Integrated Mitochondrial Genome and Transcriptomic Analyses Reveal Long Non-Coding RNAs Associated with Drought Tolerance in Sophora moorcroftiana
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials, DNA Isolation, and DNA Sequencing
2.2. Assembly and Annotation of the S. moorcroftiana Mitogenome
2.3. Detection of Repeats and Codon Usage Analysis
2.4. Phylogenetic Analysis
2.5. Transfer Fragment and Colinear Analysis
2.6. Identification and Distribution Analysis of RNA Editing Sites
2.7. Drought Treatments and Long Non-Coding RNA Transcriptome Assay
2.8. Measurement of Various Physiological Indices of S. moorcroftiana Seedlings Under Drought Stress
2.9. Statistical Analysis
3. Results
3.1. Mitogenome Assembly and Characterization
3.2. Repeat Sequences and DNA Transfer Analysis
3.3. Codon Usage Analysis of S. moorcroftiana Mitochondrial Genome
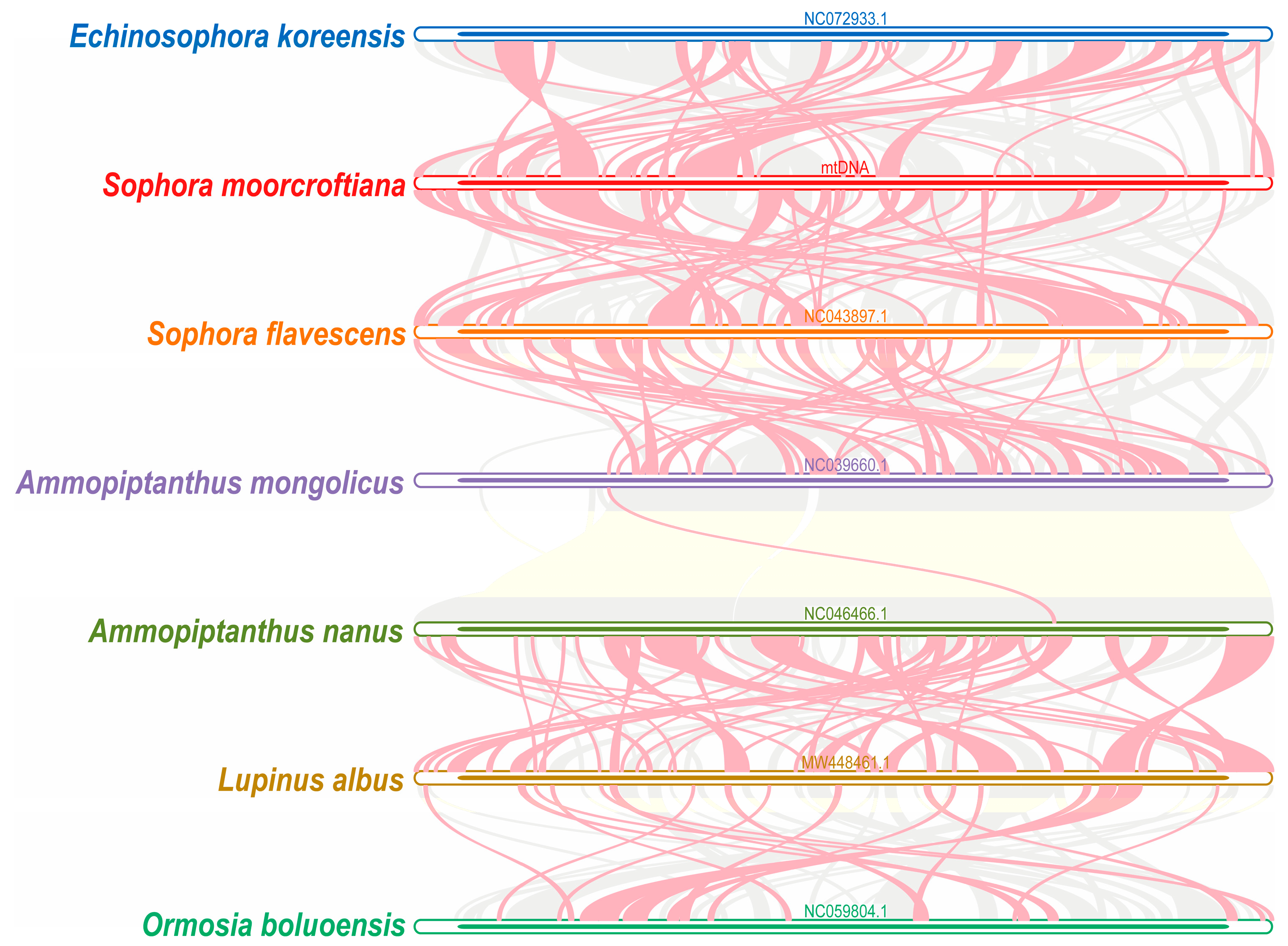
3.4. Phylogenetic and Collinearity Analysis
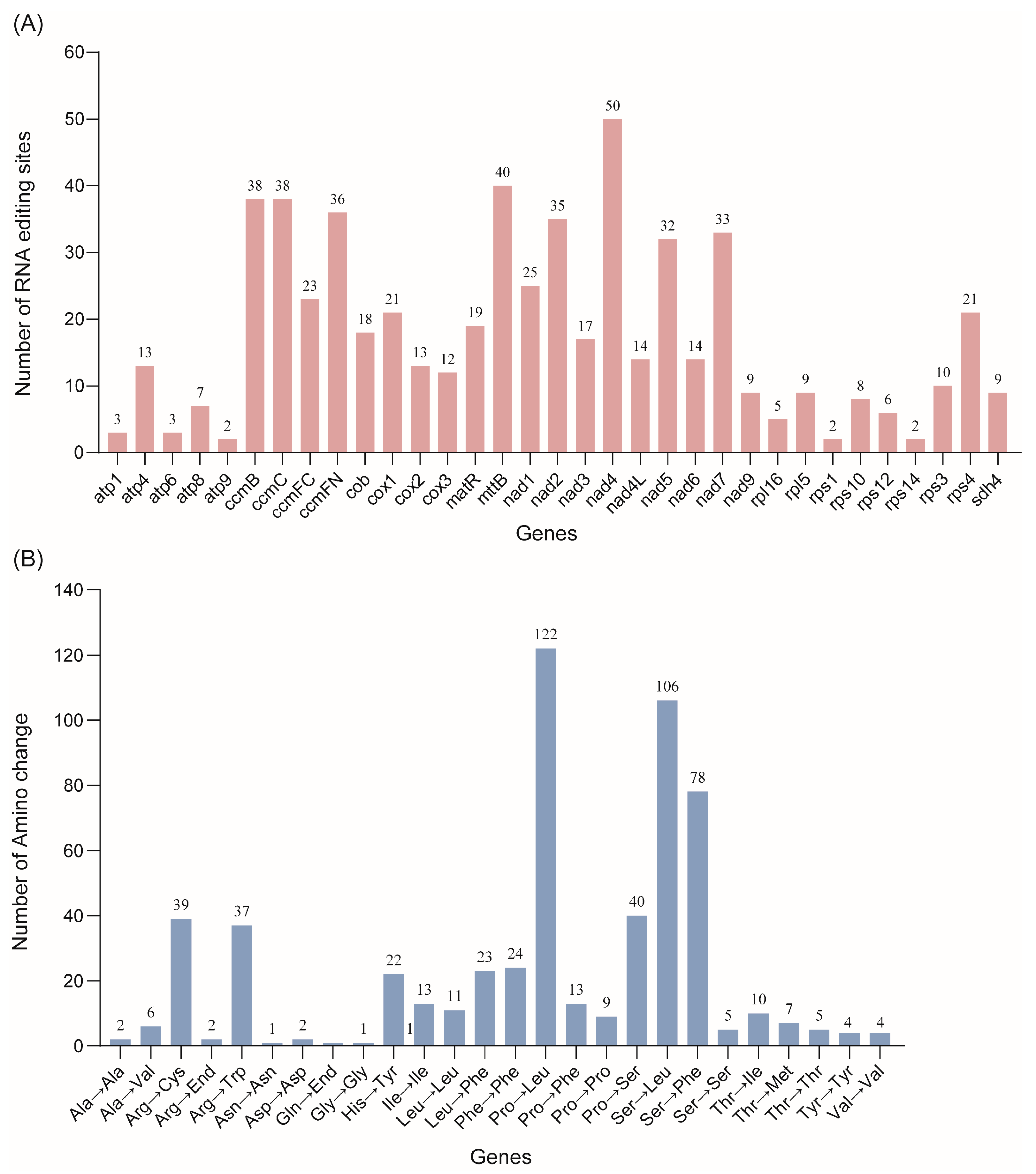
3.5. Characteristics and Distribution of RNA Editing Sites
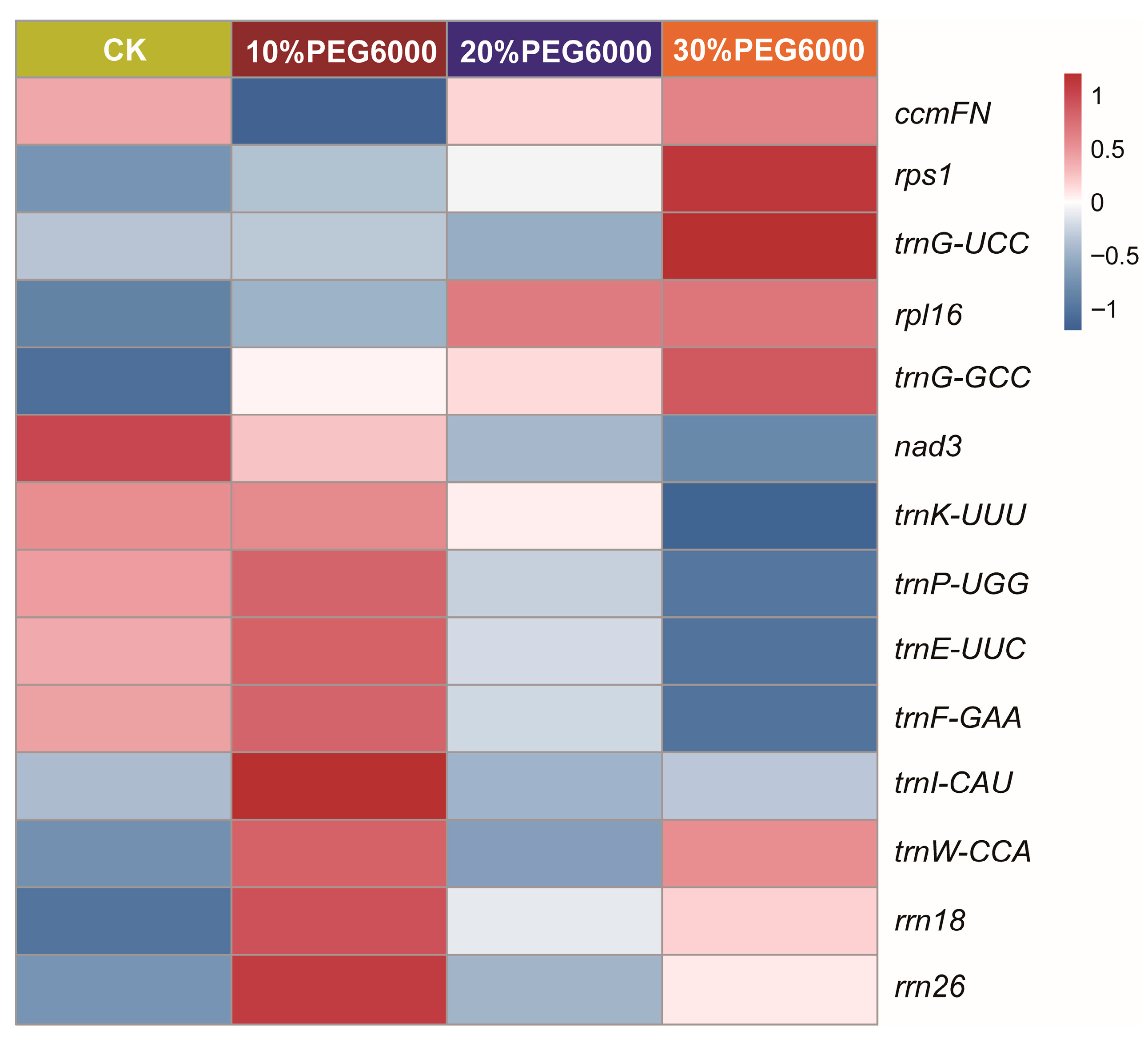
3.6. Comprehensive Analysis of the Mitogenome and lncRNAs Involved in Drought Tolerance of S. moorcroftiana
3.7. Physiological Responses of S. moorcroftiana Seedlings Under Drought Stress
4. Discussion
4.1. Characteristic of S. moorcroftiana and Its Mitogenome
4.2. RNA Editing Events in the S. moorcroftiana Mitogenome
4.3. Plant Mitochondria and the RNA Editing Process Regulate Drought Tolerance
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, Y.; Yi, F.; Yang, G.J.; Wang, Y.T.; Pubu, C.; He, R.H.; Xiao, Y.; Wang, J.C.; Lu, N.; Wang, J.H.; et al. Geographic population genetic structure and diversity of Sophora moorcroftiana based on genotyping-by-sequencing (GBS). PeerJ 2020, 8, e9609. [Google Scholar] [CrossRef]
- Dong, R.; Yang, S.; Wang, X.; Xie, L.; Ma, Y.; Wang, Y.; Zhang, L.; Zhang, M.; Qin, J. C:N:P stoichiometry in plant, soil and microbe in Sophora moorcroftiana shrubs across three sandy dune types in the middle reaches of the Yarlung Zangbo River. Front. Plant Sci. 2023, 13, 1060686. [Google Scholar] [CrossRef]
- Dong, R.; Guo, Q.Q.; Li, H.E.; Li, J.R.; Zuo, W.W.; Long, C. Estimation of morphological variation in seed traits of Sophora moorcroftiana using digital image analysis. Front. Plant Sci. 2023, 14, 1185393. [Google Scholar] [CrossRef]
- Liu, Y.; Ge, W.; Danzeng, L.; Wang, J.; Danzeng, N.; Ma, W.; Wang, Y.; Gesang, Q. Population genomics provides new insights into the genetic variation patterns, population demographic history, and high-altitude adaptation of Sophora moorcroftiana. BMC Plant Biol. 2025, 25, 899. [Google Scholar] [CrossRef]
- Seleiman, M.F.; Al-Suhaibani, N.; Ali, N.; Akmal, M.; Alotaibi, M.; Refay, Y.; Dindaroglu, T.; Abdul-Wajid, H.H.; Battaglia, M.L. Drought Stress Impacts on Plants and Different Approaches to Alleviate Its Adverse Effects. Plants 2021, 10, 259. [Google Scholar] [CrossRef] [PubMed]
- Sallam, A.; Alqudah, A.M.; Dawood, M.F.; Baenziger, P.S.; Börner, A. Drought stress tolerance in wheat and barley: Advances in physiology, breeding and genetics research. Int. J. Mol. Sci. 2019, 20, 3137. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.Q.; Zhang, W.H.; Li, H.E. Comparison of photosynthesis and antioxidative protection in Sophora moorcroftiana and Caragana maximovicziana under water stress. J. Arid. Land 2014, 6, 637–645. [Google Scholar] [CrossRef][Green Version]
- Li, H.E.; Zhang, Y.F.; Guo, Q.Q.; Yao, W.J. Molecular characterisation of a DREB gene from Sophora moorcroftiana, an endemic species of plateau. Protoplasma 2017, 254, 1735–1741. [Google Scholar] [CrossRef]
- Li, H.E.; Yao, W.J.; Fu, Y.R.; Li, S.K.; Guo, Q.Q. De Novo Assembly and Discovery of Genes That Are Involved in Drought Tolerance in Tibetan Sophora moorcroftiana. PLoS ONE 2015, 10, e111054. [Google Scholar] [CrossRef]
- Yin, X.; Yang, D.N.; Liu, Y.M.; Yang, S.H.; Zhang, R.; Sun, X.L.; Liu, H.X.; Duan, Y.W.; Yang, Y.Q.; Yang, Y.P. Sophora moorcroftiana genome analysis suggests association between sucrose metabolism and drought adaptation. Plant Physiol. 2023, 191, 844–848. [Google Scholar] [CrossRef]
- Postiglione, A.E.; Muday, G.K. Abscisic acid increases hydrogen peroxide in mitochondria to facilitate stomatal closure. Plant Physiol. 2023, 192, 469–487. [Google Scholar] [CrossRef]
- Bao, L.F.; Liu, J.H.; Mao, T.Y.; Zhao, L.B.; Wang, D.S.; Zhai, Y.L. Nanobiotechnology-mediated regulation of reactive oxygen species homeostasis under heat and drought stress in plants. Front. Plant Sci. 2024, 15, 1418515. [Google Scholar] [CrossRef]
- Igamberdiev, A.U.; Bykova, N.V. Mitochondria in photosynthetic cells: Coordinating redox control and energy balance. Plant Physiol. 2023, 191, 2104–2119. [Google Scholar] [CrossRef]
- Siqueira, J.A.; Hardoim, P.; Ferreira, P.C.G.; Nunes-Nesi, A.; Hemerly, A.S. Unraveling Interfaces between Energy Metabolism and Cell Cycle in Plants. Trends Plant Sci. 2018, 23, 731–747. [Google Scholar] [CrossRef] [PubMed]
- Van Aken, O. Mitochondrial redox systems as central hubs in plant metabolism and signaling. Plant Physiol. 2021, 186, 36–52. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.S.; Zhou, P.Y.; Tong, C.F.; Bi, C.W.; Xu, L.A. Assembly and analysis of the Populus deltoides mitochondrial genome: The first report of a multicircular mitochondrial conformation for the genus Populus. J. For. Res. 2023, 34, 717–733. [Google Scholar] [CrossRef]
- Wang, M.T.; Yang, J.P.; Hou, Z.Y.; Li, C.; Niu, Z.T.; Zhang, B.H.; Xue, Q.Y.; Liu, W.; Ding, X.Y. The multi-chromosomal structure of mitogenomes provided new insights into the accurate authentication of medicinal Dendrobium species. BMC Plant Biol. 2025, 25, 202. [Google Scholar] [CrossRef]
- Lu, G.L.; Li, Q. Complete mitochondrial genome of Syzygium samarangense reveals genomic recombination, gene transfer, and RNA editing events. Front. Plant Sci. 2024, 14, 1301164. [Google Scholar] [CrossRef]
- Lukeš, J.; Kaur, B.; Speijer, D. RNA Editing in Mitochondria and Plastids: Weird and Widespread. Trends Genet. 2021, 37, 99–102. [Google Scholar] [CrossRef]
- Wu, D.; Fu, W.T.; Fan, G.L.; Huang, D.F.; Wu, K.Y.; Zhan, Y.F.; Tu, X.M.; He, J.W. Characteristics and Comparative Analysis of the Special-Structure (Non-Single-Circle) Mitochondrial Genome of Capsicum pubescens Ruiz & Pav. Genes 2024, 15, 152. [Google Scholar]
- Li, J.L.; Zhang, L.H.; Li, C.Y.; Chen, W.J.; Wang, T.K.; Tan, L.; Qiu, Y.X.; Song, S.F.; Li, B.; Li, L. The Pentatricopeptide Repeat Protein OsPPR674 Regulates Rice Growth and Drought Sensitivity by Modulating RNA Editing of the Mitochondrial Transcript ccmC. Int. J. Mol. Sci. 2025, 26, 2646. [Google Scholar] [CrossRef] [PubMed]
- Che, R.H.; Tan, X.T.; Meng, X.N.; Li, H. ZmCYB5-1, a cytochrome b5 Gene, negatively regulates drought stress tolerance in maize. Gene 2025, 954, 149422. [Google Scholar] [CrossRef] [PubMed]
- Tel-Zur, N.; Abbo, S.; Myslabodski, D.; Mizrahi, Y. Modified CTAB procedure for DNA isolation from epiphytic cacti of the genera Hylocereus and Selenicereus (Cactaceae). Plant Mol. Biol. Rep. 1999, 17, 249–254. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Li, J.; Ni, Y.; Lu, Q.; Chen, H.; Liu, C. PMGA: A plant mitochondrial genome annotator. Plant Commun. 2025, 6, 101191. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Lewis, S.E.; Searle, S.M.J.; Harris, N.; Gibson, M.; Lyer, V.; Richter, J.; Wiel, C.; Bayraktaroglu, L.; Birney, E.; Crosby, M.A.; et al. Apollo: A sequence annotation editor. Genome Biol. 2002, 3, research0082.1. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Wynn, E.L.; Christensen, A.C. Repeats of Unusual Size in Plant Mitochondrial Genomes: Identification, Incidence and Evolution. G3 Genes Genomes Genet. 2019, 9, 549–559. [Google Scholar] [CrossRef]
- Behboudi, R.; Nouri-Baygi, M.; Naghibzadeh, M. RPTRF: A rapid perfect tandem repeat finder tool for DNA sequences. Biosystems 2023, 226, 104869. [Google Scholar] [CrossRef]
- Zhang, H.E.; Meltzer, P.; Davis, S. RCircos: An R package for Circos 2D track plots. BMC Bioinform. 2013, 14, 244. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.L.; Jakovlic, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Feng, Y.L.; Wang, J.; Zhu, S.S.; Jin, X.J.; Wu, Z.Q.; Zhang, Y.H. Comprehensive Analysis of the Complete Mitochondrial Genome of Rehmannia chingii: An Autotrophic Species in the Orobanchaceae Family. Genes 2024, 15, 98. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; dePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Shi, L.C.; Chen, H.M.; Jiang, M.; Wang, L.Q.; Wu, X.; Huang, L.F.; Liu, C. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 2019, 47, W65–W73. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; DeBarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.H.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Edera, A.A.; Small, I.; Milone, D.H.; Sanchez-Puerta, M.V. Deepred-Mt: Deep representation learning for predicting C-to-U RNA editing in plant mitochondria. Comput. Biol. Med. 2021, 136, 104682. [Google Scholar] [CrossRef] [PubMed]
- Picardi, E.; Pesole, G. REDItools: High-throughput RNA editing detection made easy. Bioinformatics 2013, 29, 1813–1814. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMB Net J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Frazee, A.C.; Pertea, G.; Jaffe, A.E.; Langmead, B.; Salzberg, S.L.; Leek, J.T. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat. Biotechnol. 2015, 33, 243–246. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 354–357. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef]
- Kang, Y.J.; Yang, D.C.; Kong, L.; Hou, M.; Meng, Y.Q.; Wei, L.; Gao, G. CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef]
- Li, A.; Zhang, J.; Zhou, Z. PLEK: A tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinform. 2014, 15, 311. [Google Scholar] [CrossRef]
- Punta, M.; Coggill, P.C.; Eberhardt, R.Y.; Mistry, J.; Tate, J.; Boursnell, C.; Pang, N.; Forslund, K.; Ceric, G.; Clements, J.; et al. The Pfam protein families database. Nucleic Acids Res. 2012, 40, D290–D301. [Google Scholar] [CrossRef]
- Lorenz, R.; Bernhart, S.H.; Höner Zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef]
- Tafer, H.; Hofacker, I.L. RNAplex: A fast tool for RNA-RNA interaction search. Bioinformatics 2008, 24, 2657–2663. [Google Scholar] [CrossRef] [PubMed]
- Gregori, J.; Villarreal, L.; Sánchez, A.; Baselga, J.; Villanueva, J. An effect size filter improves the reproducibility in spectral counting-based comparative proteomics. J. Proteom. 2013, 95, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Wu, Y.; Li, J.W.; Wang, X.; Zeng, Z.H.; Xu, J.; Liu, Y.L.; Feng, J.T.; Chen, H.; He, Y.H.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Kielkopf, C.L.; Bauer, W.; Urbatsch, I.L. Methods for Measuring the Concentrations of Proteins. Cold Spring Harb. Protoc. 2020, 2020, 102277. [Google Scholar] [CrossRef]
- Rashidi, M.A.; Falahi, S.; Farhang Dehghan, S.; Ebrahimzadeh, H.; Ghaneialvar, H.; Zendehdel, R. Green synthesis of silver nanoparticles by Smyrnium cordifolium plant and its application for colorimetric detection of ammonia. Sci. Rep. 2024, 14, 24161. [Google Scholar] [CrossRef]
- Ng, S.; De Clercq, I.; Van Aken, O.; Law, S.R.; Ivanova, A.; Willems, P.; Giraud, E.; Van Breusegem, F.; Whelan, J. Anterograde and Retrograde Regulation of Nuclear Genes Encoding Mitochondrial Proteins during Growth, Development, and Stress. Mol. Plant 2014, 7, 1075–1093. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- He, J.N.; Duan, Y.; Hua, D.P.; Fan, G.J.; Wang, L.; Liu, Y.; Chen, Z.Z.; Han, L.H.; Qu, L.J.; Gong, Z.Z. DEXH Box RNA Helicase-Mediated Mitochondrial Reactive Oxygen Species Production in Arabidopsis Mediates Crosstalk between Abscisic Acid and Auxin Signaling. Plant Cell 2012, 24, 1815–1833. [Google Scholar] [CrossRef]
- Lee, B.H.; Lee, H.J.; Xiong, L.M.; Zhu, J.K. A mitochondrial complex I defect impairs cold-regulated nuclear gene expression. Plant Cell 2002, 14, 1235–1251. [Google Scholar] [CrossRef] [PubMed]
- Dutilleul, C.; Garmier, M.; Noctor, G.; Mathieu, C.; Chétrit, P.; Foyer, C.H.; de Paepe, R. Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. Plant Cell 2003, 15, 1212–1226. [Google Scholar] [CrossRef] [PubMed]
- Small, I.D.; Schallenberg-Rüdinger, M.; Takenaka, M.; Mireau, H.; Ostersetzer-Biran, O. Plant organellar RNA editing: What 30 years of research has revealed. Plant J. 2020, 101, 1040–1056. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, A.; Wallet, C.; Iqbal, R.K.; Gualberto, J.M.; Lotfi, F. Organellar non-coding RNAs: Emerging regulation mechanisms. Biochimie 2015, 117, 48–62. [Google Scholar] [CrossRef]
- Mower, J.P. The PREP suite: Predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res. 2009, 37, W253–W259. [Google Scholar] [CrossRef]
- Salmans, M.L.; Chaw, S.M.; Lin, C.P.; Shih, A.C.C.; Wu, Y.W.; Mulligan, R.M. Editing site analysis in a gymnosperm mitochondrial genome reveals similarities with angiosperm mitochondrial genomes. Curr. Genet. 2010, 56, 439–446. [Google Scholar] [CrossRef]
- Meng, C.; Yang, M.; Wang, Y.; Chen, C.; Sui, N.; Meng, Q.; Zhuang, K.; Lv, W. SlWHY2 interacts with SlRECA2 to maintain mitochondrial function under drought stress in tomato. Plant Sci. 2020, 301, 110674. [Google Scholar] [CrossRef]
- Yang, Y.Z.; Ding, S.; Wang, H.C.; Sun, F.; Huang, W.L.; Song, S.; Xu, C.; Tan, B.C. The pentatricopeptide repeat protein EMP9 is required for mitochondrial ccmB and rps4 transcript editing, mitochondrial complex biogenesis and seed development in maize. New Phytol. 2017, 214, 782–795. [Google Scholar] [CrossRef]
- Ichinose, M.; Sugita, M. RNA Editing and Its Molecular Mechanism in Plant Organelles. Genes 2017, 8, 5. [Google Scholar] [CrossRef]
- Tang, W.; Luo, C. Molecular and Functional Diversity of RNA Editing in Plant Mitochondria. Mol. Biotechnol. 2018, 60, 935–945. [Google Scholar] [CrossRef]
- Guo, S.; Li, Z.Y.; Li, C.L.; Liu, Y.; Liang, X.L.; Qin, Y.M. Assembly and characterization of the complete mitochondrial genome of Ventilago leiocarpa. Plant Cell Rep. 2024, 43, 77. [Google Scholar] [CrossRef] [PubMed]
- Fujii, S.; Small, I. The evolution of RNA editing and pentatricopeptide repeat genes. New Phytol. 2011, 191, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Gommans, W.M.; Mullen, S.P.; Maas, S. RNA editing: A driving force for adaptive evolution? Bioessays 2009, 31, 1137–1145. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.L.; Liu, J.; Guo, W.J.; Zheng, Z.H.; Xu, Y.F.; Xia, H.J.; Xiao, T. Insights into the multi-chromosomal mitochondrial genome structure of the xero-halophytic plant Haloxylon Ammodendron (C.A.Mey.) Bunge ex Fenzl. BMC Genom. 2024, 25, 123. [Google Scholar] [CrossRef]
- Mohamed, N.G.; Ramadan, A.M.; Amer, M.; Morsy, Y.; Mohamed, R.A.; Said, O.A.M.; Alnufaei, A.A.; Ibrahim, M.I.M.; Hassanein, S.E.; Eissa, H.F. RNA editing-induced structural and functional adaptations of NAD9 in Triticum aestivum under drought stress. Front. Plant Sci. 2024, 15, 1490288. [Google Scholar] [CrossRef]
- Luo, Z.; Xiong, J.; Xia, H.; Wang, L.; Hou, G.; Li, Z.; Li, J.; Zhou, H.; Li, T.; Luo, L. Pentatricopeptide Repeat Gene-Mediated Mitochondrial RNA Editing Impacts on Rice Drought Tolerance. Front. Plant Sci. 2022, 13, 926285. [Google Scholar] [CrossRef]
- Fahad, S.; Bajwa, A.A.; Nazir, U.; Anjum, S.A.; Farooq, A.; Zohaib, A.; Sadia, S.; Nasim, W.; Adkins, S.; Saud, S.; et al. Crop Production under Drought and Heat Stress: Plant Responses and Management Options. Front. Plant Sci. 2017, 8, 1147. [Google Scholar] [CrossRef]
- Lamaoui, M.; Jemo, M.; Datla, R.; Bekkaoui, F. Heat and Drought Stresses in Crops and Approaches for Their Mitigation. Front. Chem. 2018, 6, 26. [Google Scholar] [CrossRef]
- Ghifari, A.S.; Saha, S.; Murcha, M.W. The biogenesis and regulation of the plant oxidative phosphorylation system. Plant Physiol. 2023, 192, 728–747. [Google Scholar] [CrossRef]
- Hu, Y.X.; Huang, A.; Li, Y.; Molloy, D.P.; Huang, C. Emerging roles of the C-to-U RNA editing in plant stress responses. Plant Sci. 2024, 349, 112263. [Google Scholar] [CrossRef]
- Ahmad, F.; Abdullah, M.; Khan, Z.; Stepien, P.; Rehman, S.U.; Akram, U.; Rahman, M.H.U.; Ali, Z.; Ahmad, D.; Gulzar, R.M.A.; et al. Genome-wide analysis and prediction of chloroplast and mitochondrial RNA editing sites of AGC gene family in cotton (Gossypium hirsutum L.) for abiotic stress tolerance. BMC Plant Biol. 2024, 24, 888. [Google Scholar] [CrossRef]
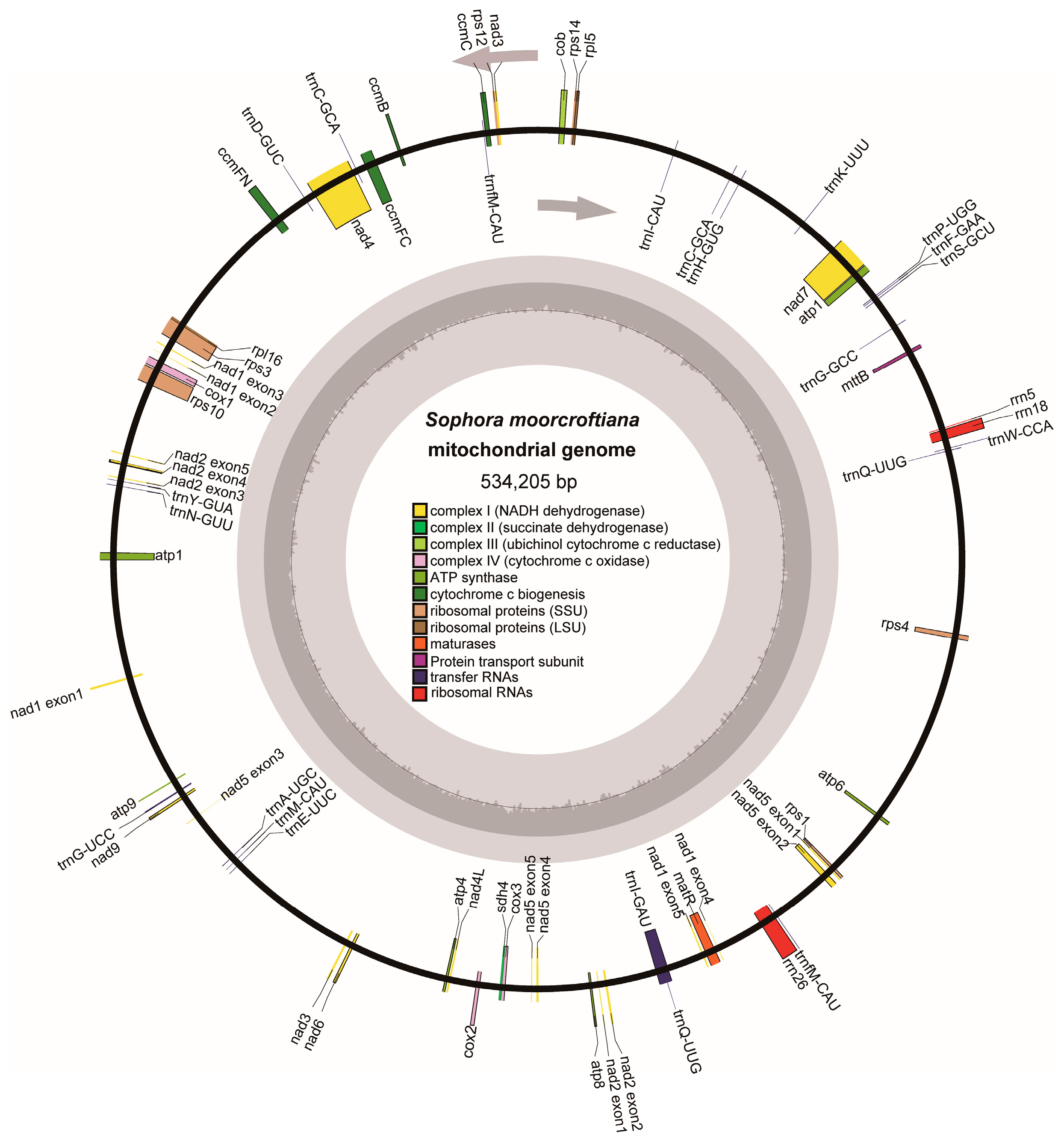
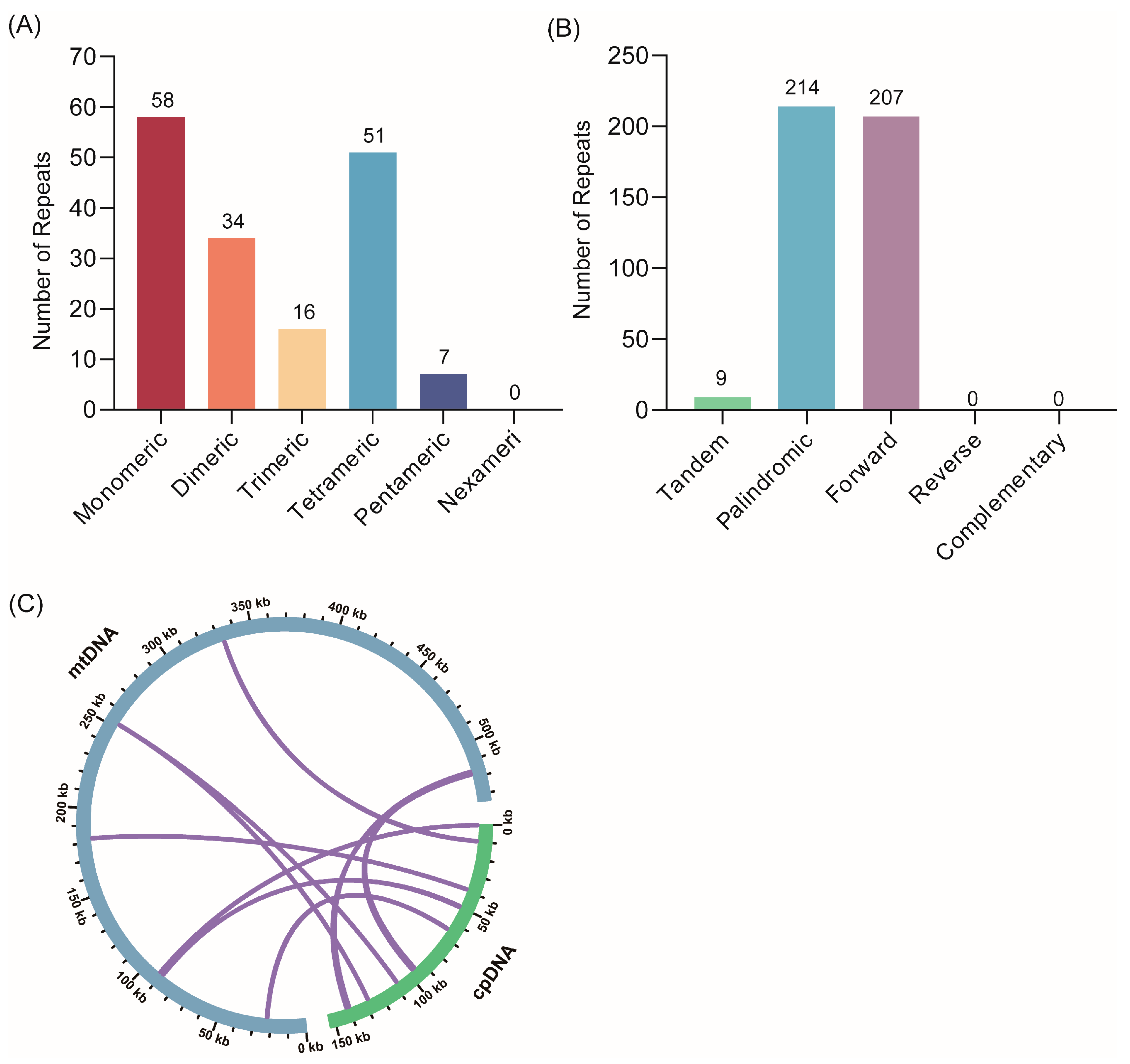
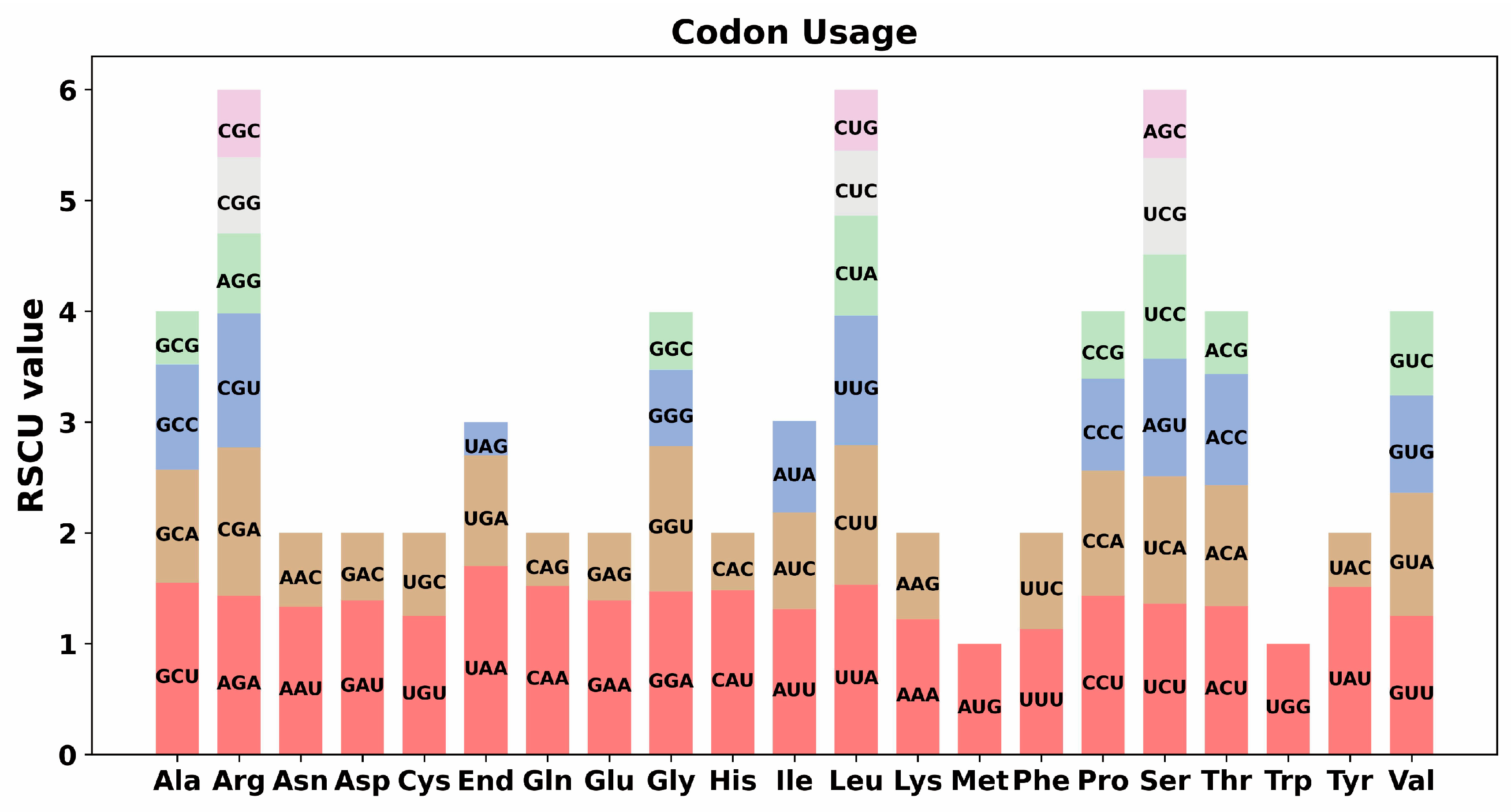
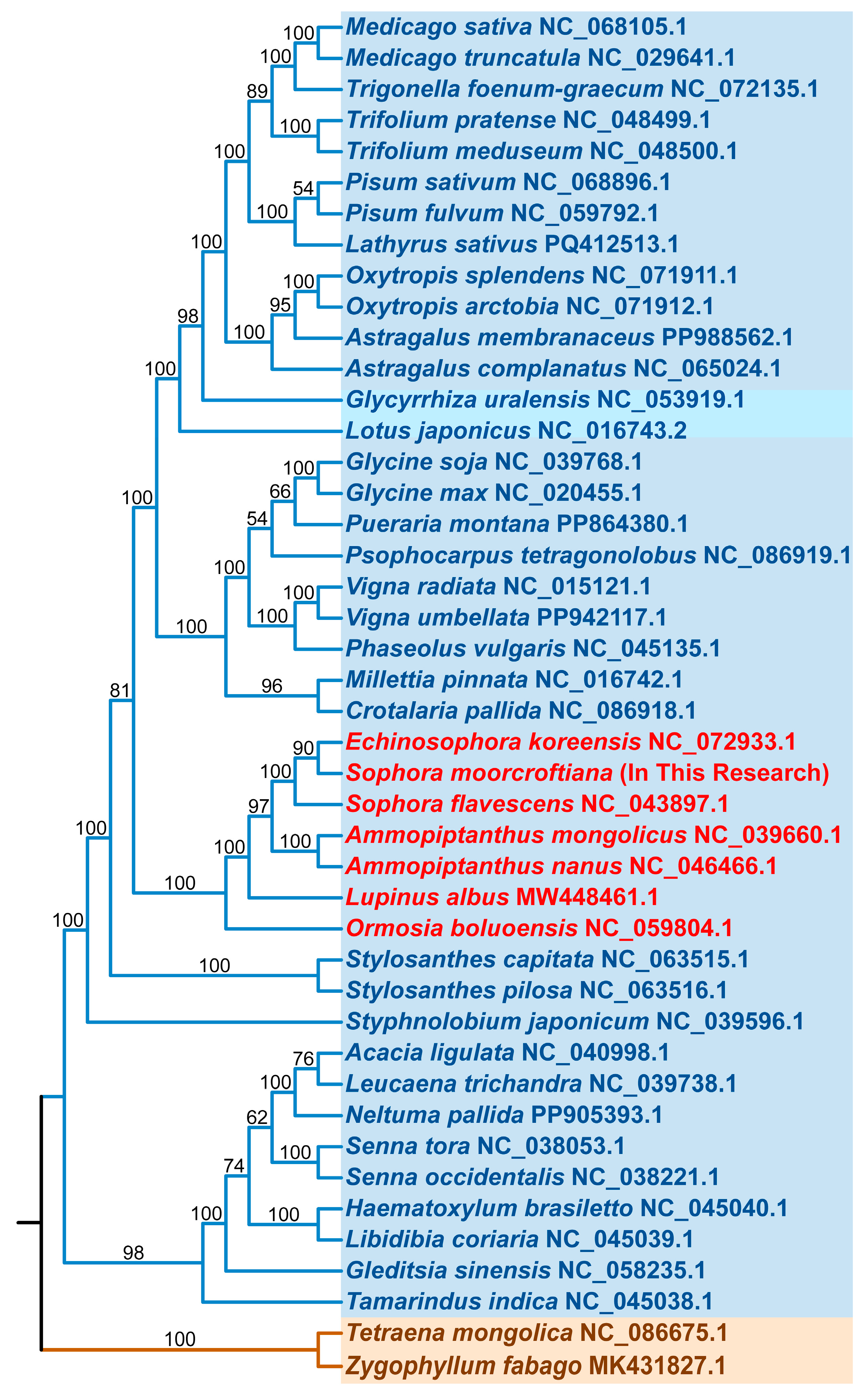
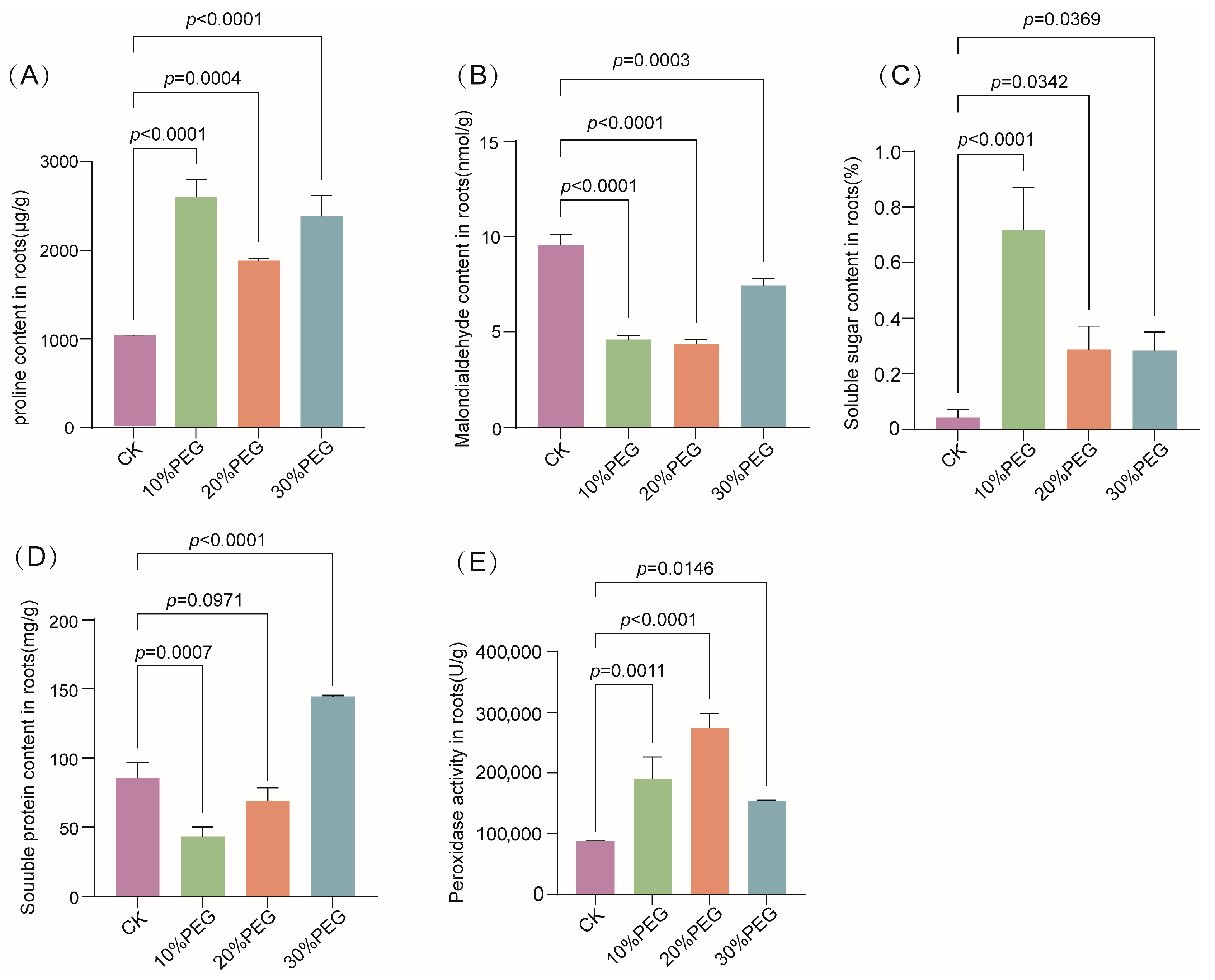
| Type | Mitochondrial Genome |
|---|---|
| Circular molecular number | 1 |
| Structure | circular |
| Total length | 534,205 bp |
| GC content | 44.93% |
| Depth (×) | 104 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, J.; Sun, Y.; Wang, Y.; Nan, J.; Gesang, Q.; Li, B. Integrated Mitochondrial Genome and Transcriptomic Analyses Reveal Long Non-Coding RNAs Associated with Drought Tolerance in Sophora moorcroftiana. Biology 2025, 14, 1711. https://doi.org/10.3390/biology14121711
Xu J, Sun Y, Wang Y, Nan J, Gesang Q, Li B. Integrated Mitochondrial Genome and Transcriptomic Analyses Reveal Long Non-Coding RNAs Associated with Drought Tolerance in Sophora moorcroftiana. Biology. 2025; 14(12):1711. https://doi.org/10.3390/biology14121711
Chicago/Turabian StyleXu, Jun, Yan Sun, Yuting Wang, Jibin Nan, Quzhen Gesang, and Bingzhang Li. 2025. "Integrated Mitochondrial Genome and Transcriptomic Analyses Reveal Long Non-Coding RNAs Associated with Drought Tolerance in Sophora moorcroftiana" Biology 14, no. 12: 1711. https://doi.org/10.3390/biology14121711
APA StyleXu, J., Sun, Y., Wang, Y., Nan, J., Gesang, Q., & Li, B. (2025). Integrated Mitochondrial Genome and Transcriptomic Analyses Reveal Long Non-Coding RNAs Associated with Drought Tolerance in Sophora moorcroftiana. Biology, 14(12), 1711. https://doi.org/10.3390/biology14121711






