A Deep Autoencoder Compression-Based Genomic Prediction Method for Whole-Genome Sequencing Data
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
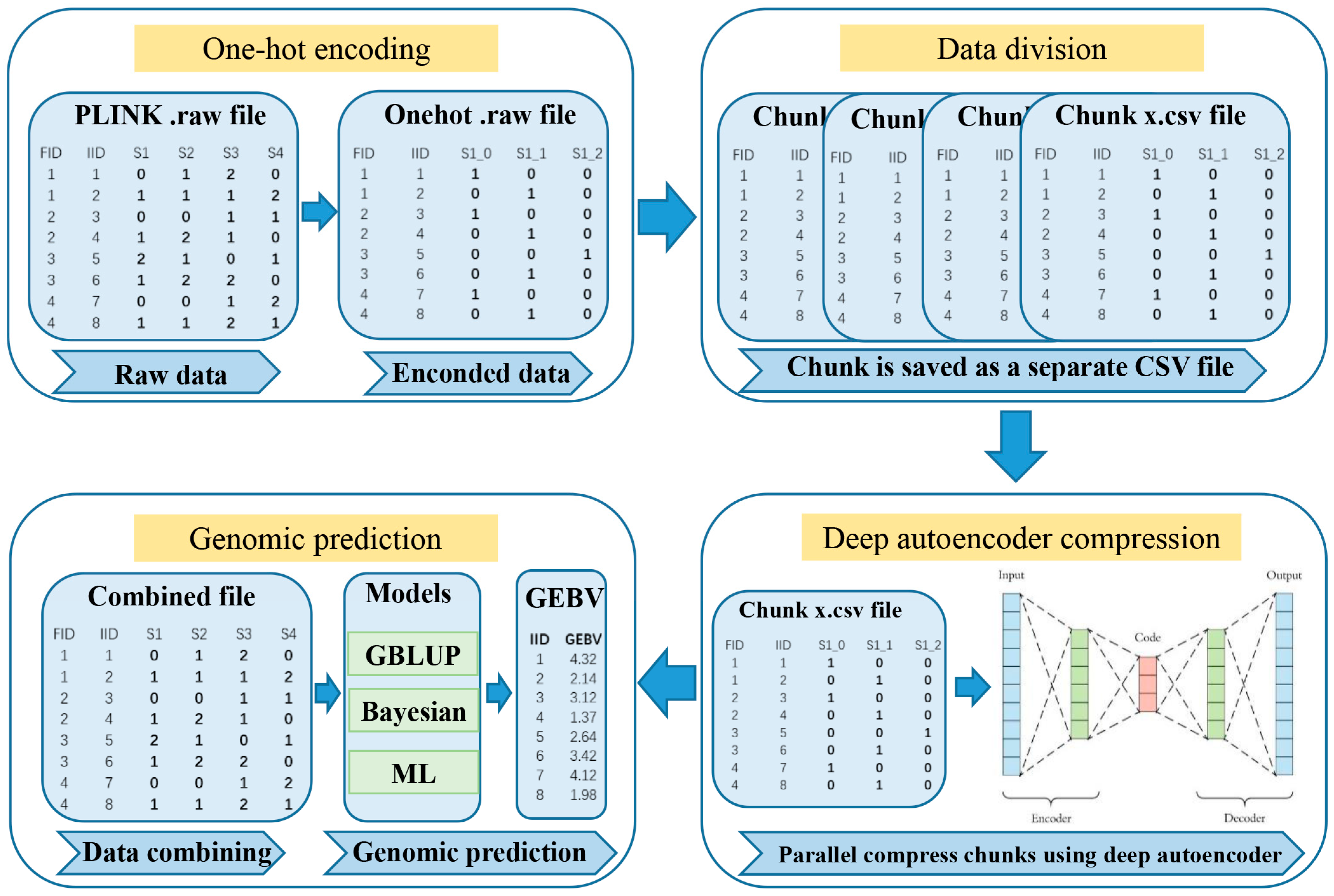
2.1. DAGP Framework
2.1.1. One-Hot Encoding and Data Division
2.1.2. Deep Autoencoder Compression
2.1.3. Genomic Prediction
GBLUP
Bayesian and Machine Learning Methods
2.2. Evaluation of the Performance of the DAGP Method
2.2.1. Sturgeon Dataset
2.2.2. Maize Dataset
2.2.3. Assessing Prediction Efficiency
3. Results
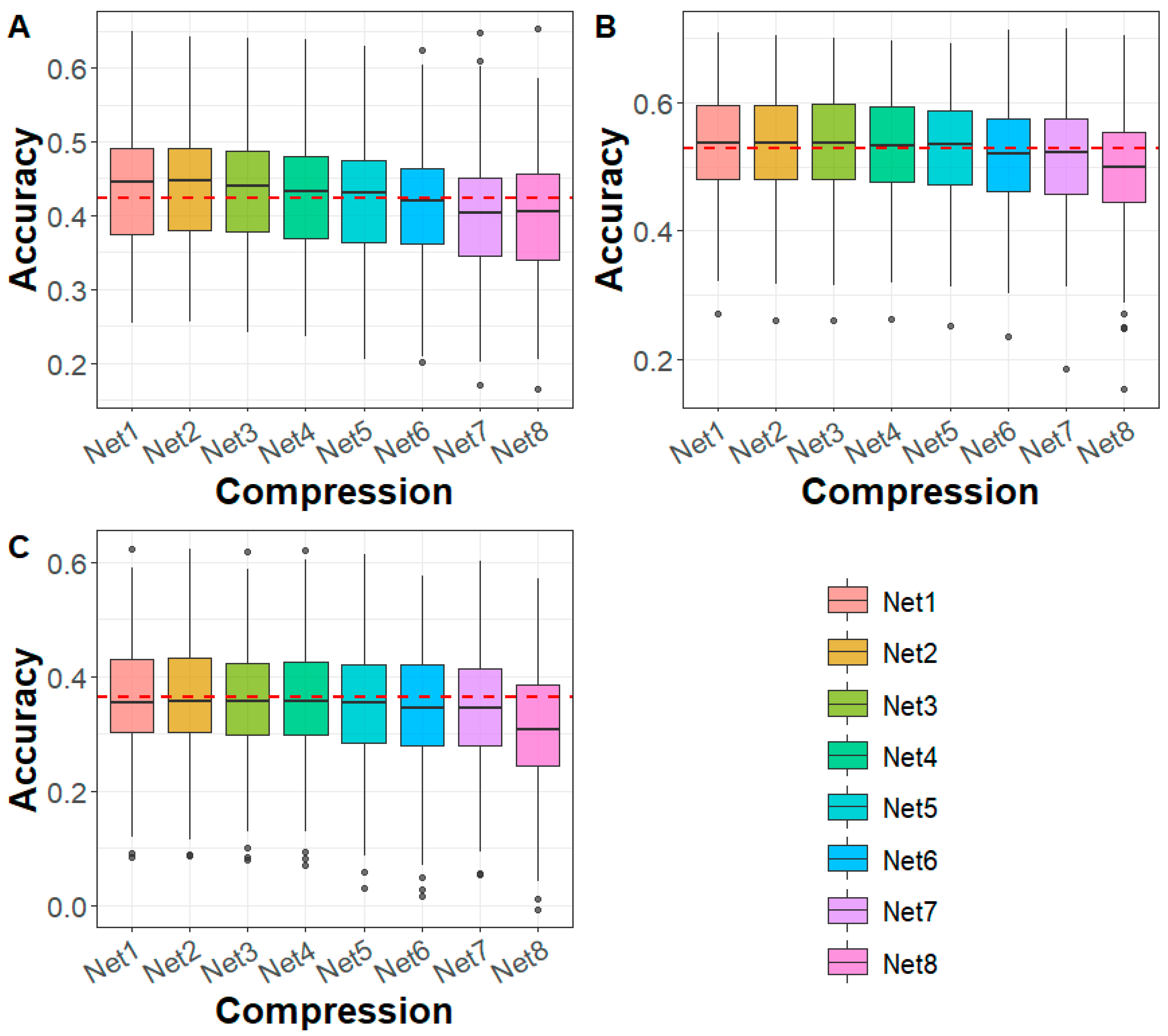
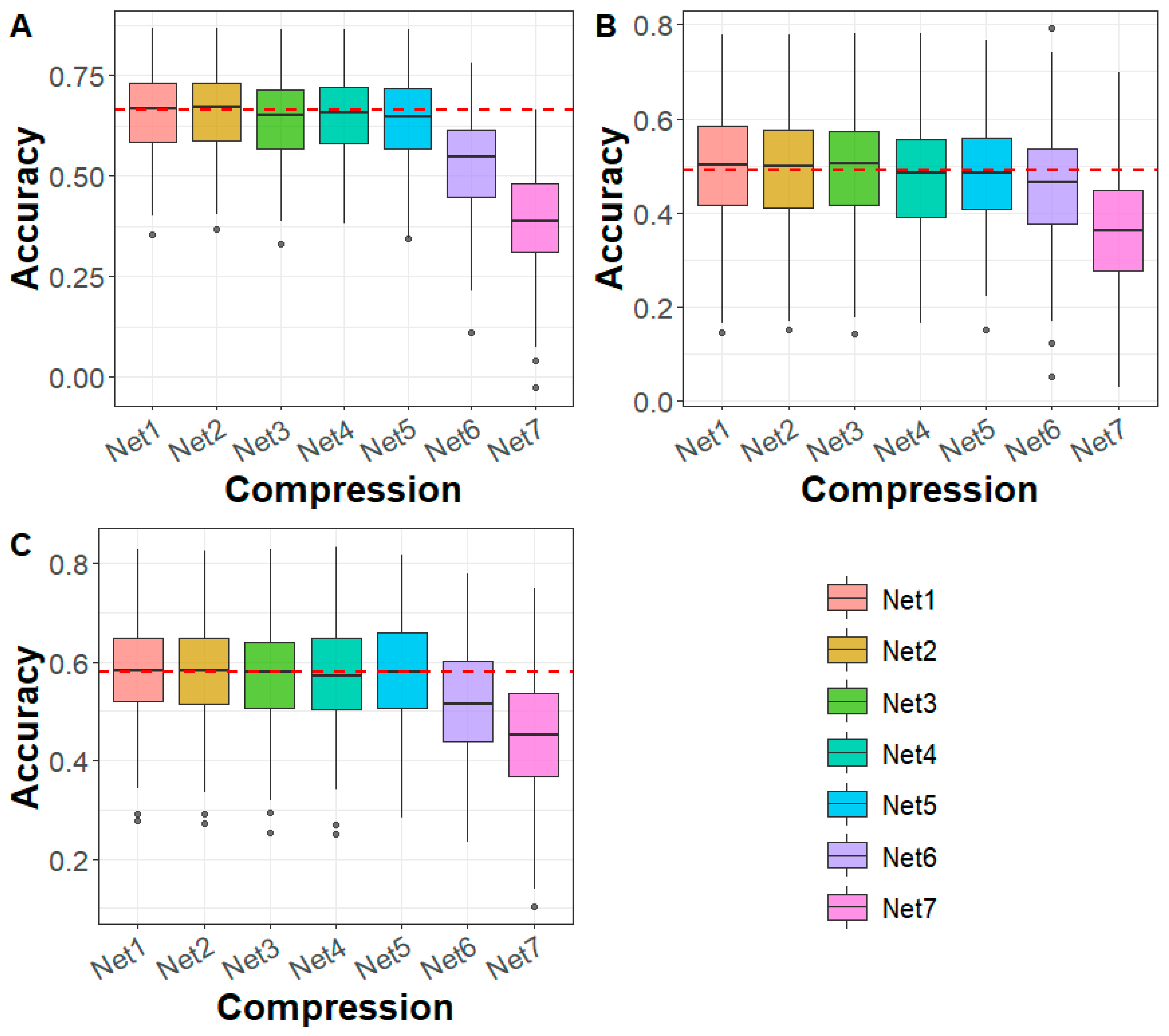
3.1. Compression of Whole-Genome Sequencing Data
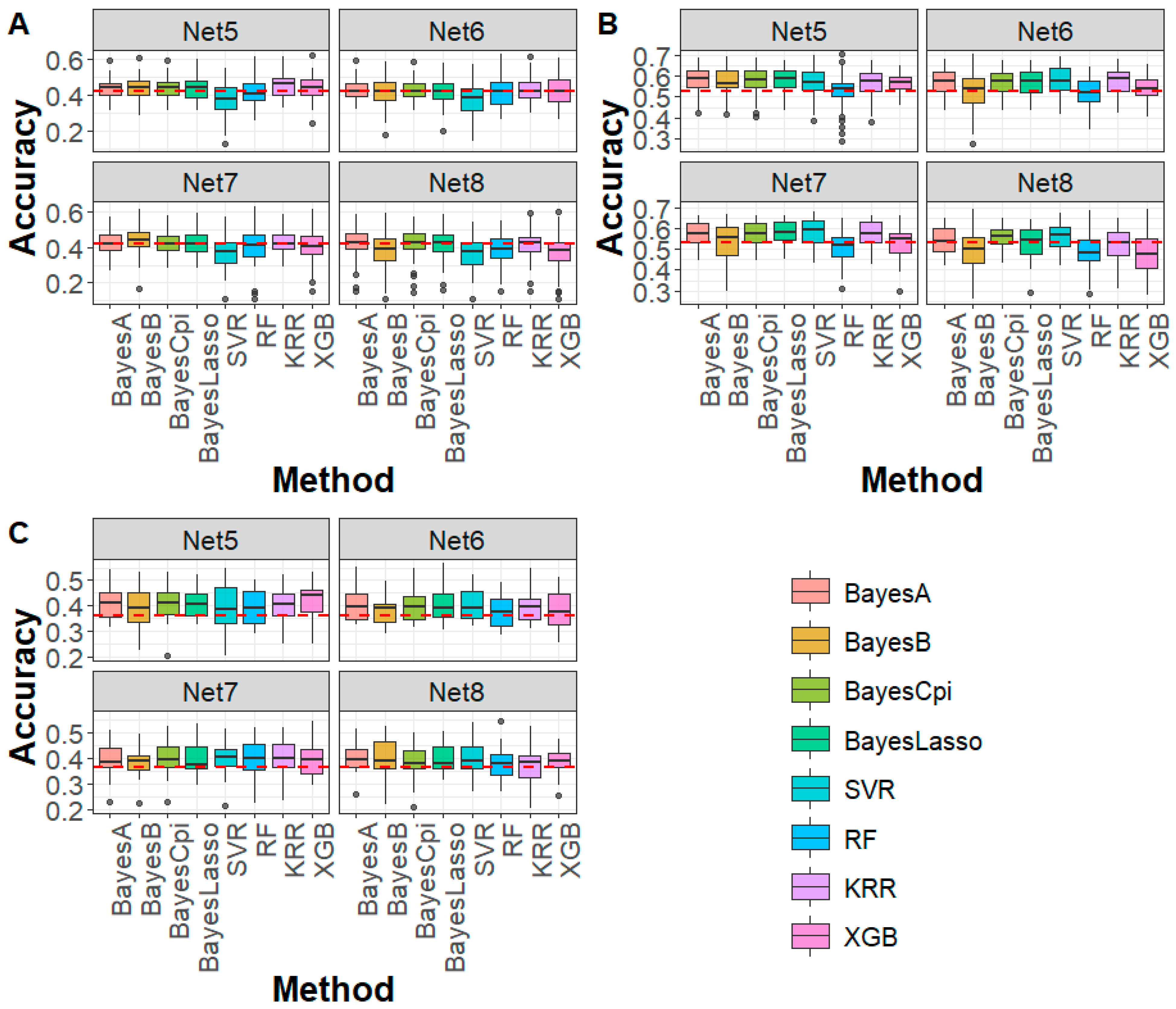
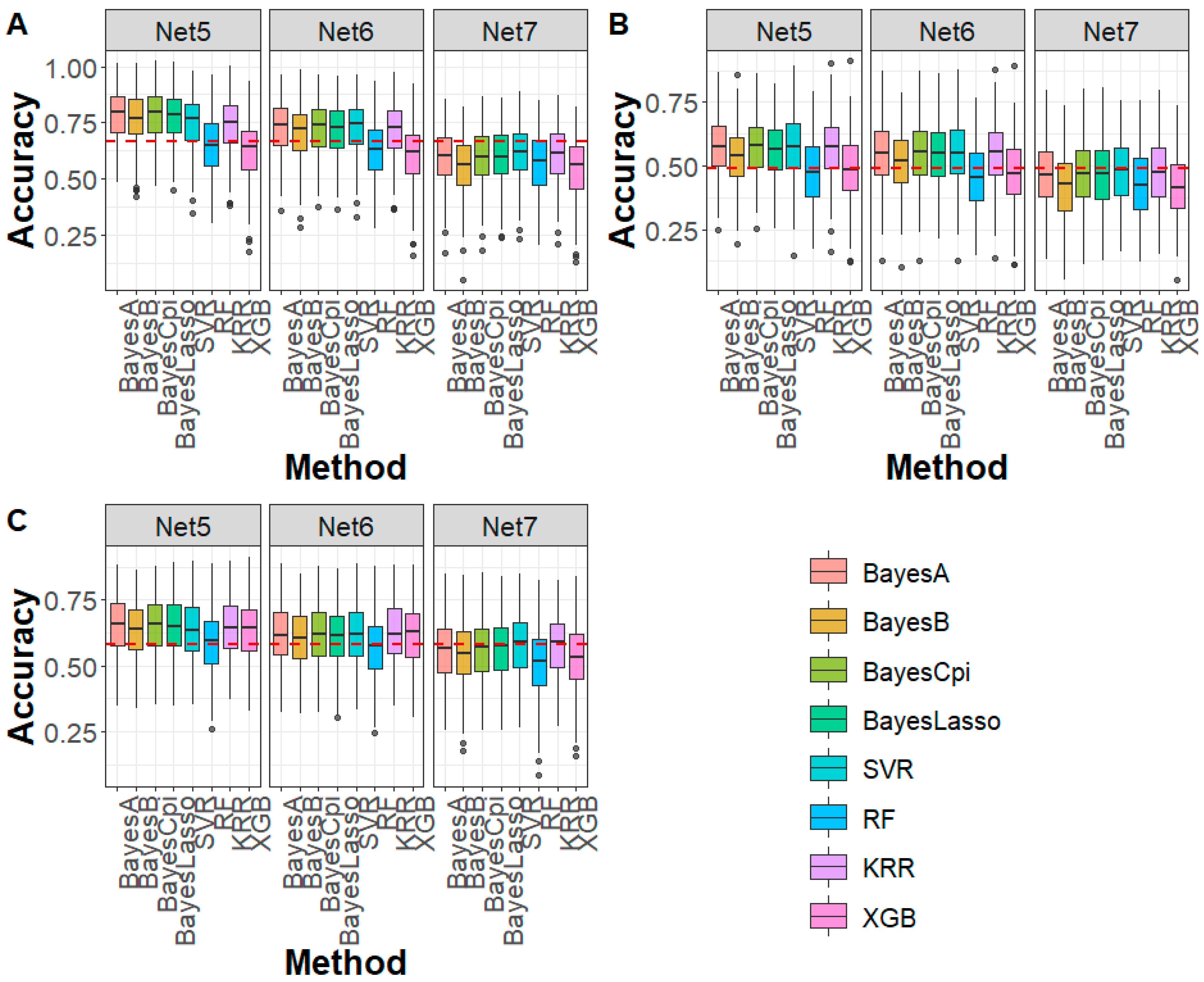
3.2. Performance Evaluation of DAGP
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Meuwissen, T.H.E.; Hayes, B.J.; Goddard, M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef]
- VanRaden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef] [PubMed]
- Montesinos-Lopez, O.A.; Montesinos-Lopez, A.; Perez-Rodriguez, P.; Barron-Lopez, J.A.; Martini, J.W.R.; Fajardo-Flores, S.B.; Gaytan-Lugo, L.S.; Santana-Mancilla, P.C.; Crossa, J. A review of deep learning applications for genomic selection. BMC Genom. 2021, 22, 19. [Google Scholar] [CrossRef]
- Meuwissen, T.; Hayes, B.; Goddard, M. Accelerating improvement of livestock with genomic selection. Annu. Rev. Anim. Biosci. 2013, 1, 221–237. [Google Scholar] [CrossRef]
- Raymond, B.; Bouwman, A.C.; Schrooten, C.; Houwing-Duistermaat, J.; Veerkamp, R.F. Utility of whole-genome sequence data for across-breed genomic prediction. Genet. Sel. Evol. 2018, 50, 27. [Google Scholar] [CrossRef]
- Ni, G.Y.; Cavero, D.; Fangmann, A.; Erbe, M.; Simianer, H. Whole-genome sequence-based genomic prediction in laying chickens with different genomic relationship matrices to account for genetic architecture. Genet. Sel. Evol. 2017, 49, 8. [Google Scholar] [CrossRef]
- Song, H.L.; Dong, T.; Yan, X.Y.; Wang, W.; Tian, Z.H.; Sun, A.; Dong, Y.; Zhu, H.; Hu, H.X. Genomic selection and its research progress in aquaculture breeding. Rev. Aquacult. 2023, 15, 274–291. [Google Scholar] [CrossRef]
- Alemu, A.; Astrand, J.; Montesinos-Lopez, O.A.; Isidro, Y.S.J.; Fernandez-Gonzalez, J.; Tadesse, W.; Vetukuri, R.R.; Carlsson, A.S.; Ceplitis, A.; Crossa, J.; et al. Genomic selection in plant breeding: Key factors shaping two decades of progress. Mol. Plant 2024, 17, 552–578. [Google Scholar] [CrossRef]
- Kramer, M.A. Nonlinear principal component analysis using autoassociative neural networks. AIChE J. 1991, 37, 233–243. [Google Scholar] [CrossRef]
- Lange, S.; Riedmiller, M. Deep auto-encoder neural networks in reinforcement learning. In Proceedings of the the 2010 International Joint Conference on Neural Networks (IJCNN), Barcelona, Spain, 18–23 July 2010; pp. 1–8. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Habier, D.; Fernando, R.L.; Kizilkaya, K.; Garrick, D.J. Extension of the bayesian alphabet for genomic selection. BMC Bioinform. 2011, 12, 186. [Google Scholar] [CrossRef]
- de los Campos, G.; Naya, H.; Gianola, D.; Crossa, J.; Legarra, A.; Manfredi, E.; Weigel, K.; Cotes, J.M. Predicting quantitative traits with regression models for dense molecular markers and pedigree. Genetics 2009, 182, 375–385. [Google Scholar] [CrossRef]
- Long, N.; Gianola, D.; Rosa, G.J.M.; Weigel, K.A. Application of support vector regression to genome-assisted prediction of quantitative traits. Theor. Appl. Genet. 2011, 123, 1065. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Douak, F.; Melgani, F.; Benoudjit, N. Kernel ridge regression with active learning for wind speed prediction. Appl. Energy 2013, 103, 328–340. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; Association for Computing Machinery: New York, NY, USA, 2016; pp. 785–794. [Google Scholar]
- Pérez, P.; de Los Campos, G. Genome-wide regression and prediction with the BGLR statistical package. Genetics 2014, 198, 483–495. [Google Scholar] [CrossRef]
- Song, H.; Dong, T.; Wang, W.; Jiang, B.; Yan, X.; Geng, C.; Bai, S.; Xu, S.; Hu, H. Cost-effective genomic prediction of critical economic traits in sturgeons through low-coverage sequencing. Genomics 2024, 116, 110874. [Google Scholar] [CrossRef]
- Du, K.; Stock, M.; Kneitz, S.; Klopp, C.; Woltering, J.M.; Adolfi, M.C.; Feron, R.; Prokopov, D.; Makunin, A.; Kichigin, I.; et al. The sterlet sturgeon genome sequence and the mechanisms of segmental rediploidization. Nat. Ecol. Evol. 2020, 4, 841–852. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Browning, B.L.; Zhou, Y.; Browning, S.R. A One-Penny Imputed Genome from Next-Generation Reference Panels. Am. J. Hum. Genet. 2018, 103, 338–348. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Lin, Z.; Li, X.; Zhao, Y.; Zhao, B.; Wu, G.; Ma, X.; Wang, H.; Xie, Y.; Li, Q.; et al. Genome-wide selection and genetic improvement during modern maize breeding. Nat. Genet. 2020, 52, 565–571. [Google Scholar] [CrossRef]
- Wang, T.; Chen, Y.P.; Goddard, M.E.; Meuwissen, T.H.; Kemper, K.E.; Hayes, B.J. A computationally efficient algorithm for genomic prediction using a Bayesian model. Genet. Sel. Evol. 2015, 47, 34. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Guo, T.; Yuan, C.; Liu, J.; Li, J.; Han, M.; Zhao, H.; Wu, Y.; Sun, W.; Wang, X.; et al. Evaluation of Bayesian alphabet and GBLUP based on different marker density for genomic prediction in Alpine Merino sheep. G3 (Bethesda) 2021, 11, jkab206. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Su, G.; Janss, L.; Zhang, Y.; Lund, M.S. Model comparison on genomic predictions using high-density markers for different groups of bulls in the Nordic Holstein population. J. Dairy Sci. 2013, 96, 4678–4687. [Google Scholar] [CrossRef]
- Joshi, R.; Skaarud, A.; Alvarez, A.T.; Moen, T.; Odegard, J. Bayesian genomic models boost prediction accuracy for survival to Streptococcus agalactiae infection in Nile tilapia (Oreochromus nilioticus). Genet. Sel. Evol. 2021, 53, 37. [Google Scholar] [CrossRef]
- Song, H.; Dong, T.; Yan, X.; Wang, W.; Tian, Z.; Hu, H. Using Bayesian threshold model and machine learning method to improve the accuracy of genomic prediction for ordered categorical traits in fish. Agric. Commun. 2023, 1, 100005. [Google Scholar] [CrossRef]
- Chafai, N.; Hayah, I.; Houaga, I.; Badaoui, B. A review of machine learning models applied to genomic prediction in animal breeding. Front. Genet. 2023, 14, 1150596. [Google Scholar] [CrossRef]
- Wang, X.; Shi, S.L.; Wang, G.J.; Luo, W.X.; Wei, X.; Qiu, A.; Luo, F.; Ding, X.D. Using machine learning to improve the accuracy of genomic prediction of reproduction traits in pigs. J. Anim. Sci. Biotechno. 2022, 13, 60. [Google Scholar] [CrossRef]
- Nazzicari, N.; Biscarini, F. Stacked kinship CNN vs. GBLUP for genomic predictions of additive and complex continuous phenotypes. Sci. Rep. 2022, 12, 19889. [Google Scholar] [CrossRef]
- Kick, D.R.; Wallace, J.G.; Schnable, J.C.; Kolkman, J.M.; Alaca, B.; Beissinger, T.M.; Edwards, J.; Ertl, D.; Flint-Garcia, S.; Gage, J.L.; et al. Yield prediction through integration of genetic, environment, and management data through deep learning. G3 (Bethesda) 2023, 13, jkad006. [Google Scholar] [CrossRef]
- Song, H.; Dong, T.; Wang, W.; Yan, X.; Geng, C.; Bai, S.; Hu, H. GWAS Enhances Genomic Prediction Accuracy of Caviar Yield, Caviar Color and Body Weight Traits in Sturgeons Using Whole-Genome Sequencing Data. Int. J. Mol. Sci. 2024, 25, 9756. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Wang, W.; Dong, T.; Yan, X.; Geng, C.; Bai, S.; Hu, H. Prioritized SNP Selection from Whole-Genome Sequencing Improves Genomic Prediction Accuracy in Sturgeons Using Linear and Machine Learning Models. Int. J. Mol. Sci. 2025, 26, 7007. [Google Scholar] [CrossRef] [PubMed]
| Datasets | Trait a | Sample Size | Mean | Standard Deviation | Coefficient of Variation (%) |
|---|---|---|---|---|---|
| Sturgeon | CY | 673 | 0.190 | 0.057 | 30.000 |
| CC | 673 | 2.453 | 0.653 | 26.620 | |
| BW | 673 | 19.933 | 4.029 | 20.213 | |
| Maize | DTA | 350 | 67.319 | 4.408 | 6.548 |
| EP | 350 | 36.084 | 5.304 | 14.699 | |
| TBN | 350 | 10.207 | 4.441 | 43.511 |
| Datasets | Compression | Hyperparameters | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Neurons | Batch Size | Epochs | Number of Split Files | Original Dimension (p) | Compressed Dimension (d) | MSE | Compression Ratio (%) | ||
| Sturgeon | Net 1 | 312, 250, 200, 100 | 52 | 200 | 100,000 | 10,409,793 | 10,000,000 | 0.027 | 3.94 |
| Net 2 | 100, 80, 64, 20 | 32 | 200 | 100,000 | 10,409,793 | 2,000,000 | 0.022 | 80.79 | |
| Net 3 | 20, 16, 12, 5 | 32 | 200 | 100,000 | 10,409,793 | 500,000 | 0.110 | 95.20 | |
| Net 4 | 50, 40, 30, 20 | 32 | 200 | 10,000 | 10,409,793 | 200,000 | 0.091 | 98.08 | |
| Net 5 | 20, 16, 12, 5 | 32 | 200 | 10,000 | 10,409,793 | 50,000 | 0.124 | 99.52 | |
| Net 6 | 50, 40, 30, 20 | 32 | 200 | 1000 | 10,409,793 | 20,000 | 0.098 | 99.81 | |
| Net 7 | 20, 16, 12, 10 | 32 | 200 | 1000 | 10,409,793 | 10,000 | 0.105 | 99.90 | |
| Net 8 | 10, 8, 6, 5 | 32 | 200 | 1000 | 10,409,793 | 5000 | 0.113 | 99.95 | |
| Maize | Net 1 | 79, 63, 50, 20 | 32 | 200 | 100,000 | 2,663,873 | 2,000,000 | 0.036 | 24.92 |
| Net 2 | 20, 16, 12, 5 | 32 | 200 | 100,000 | 2,663,873 | 500,000 | 0.060 | 81.23 | |
| Net 3 | 50, 40, 30, 20 | 32 | 200 | 10,000 | 2,663,873 | 200,000 | 0.107 | 92.49 | |
| Net 4 | 20, 16, 12, 5 | 32 | 200 | 10,000 | 2,663,873 | 50,000 | 0.130 | 98.12 | |
| Net 5 | 50, 40, 30, 20 | 32 | 200 | 1000 | 2,663,873 | 20,000 | 0.129 | 99.25 | |
| Net 6 | 20, 16, 12, 10 | 32 | 200 | 1000 | 2,663,873 | 10,000 | 0.124 | 99.62 | |
| Net 7 | 10, 8, 6, 5 | 32 | 200 | 1000 | 2,663,873 | 5000 | 0.123 | 99.81 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, H.; Dong, T.; Wang, W.; Yan, X.; Geng, C.; Bai, S.; Hu, H. A Deep Autoencoder Compression-Based Genomic Prediction Method for Whole-Genome Sequencing Data. Biology 2025, 14, 1622. https://doi.org/10.3390/biology14111622
Song H, Dong T, Wang W, Yan X, Geng C, Bai S, Hu H. A Deep Autoencoder Compression-Based Genomic Prediction Method for Whole-Genome Sequencing Data. Biology. 2025; 14(11):1622. https://doi.org/10.3390/biology14111622
Chicago/Turabian StyleSong, Hailiang, Tian Dong, Wei Wang, Xiaoyu Yan, Chenfan Geng, Song Bai, and Hongxia Hu. 2025. "A Deep Autoencoder Compression-Based Genomic Prediction Method for Whole-Genome Sequencing Data" Biology 14, no. 11: 1622. https://doi.org/10.3390/biology14111622
APA StyleSong, H., Dong, T., Wang, W., Yan, X., Geng, C., Bai, S., & Hu, H. (2025). A Deep Autoencoder Compression-Based Genomic Prediction Method for Whole-Genome Sequencing Data. Biology, 14(11), 1622. https://doi.org/10.3390/biology14111622






