrossRef]
- Cai, W.; Zhang, Y.; Chang, T.; Wang, Z.; Zhu, B.; Chen, Y.; Gao, X.; Xu, L.; Zhang, L.; Gao, H.; et al. The eQTL colocalization and transcriptome-wide association study identify potentially causal genes responsible for economic traits in Simmental beef cattle. J. Anim. Sci. Biotechnol. 2023, 14, 78. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Lu, Y.; Chong, Y.; Li, M.; Hong, J.; Wu, J.; Wu, D.; Xi, D.; Deng, W. Beef cattle genome project: Advances in genome sequencing, assembly, and functional genes discovery. Int. J. Mol. Sci. 2024, 25, 7147. [Google Scholar] [CrossRef]
- Jitjumnong, J.; Taweechaipaisankul, A.; Lin, J.C.; Wongchanla, S.; Chuwatthanakhajorn, S.; Lin, C.J.; Khang, L.T.P.; Linh, N.V.; Sangsawad, P.; Dinh-Hung, N.; et al. An overview of advancements in proteomic approaches to enhance livestock production and aquaculture. Animals 2025, 15, 1946. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.L.; Wilson, M.L.; Nilson, S.M.; Rowan, T.N.; Schnabel, R.D.; Decker, J.E.; Seabury, C.M. Genome-wide association and genotype by environment interactions for growth traits in U.S. Red Angus cattle. BMC Genom. 2022, 23, 926. [Google Scholar] [CrossRef]
- Fulco, C.P.; Munschauer, M.; Anyoha, R.; Munson, G.; Grossman, S.R.; Perez, E.M.; Kane, M.; Cleary, B.; Lander, E.S.; Engreitz, J.M. Systematic mapping of functional enhancer–promoter connections with CRISPR interference. Science 2016, 354, 769–773. [Google Scholar] [CrossRef]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef] [PubMed]
- Giambartolomei, C.; Vukcevic, D.; E Schadt, E.; Franke, L.; Hingorani, A.D.; Wallace, C.; Plagnol, V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014, 10, e1004383. [Google Scholar] [CrossRef]
- Barbeira, A.N.; Pividori, M.; Zheng, J.; E Wheeler, H.; Nicolae, D.L.; Im, H.K. Integrating predicted transcriptome from multiple tissues improves association detection. PLoS Genet. 2019, 15, e1007889. [Google Scholar] [CrossRef]
- Gusev, A.; Ko, A.; Shi, H.; Bhatia, G.; Chung, W.; Penninx, B.W.J.H.; Jansen, R.; De Geus, E.J.C.; Boomsma, D.I.; Wright, F.A.; et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016, 48, 145–152. [Google Scholar] [CrossRef]
- Takasuga, A. PLAG1 and NCAPG-LCORL in livestock. Anim. Sci. J. 2015, 87, 159–167. [Google Scholar] [CrossRef]
- Kenny, D.A.; Fitzsimons, C.; Waters, S.M.; McGee, M. Invited review: Improving feed efficiency of beef cattle—The current state of the art and future challenges. Animal 2018, 12, 1815–1826. [Google Scholar] [CrossRef]
- Berry, D.P.; Crowley, J.J. Residual intake and body weight gain: A new measure of efficiency in growing cattle. J. Anim. Sci. 2012, 90, 109–115. [Google Scholar] [CrossRef]
- Berry, D.P.; Crowley, J.J. Cell biology symposium: Genetics of feed efficiency in dairy and beef cattle. J. Anim. Sci. 2013, 91, 1594–1613. [Google Scholar] [CrossRef]
- Mulhall, S.A.; Sleator, R.D.; Evans, R.D.; Berry, D.P.; Twomey, A.J. Effect on prime animal beef merit from breeding solely for lighter dairy cows. J. Dairy Sci. 2024, 107, 8150–8156. [Google Scholar] [CrossRef] [PubMed]
- Basiel, B.L.; Felix, T.L. Board Invited Review: Crossbreeding beef × dairy cattle for the modern beef production system. Transl. Anim. Sci. 2022, 6, txac025. [Google Scholar] [CrossRef] [PubMed]
- Weik, F.; Hickson, R.E.; Morris, S.T.; Garrick, D.J.; Archer, J.A. Genetic parameters for growth, ultrasound and carcass traits in New Zealand beef cattle and their correlations with maternal performance. Animals 2021, 12, 25. [Google Scholar] [CrossRef]
- Do, C.; Park, B.; Kim, S.; Choi, T.; Yang, B.; Park, S.; HyungJun, S. Genetic parameter estimates of carcass traits under national scale breeding scheme for beef cattle. Asian Australas. J. Anim. Sci. 2016, 29, 1083–1094. [Google Scholar] [CrossRef]
- Majeres, L.E.; Dilger, A.C.; Shike, D.W.; McCann, J.C.; Beever, J.E. Defining a haplotype encompassing the LCORL-NCAPG locus associated with increased lean growth in beef cattle. Genes 2024, 15, 576. [Google Scholar] [CrossRef] [PubMed]
- An, B.; Xia, J.; Chang, T.; Wang, X.; Xu, L.; Zhang, L.; Gao, X.; Chen, Y.; Li, J.; Gao, H. Genome-wide association study reveals candidate genes associated with body measurement traits in Chinese wagyu beef cattle. Anim. Genet. 2019, 50, 386–390. [Google Scholar] [CrossRef]
- Wu, D.-D.; Yang, C.-P.; Wang, M.-S.; Dong, K.-Z.; Yan, D.-W.; Hao, Z.-Q.; Fan, S.-Q.; Chu, S.-Z.; Shen, Q.-S.; Jiang, L.-P.; et al. Convergent genomic signatures of high-altitude adaptation among domestic mammals. Natl. Sci. Rev. 2020, 7, 952–963. [Google Scholar] [CrossRef]
- Yan, S.; Pei, F.; Si, J.; Khan, M.Y.A.; Ou, S.; Yang, Y.; Zhao, Z.; Pauciullo, A.; Zhang, Y. Gene co-expression network and differential expression analyses reveal key genes for weaning weight in Simmental-Holstein crossbred cattle. Anim. Biotechnol. 2024, 35, 2404042. [Google Scholar] [CrossRef]
- Zhang, S.; Yao, Z.; Li, X.; Zhang, Z.; Liu, X.; Yang, P.; Chen, N.; Xia, X.; Lyu, S.; Shi, Q.; et al. Assessing genomic diversity and signatures of selection in Pinan cattle using whole-genome sequencing data. BMC Genom. 2022, 23, 460. [Google Scholar] [CrossRef]
- Tan, Z.; Jiang, H. Molecular and cellular mechanisms of intramuscular fat development and growth in cattle. Int. J. Mol. Sci. 2024, 25, 2520. [Google Scholar] [CrossRef]
- Martins, R.; Machado, P.C.; Pinto, L.F.B.; Silva, M.R.; Schenkel, F.S.; Brito, L.F.; Pedrosa, V.B. Genome-wide association study and pathway analysis for fat deposition traits innellorecattle raised in pasture-based systems. J. Anim. Breed. Genet. 2021, 138, 360–378. [Google Scholar] [CrossRef] [PubMed]
- Silva-Vignato, B.; Cesar, A.S.M.; Afonso, J.; Moreira, G.C.M.; Poleti, M.D.; Petrini, J.; Garcia, I.S.; Clemente, L.G.; Mourao, G.B.; Regitano, L.C.A.; et al. Integrative analysis between genome-wide association study and expression quantitative trait loci reveals bovine muscle gene expression regulatory polymorphisms associated with intramuscular fat and backfat thickness. Front. Genet. 2022, 13, 935238. [Google Scholar] [CrossRef] [PubMed]
- Cesar, A.S.; Regitano, L.C.; Mourão, G.B.; Tullio, R.R.; Lanna, D.P.; Nassu, R.T.; A Mudado, M.; Oliveira, P.S.; Nascimento, M.L.D.; Chaves, A.S.; et al. Genome-wide association study for intramuscular fat deposition and composition in Nellore cattle. BMC Genet. 2014, 15, 39. [Google Scholar] [CrossRef] [PubMed]
- Nasab, S.E.; Dashab, G.R.; Rokouei, M.; Roudbari, Z.; Sadkowski, T. Unveiling Conserved Molecular Pathways of Intramuscular Fat Deposition and Shared Metabolic Processes in Semitendinosus Muscle of Hereford, Holstein, and Limousine Cattle via RNA-Seq Analysis. Genes 2025, 16, 984. [Google Scholar] [CrossRef]
- Zhou, D.; Wang, Y.; Yang, R.; Wang, F.; Zhao, Z.; Wang, X.; Xie, L.; Tian, X.; Wang, G.; Li, B.; et al. The MyoD1 promoted muscle differentiation and generation by activating CCND2 in Guanling cattle. Animals 2022, 12, 2571. [Google Scholar] [CrossRef]
- Flowers, S.; Hamblen, H.; Joel, D.L.-G.; Elzo, M.A.; Johnson, D.D.; Mateescu, R.G. Fatty acid profile, mineral content, and palatability of beef from a multibreed Angus–Brahman population1. J. Anim. Sci. 2018, 96, 4264–4275. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, J.; Gong, H.; Cui, L.; Zhang, W.; Ma, J.; Chen, C.; Ai, H.; Xiao, S.; Huang, L.; et al. Genetic correlation of fatty acid composition with growth, carcass, fat deposition and meat quality traits based on GWAS data in six pig populations. Meat Sci. 2019, 150, 47–55. [Google Scholar] [CrossRef]
- Li, R.; Chen, S.; Li, C.; Xiao, H.; Costa, V.; Bhuiyan, M.S.A.; Baig, M.; Beja-Pereira, A. Whole-Genome analysis deciphers population structure and genetic introgression among bovine species. Front. Genet. 2022, 13, 847492. [Google Scholar] [CrossRef]
- Neto, L.R.P.; Bunch, R.J.; Harrison, B.E.; Barendse, W. Variation in the XKR4 gene was significantly associated with subcutaneous rump fat thickness in indicine and composite cattle. Anim. Genet. 2012, 43, 785–789. [Google Scholar] [CrossRef]
- Cammack, K.M.; Thomas, M.G.; Enns, R.M. Reproductive traits and their heritabilities in beef cattle. Prof. Anim. Sci. 2009, 25, 517–528. [Google Scholar] [CrossRef]
- Sánchez, J.M.; Keogh, K.; Kelly, A.K.; Byrne, C.J.; Lonergan, P.; Kenny, D.A. A high plane of nutrition during early life alters the hypothalamic transcriptome of heifer calves. Sci. Rep. 2021, 11, 13978. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, Z.; Chen, Y.; Ding, H.; Fang, Y.; Fang, X.; Liu, H.; Guo, J.; Zhao, J.; Wang, J.; et al. ALKBH5 reduces BMP15 mRNA stability and regulates bovine puberty initiation through an m6A-dependent pathway. Int. J. Mol. Sci. 2024, 25, 11605. [Google Scholar] [CrossRef]
- Seminara, S.B.; Messager, S.; Chatzidaki, E.E.; Thresher, R.R.; Acierno, J.S.; Shagoury, J.K.; Bo-Abbas, Y.; Kuohung, W.; Schwinof, K.M.; Hendrick, A.G.; et al. The GPR54 gene as a regulator of puberty. N. Engl. J. Med. 2003, 349, 1614–1627. [Google Scholar] [CrossRef]
- Roa, J.; Tena-Sempere, M. Connecting metabolism and reproduction: Roles of central energy sensors and key molecular mediators. Mol. Cell. Endocrinol. 2014, 397, 4–14. [Google Scholar] [CrossRef]
- Hu, B.; Jin, H.; Shi, Y.; Yu, H.; Wu, X.; Wang, S.; Zhang, K. Single-cell RNA-Seq reveals the earliest lineage specification and X chromosome dosage compensation in bovine preimplantation embryos. FASEB J. 2024, 38, e23492. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Iii, W.M.M.; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M. Integrated analysis of multimodal single-cell data—ScienceDirect. Cell 2021, 184, 3573–3587.e3529. [Google Scholar] [CrossRef] [PubMed]
- Gasperini, M.; Hill, A.J.; José, L.M.-F.; Martin, B.; Kim, S.; Zhang, M.D.; Jackson, D.; Leith, A.; Schreiber, J.; Noble, W.S.; et al. A Genome-wide framework for mapping gene regulation via cellular genetic screens. Cell 2019, 176, 377–390.e319. [Google Scholar] [CrossRef]
- Kaya-Okur, H.S.; Wu, S.J.; Codomo, C.A.; Pledger, E.S.; Bryson, T.D.; Henikoff, J.G.; Ahmad, K.; Henikoff, S. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 2019, 10, 1930. [Google Scholar] [CrossRef]
- Buenrostro, J.D.; Giresi, P.G.; Zaba, L.C.; Chang, H.Y.; Greenleaf, W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 2013, 10, 1213–1218. [Google Scholar] [CrossRef]
- Cusanovich, D.A.; Daza, R.; Adey, A.; Pliner, H.A.; Christiansen, L.; Gunderson, K.L.; Steemers, F.J.; Trapnell, C.; Shendure, J. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 2015, 348, 910–914. [Google Scholar] [CrossRef]
- Chen, L.L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 2020, 21, 475–490. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.M.; Liu, H.; Zhuang, J.; Khan, N.M.; Zhang, D.; Chen, J.; Xu, T.; Avalos, L.F.C.; Zhou, X.; Zhang, Y. Circular RNA Expression and Regulation Profiling in Testicular Tissues of Immature and Mature Wandong Cattle. Front. Genet. 2021, 12, 685541. [Google Scholar] [CrossRef]
- Freitas, P.H.F.; Wang, Y.; Yan, P.; Oliveira, H.R.; Schenkel, F.S.; Zhang, Y.; Xu, Q.; Brito, L.F. Genetic diversity and signatures of selection for thermal stress in cattle and other two Bos species adapted to divergent climatic conditions. Front. Genet. 2021, 12, 604823. [Google Scholar] [CrossRef] [PubMed]
- Colombi, D.; Perini, F.; Bettini, S.; Mastrangelo, S.; Abeni, F.; Conte, G.; Marletta, D.; Cassandro, M.; Bernabucci, U.; Ciampolini, R.; et al. Genomic responses to climatic challenges in beef cattle: A review. Anim. Genet. 2024, 55, 854–870. [Google Scholar] [CrossRef]
- Low, W.Y.; Tearle, R.; Liu, R.; Koren, S.; Rhie, A.; Bickhart, D.M.; Rosen, B.D.; Kronenberg, Z.N.; Kingan, S.B.; Tseng, E.; et al. Haplotype-resolved genomes provide insights into structural variation and gene content in Angus and Brahman cattle. Nat. Commun. 2020, 11, 2071. [Google Scholar] [CrossRef]
- Tijjani, A.; Salim, B.; da Silva, M.V.B.; Eltahir, H.A.; Musa, T.H.; Marshall, K.; Hanotte, O.; Musa, H.H. Genomic signatures for drylands adaptation at gene-rich regions in African zebu cattle. Genomics 2022, 114, 110423. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Ayala, L.C.; Rocha, D.; Ramos-Onsins, S.E.; Leno-Colorado, J.; Charles, M.; Bouchez, O.; Rodríguez-Valera, Y.; Pérez-Enciso, M.; Ramayo-Caldas, Y. Whole-genome sequencing reveals insights into the adaptation of French Charolais cattle to Cuban tropical conditions. Genet. Sel. Evol. 2021, 53, 3. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Yang, L.; Zhu, B.; Zhang, W.; Wang, Z.; Chen, Y.; Zhang, L.; Gao, X.; Gao, H.; Liu, G.E.; et al. Genome-wide scan reveals genetic divergence and diverse adaptive selection in Chinese local cattle. BMC Genom. 2019, 20, 494. [Google Scholar] [CrossRef] [PubMed]
- Knap, P.W.; Doeschl-Wilson, A. Why breed disease-resilient livestock, and how? Genet. Sel. Evol. 2020, 52, 60. [Google Scholar] [CrossRef] [PubMed]
- Prat-Benhamou, A.; Meuwissen, M.; Slijper, T.; Bernués, A.; Gaspar-García, P.; Lizarralde, J.; Mancilla-Leytón, J.; Mandaluniz, N.; Mena, Y.; Soriano, B.; et al. A practical approach to assess the resilience attributes of livestock farms. Animal 2025, 19, 101566. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.-L.; Lin, J.; Huang, Y.-Y.; Gao, Q.-S.; Piao, Z.-Y.; Yuan, S.-L.; Chen, L.; Ren, X.; Ye, R.-C.; Dong, M.; et al. Population genomics reveals that natural variation in PRDM16 contributes to cold tolerance in domestic cattle. Zool. Res. 2022, 43, 275–284. [Google Scholar] [CrossRef]
- Ghoreishifar, S.M.; Eriksson, S.; Johansson, A.M.; Khansefid, M.; Moghaddaszadeh-Ahrabi, S.; Parna, N.; Davoudi, P.; Javanmard, A. Signatures of selection reveal candidate genes involved in economic traits and cold acclimation in five Swedish cattle breeds. Genet. Sel. Evol. 2020, 52, 52. [Google Scholar] [CrossRef]
- Huang, K.; Li, Z.; Zhong, D.; Yang, Y.; Yan, X.; Feng, T.; Wang, X.; Zhang, L.; Shen, X.; Chen, M.; et al. A circular RNA generated from Nebulin (NEB) gene splicing promotes skeletal muscle myogenesis in cattle as detected by a multi-Omics approach. Adv. Sci. 2023, 11, e2300702. [Google Scholar] [CrossRef]
- Cao, Y.; Jia, P.; Wu, Z.; Huang, M.; Chen, S.; Zhang, J.; Huang, B.; Lei, C. A novel SNP of MYO1A gene associated with heat-tolerance in Chinese cattle. Anim. Biotechnol. 2020, 33, 810–815. [Google Scholar] [CrossRef]
- Ma, X.; Liu, Y.; Sun, L.; Hanif, Q.; Qu, K.; Liu, J.; Zhang, J.; Huang, B.; Lei, C. A novel SNP of TECPR2 gene associated with heat tolerance in Chinese cattle. Anim. Biotechnol. 2021, 34, 1050–1057. [Google Scholar] [CrossRef]
- Zeng, L.; Chen, N.; Ning, Q.; Yao, Y.; Chen, H.; Dang, R.; Zhang, H.; Lei, C. PRLH and SOD1 gene variations associated with heat tolerance in Chinese cattle. Anim. Genet. 2018, 49, 447–451. [Google Scholar] [CrossRef]
- Mei, C.; Wang, H.; Liao, Q.; Wang, L.; Cheng, G.; Wang, H.; Zhao, C.; Zhao, S.; Song, J.; Guang, X.; et al. Genetic architecture and selection of Chinese cattle revealed by whole genome resequencing. Mol. Biol. Evol. 2018, 35, 688–699. [Google Scholar] [CrossRef]
- Wang, S.; Liu, J.; Zhao, W.; Wang, G.; Gao, S. Selection of candidate genes for differences in fat metabolism between cattle subcutaneous and perirenal adipose tissue based on RNA-seq. Anim. Biotechnol. 2021, 34, 633–644. [Google Scholar] [CrossRef]
- Gim, G.-M.; Uhm, K.-H.; Kwon, D.-H.; Kim, M.-J.; Jung, D.-J.; Kim, D.-H.; Yi, J.-K.; Ha, J.-J.; Yum, S.-Y.; Son, W.-J.; et al. Germline transmission of MSTN knockout cattle via CRISPR-Cas9. Theriogenology 2022, 192, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Messmer, T.; Dohmen, R.G.J.; Schaeken, L.; Melzener, L.; Hueber, R.; Godec, M.; Didoss, C.; Post, M.J.; Flack, J.E. Single-cell analysis of bovine muscle-derived cell types for cultured meat production. Front. Nutr. 2023, 10, 1212196. [Google Scholar] [CrossRef]
- Bai, F.; Cai, Y.; Qi, M.; Liang, C.; Pan, L.; Liu, Y.; Feng, Y.; Cao, X.; Yang, Q.; Ren, G.; et al. LCORL and STC2 variants increase body size and growth rate in cattle and other animals. Genom. Proteom. Bioinform. 2025, 23, qzaf025. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Zhao, R.; Chen, J.; Yan, Z.; Sui, X.; Li, H.; Li, Q.; Du, X.; Liu, Y.; Yao, S. Integrative transcriptomic, proteomic and metabolomic analyses yields insights into muscle fiber type in cattle. Food Chem. 2025, 468, 142479. [Google Scholar] [CrossRef]
- Song, Y.; Zhang, J.; Jiang, C.; Song, X.; Chen, X.; Raza, S.H.A.; Pant, S.D.; Ma, Y.; Zan, L.; Wei, D.; et al. Vitamin A mediates FABP4 to regulate intramuscular fat production: A new target and strategy for optimizing beef quality. BMC Genom. 2025, 26, 397. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.; An, B.; Deng, T.; Du, L.; Li, K.; Cao, S.; Du, Y.; Xu, L.; Zhang, L.; Gao, X.; et al. Incorporating genome-wide and transcriptome-wide association studies to identify genetic elements of longissimus dorsi muscle in Huaxi cattle. Front. Genet. 2023, 13, 982433. [Google Scholar] [CrossRef]
- Imai, K.; Keele, L.; Tingley, D. A general approach to causal mediation analysis. Psychol. Methods 2010, 15, 309–334. [Google Scholar] [CrossRef]
- Marrella, M.A.; Biase, F.H. A multi-omics analysis identifies molecular features associated with fertility in heifers (Bos taurus). Sci. Rep. 2023, 13, 12664. [Google Scholar] [CrossRef]
- Chen, N.; Cai, Y.; Chen, Q.; Li, R.; Wang, K.; Huang, Y.; Hu, S.; Huang, S.; Zhang, H.; Zheng, Z.; et al. Whole-genome resequencing reveals world-wide ancestry and adaptive introgression events of domesticated cattle in East Asia. Nat. Commun. 2018, 9, 2337. [Google Scholar] [CrossRef]
- Wang, L.; Gao, P.; Li, C.; Liu, Q.; Yao, Z.; Li, Y.; Zhang, X.; Sun, J.; Simintiras, C.; Welborn, M.; et al. A single-cell atlas of bovine skeletal muscle reveals mechanisms regulating intramuscular adipogenesis and fibrogenesis. J. Cachexia Sarcopenia Muscle 2023, 14, 2152–2167. [Google Scholar] [CrossRef]
- Jang, J.; Kim, K.; Lee, Y.H.; Kim, H. Population differentiated copy number variation of Bos taurus, Bos indicus and their African hybrids. BMC Genom. 2021, 22, 531. [Google Scholar] [CrossRef]
- Salehian-Dehkordi, H.; Xu, Y.X.; Xu, S.S.; Li, X.; Luo, L.Y.; Liu, Y.J.; Wang, D.F.; Cao, Y.H.; Shen, M.; Gao, L.; et al. Genome-wide detection of copy number variations and their association with distinct phenotypes in the world’s sheep. Front. Genet. 2021, 12, 670582. [Google Scholar] [CrossRef]
- Fodor, I.; Spoelstra, M.; Calus, M.P.L.; Kamphuis, C. A systematic review of genotype-by-climate interaction studies in cattle, pigs, and chicken. Front. Anim. Sci. 2023, 4, 1324830. [Google Scholar] [CrossRef]
- Filho, I.C.; Campos, G.S.; Lourenco, D.; Schenkel, F.S.; da Silva, D.A.; Silva, T.L.; Teixeira, C.S.; Fonseca, L.F.S.; Júnior, G.A.F.; de Albuquerque, L.G.; et al. Genotype by environment interaction for productive and reproductive traits in beef cattle using imputed whole genome sequence. J. Appl. Genet. 2025. [Google Scholar] [CrossRef] [PubMed]
- Sartori, C.; Tiezzi, F.; Guzzo, N.; Mancin, E.; Tuliozi, B.; Mantovani, R. Genotype by environment interaction and selection response for milk yield traits and conformation in a local cattle breed using a reaction norm approach. Animals 2022, 12, 839. [Google Scholar] [CrossRef] [PubMed]
- Jarquin, D.; Crossa, J.; Lacaze, X.; Du Cheyron, P.; Daucourt, J.; Lorgeou, J.; Piraux, F.; Guerreiro, L.; Perez, P.; Calus, M.; et al. A reaction norm model for genomic selection using high-dimensional genomic and environmental data. Theor. Appl. Genet. 2014, 127, 595–607. [Google Scholar] [CrossRef]
- MacLeod, I.M.; Bowman, P.J.; Vander Jagt, C.J.; Haile-Mariam, M.; Kemper, K.E.; Chamberlain, A.J.; Schrooten, C.; Hayes, B.J.; Goddard, M.E. Exploiting biological priors and sequence variants enhances QTL discovery and genomic prediction of complex traits. BMC Genom. 2016, 17, 144. [Google Scholar] [CrossRef]
- Teissier, M.; Larroque, H.; Robert-Granie, C. Weighted single-step genomic BLUP improves accuracy of genomic breeding values for protein content in French dairy goats: A quantitative trait influenced by a major gene. Genet. Sel. Evol. 2018, 50, 31. [Google Scholar] [CrossRef] [PubMed]
- Raymond, B.; Yengo, L.; Costilla, R.; Schrooten, C.; Bouwman, A.C.; Hayes, B.J.; Veerkamp, R.F.; Visscher, P.M. Using prior information from humans to prioritize genes and gene-associated variants for complex traits in livestock. PLoS Genet. 2020, 16, e1008780. [Google Scholar] [CrossRef]
- Wiggans, G.R.; Cole, J.B.; Hubbard, S.M.; Sonstegard, T.S. Genomic selection in dairy cattle: The USDA experience. Annu. Rev. Anim. Biosci. 2016, 5, 309–327. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Wang, J.; Zhang, K.; Cheng, G.; Mei, C.; Zan, L. Integrated multi-omics analysis reveals variation in intramuscular fat among muscle locations of Qinchuan cattle. BMC Genom. 2023, 24, 367. [Google Scholar] [CrossRef] [PubMed]
- Libbrecht, M.W.; Noble, W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015, 16, 321–332. [Google Scholar] [CrossRef]
- Gustavo, D.L.C.; Gianola, D.; Allison, D.B. Predicting genetic predisposition in humans: The promise of whole-genome markers. Nat. Rev. Genet. 2010, 11, 880–886. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Che, Y.; Liu, R.; Wang, Z.; Liu, W. Deep learning–driven multi-omics analysis: Enhancing cancer diagnostics and therapeutics. Brief. Bioinform. 2025, 26, bbaf440. [Google Scholar] [CrossRef]
- Montesinos-Lopez, O.A.; Montesinos-Lopez, A.; Perez-Rodriguez, P.; Barron-Lopez, J.A.; Martini, J.W.R.; Fajardo-Flores, S.B.; Gaytan-Lugo, L.S.; Santana-Mancilla, P.C.; Crossa, J. A review of deep learning applications for genomic selection. BMC Genom. 2021, 22, 19. [Google Scholar] [CrossRef]
- Jubair, S.; Domaratzki, M. Crop genomic selection with deep learning and environmental data: A survey. Front. Artif. Intell. 2022, 5, 1040295. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Y.; Li, D.; Ma, R.; Gao, Y.; Gao, Z.; Qian, Y.; Xi, D.; Deng, W.; Wu, J. Omics Evidence Chains for Complex Traits in Beef Cattle: From Cross-Layer Colocalization to Genetic Evaluation and Application. Biology 2025, 14, 1725. https://doi.org/10.3390/biology14121725
Lu Y, Li D, Ma R, Gao Y, Gao Z, Qian Y, Xi D, Deng W, Wu J. Omics Evidence Chains for Complex Traits in Beef Cattle: From Cross-Layer Colocalization to Genetic Evaluation and Application. Biology. 2025; 14(12):1725. https://doi.org/10.3390/biology14121725
Chicago/Turabian StyleLu, Ying, Dongfang Li, Ruoshan Ma, Yuyang Gao, Zhendong Gao, Yuwei Qian, Dongmei Xi, Weidong Deng, and Jiao Wu. 2025. "Omics Evidence Chains for Complex Traits in Beef Cattle: From Cross-Layer Colocalization to Genetic Evaluation and Application" Biology 14, no. 12: 1725. https://doi.org/10.3390/biology14121725
APA StyleLu, Y., Li, D., Ma, R., Gao, Y., Gao, Z., Qian, Y., Xi, D., Deng, W., & Wu, J. (2025). Omics Evidence Chains for Complex Traits in Beef Cattle: From Cross-Layer Colocalization to Genetic Evaluation and Application. Biology, 14(12), 1725. https://doi.org/10.3390/biology14121725






