Pan-Mitogenome Construction, Intraspecific Variation, and Adaptive Evolution of the Plant Pathogenic Fungus Claviceps purpurea
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Mitogenome Sequencing, Assembly, and Annotation
2.3. Comparative Analysis of Mitogenome
2.4. Phylogenetic Analysis
2.5. Pan-Mitogenome Analysis
3. Results
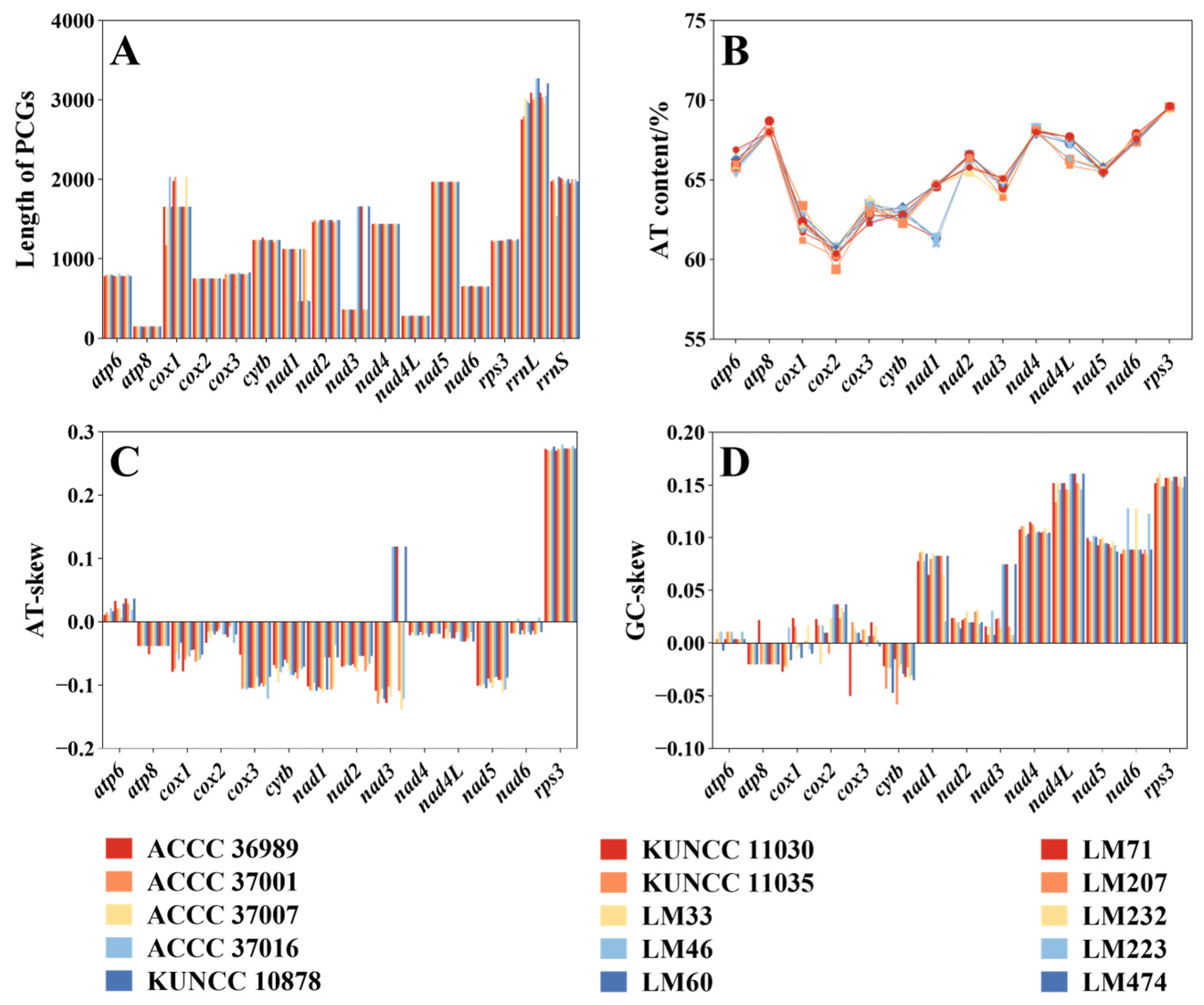
3.1. Mitogenome Characteristics
3.2. Codon Usage
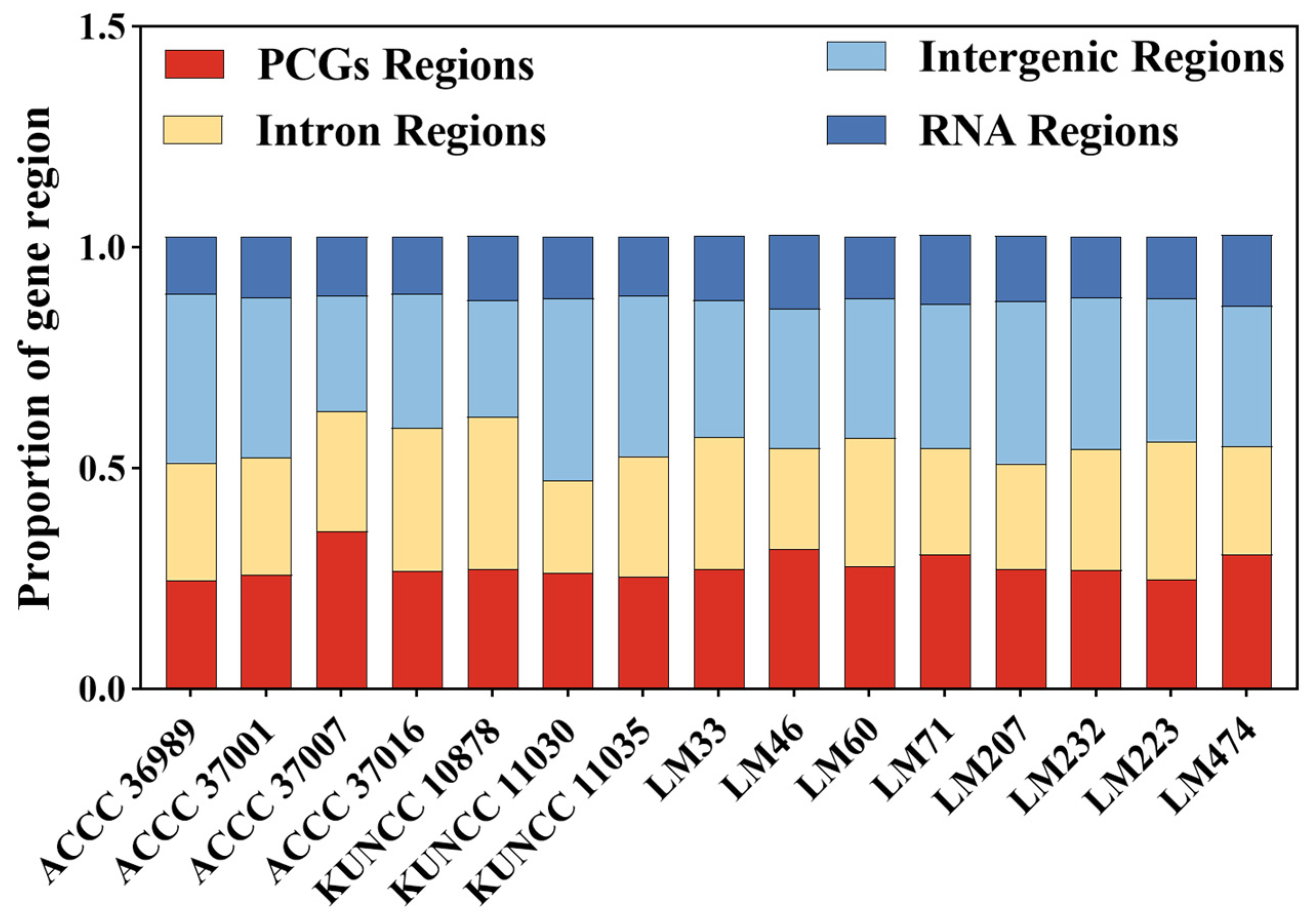
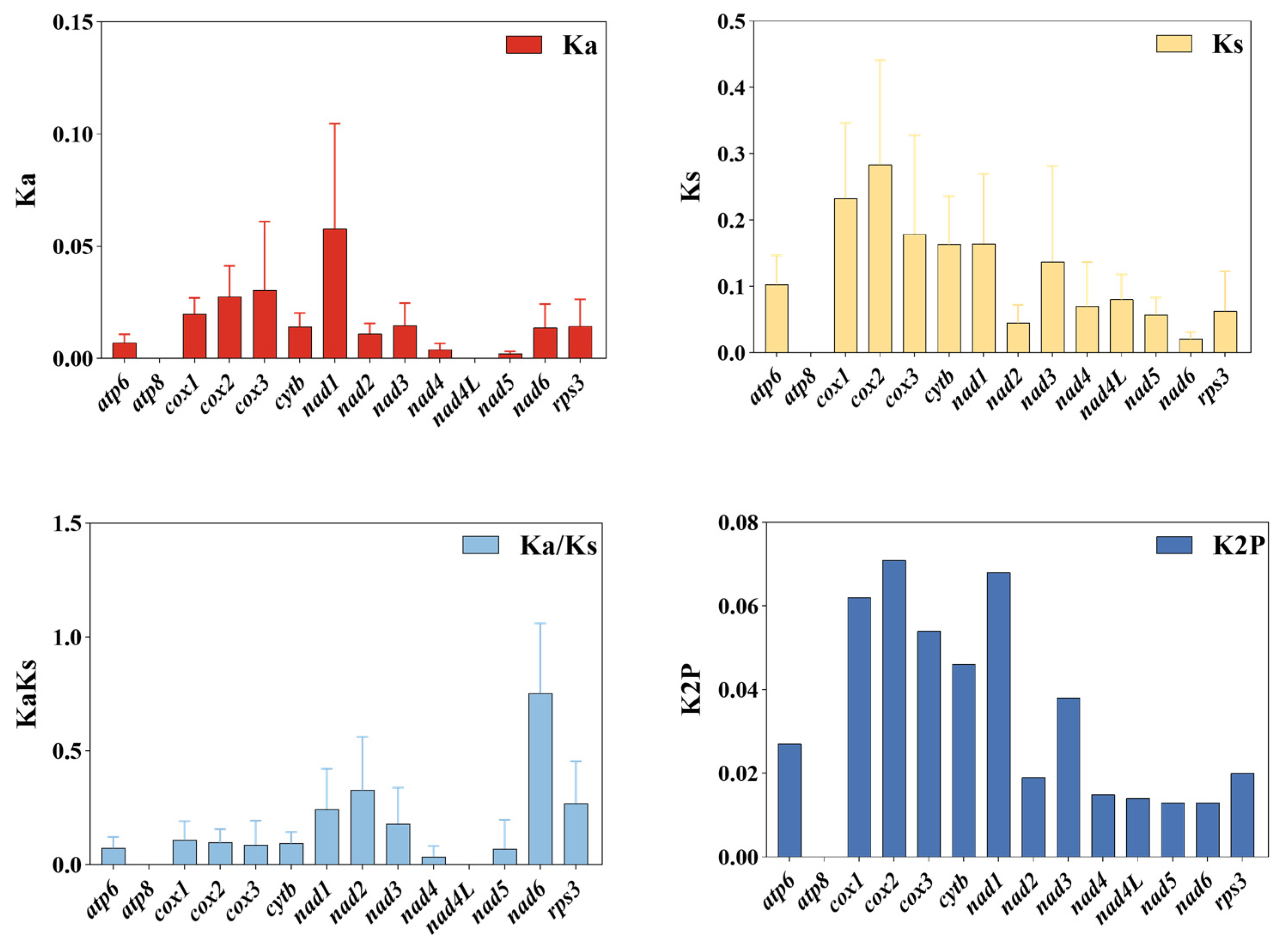
3.3. Variation in Mitogenome PCGs
3.4. Repeat Sequence
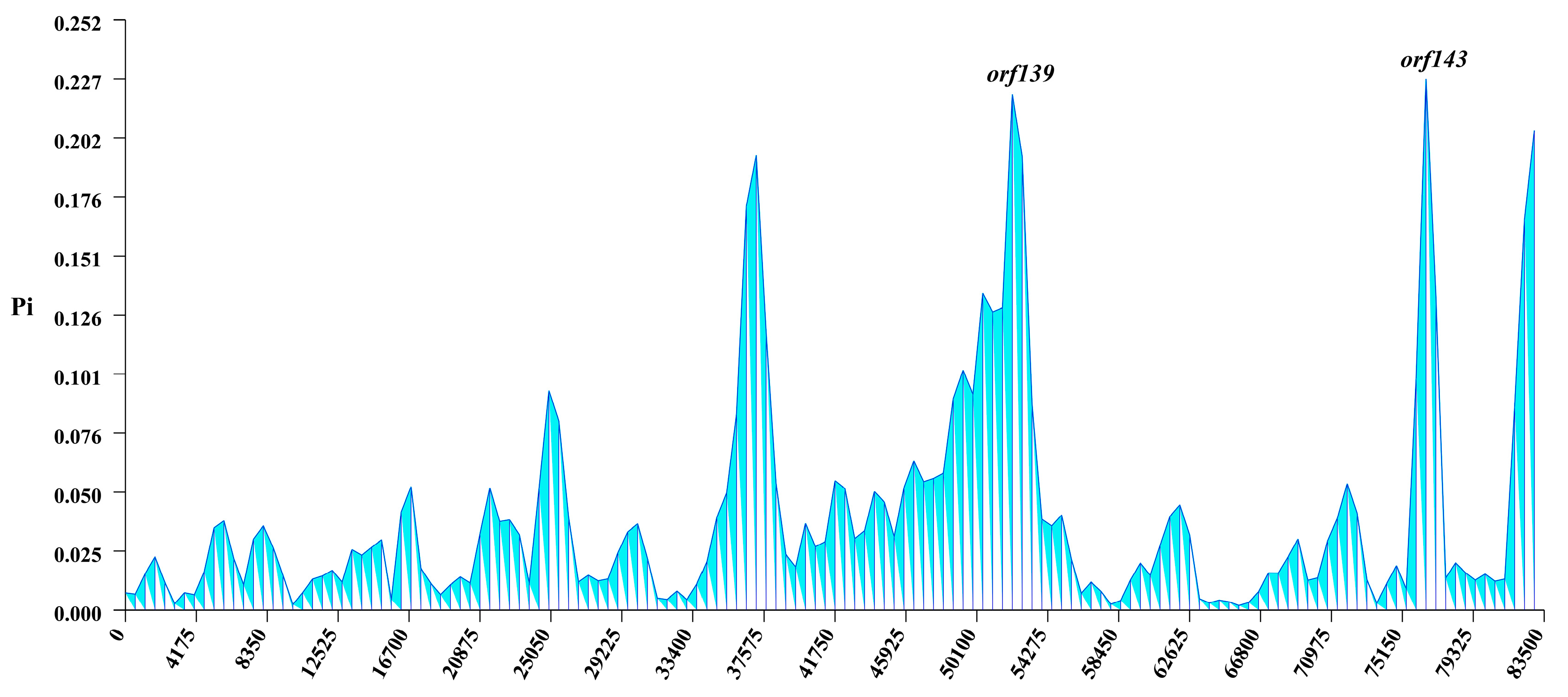
3.5. Nucleotide Diversity
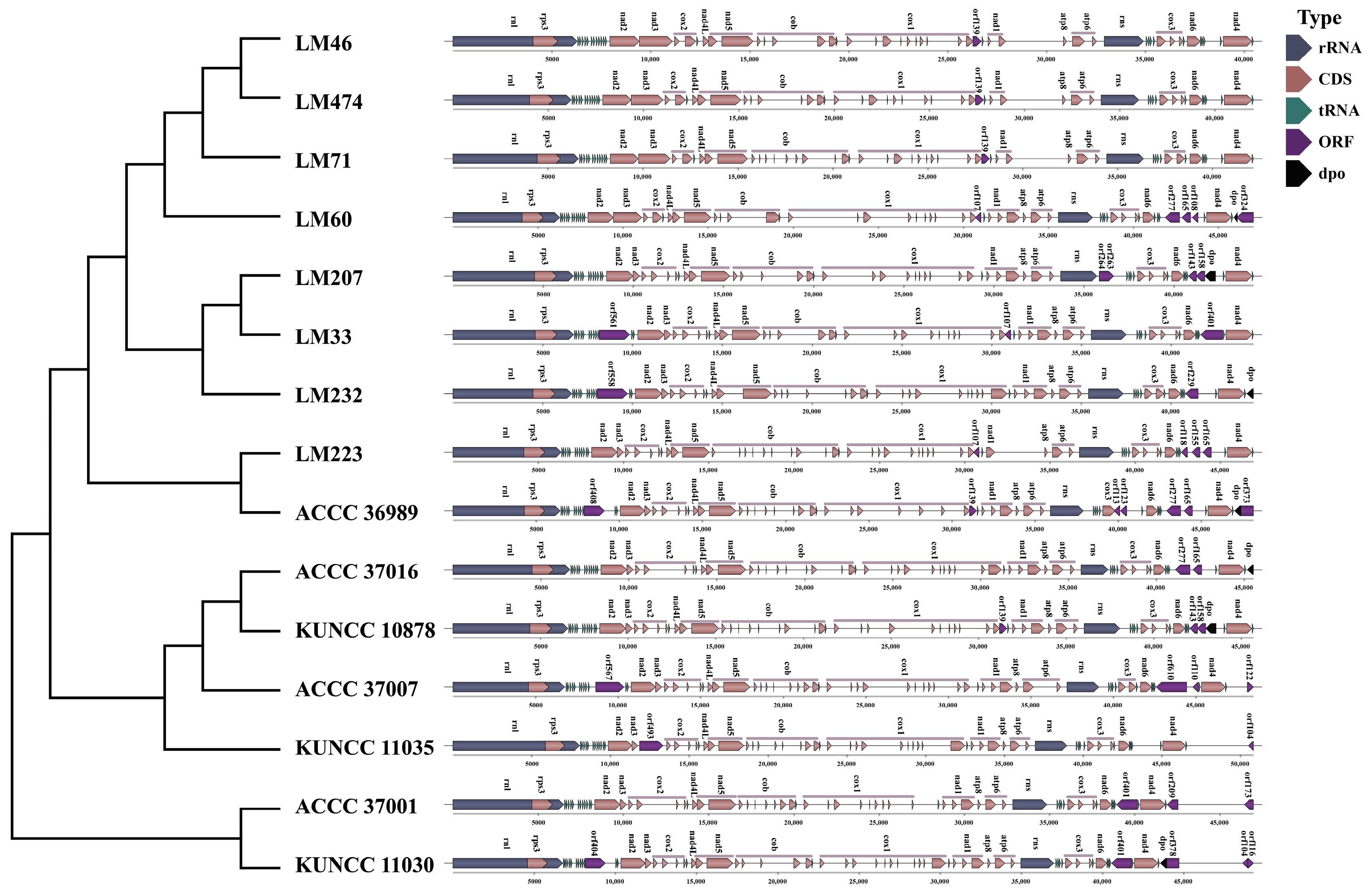
3.6. Intron Distribution
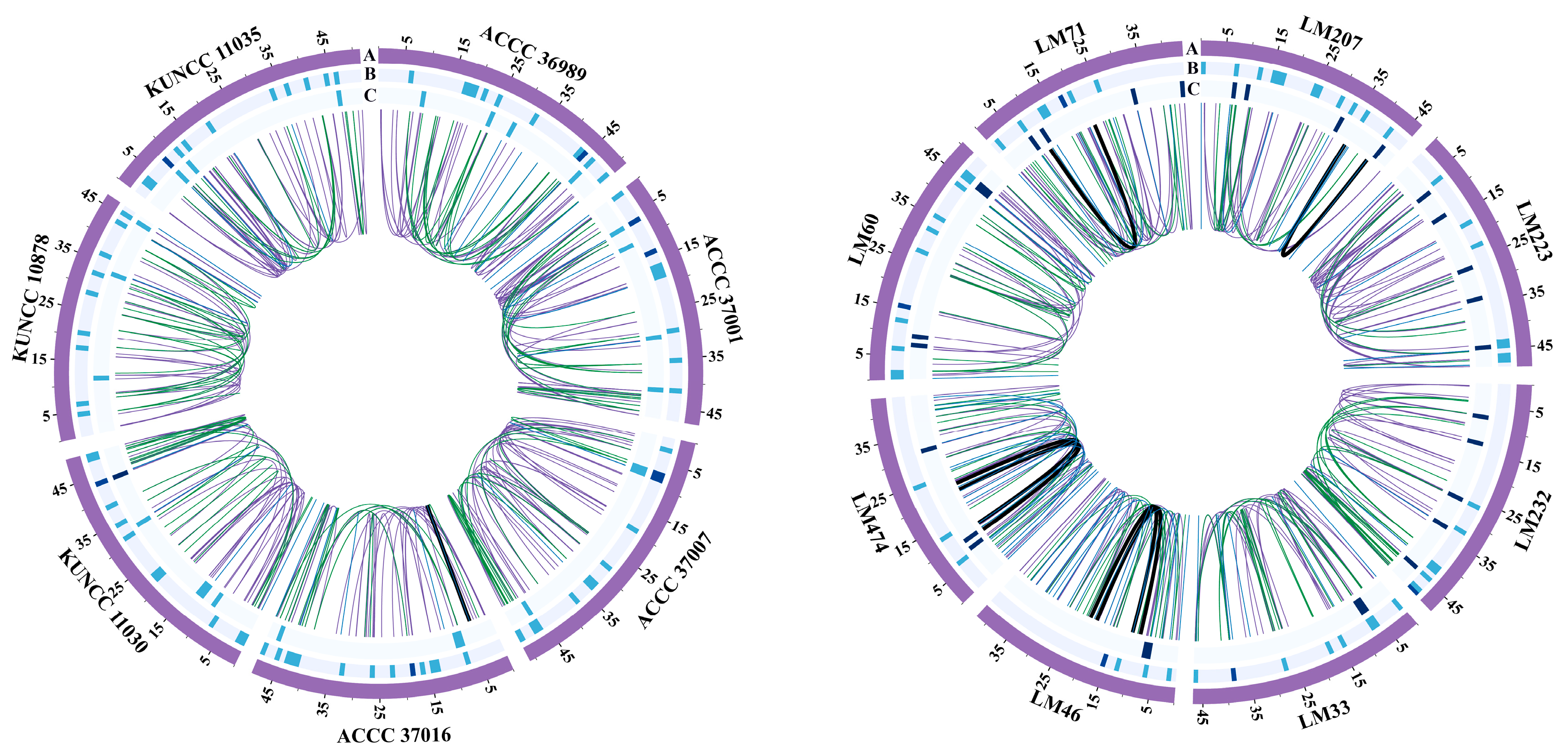
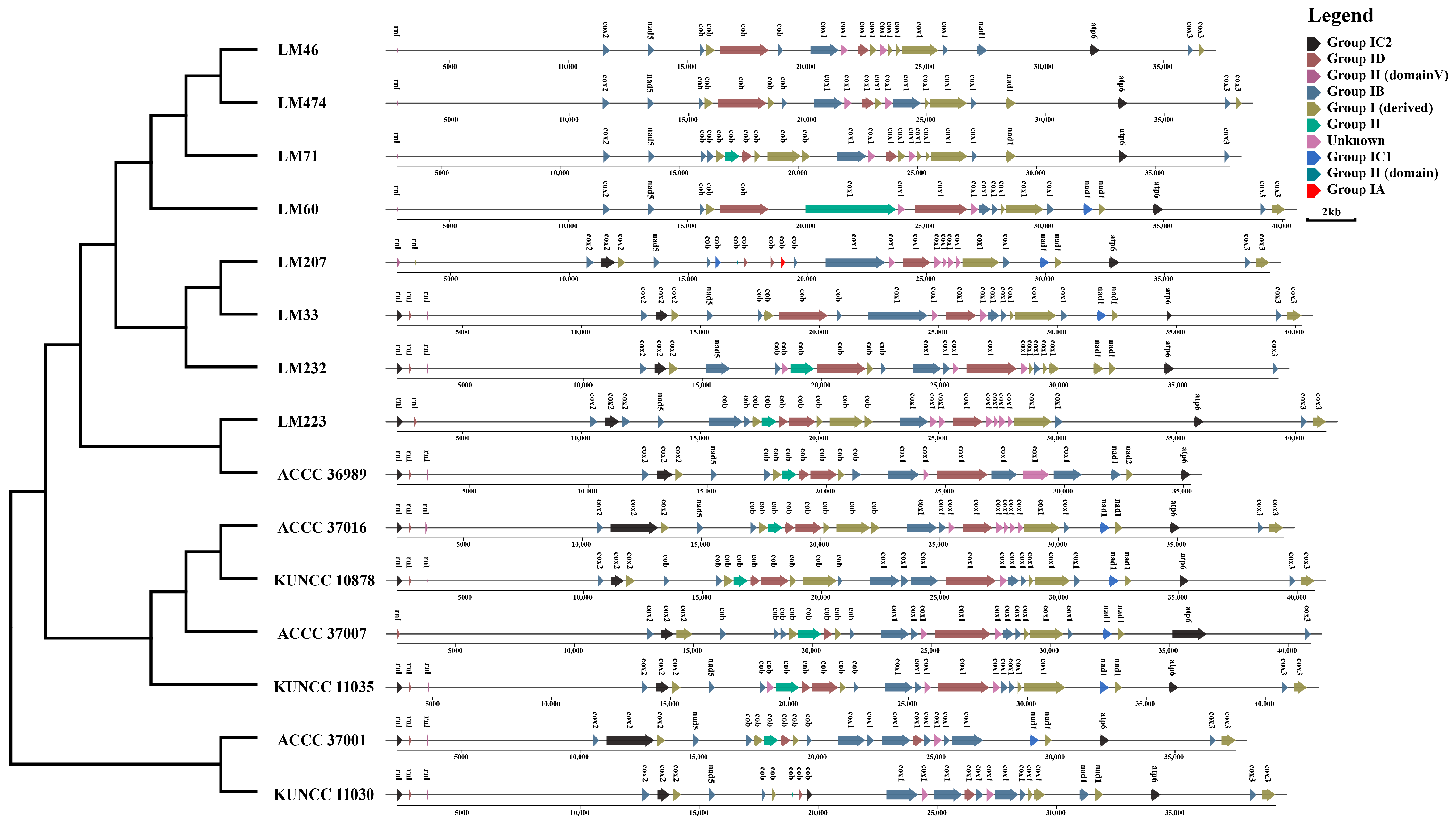
3.7. Structural Variation
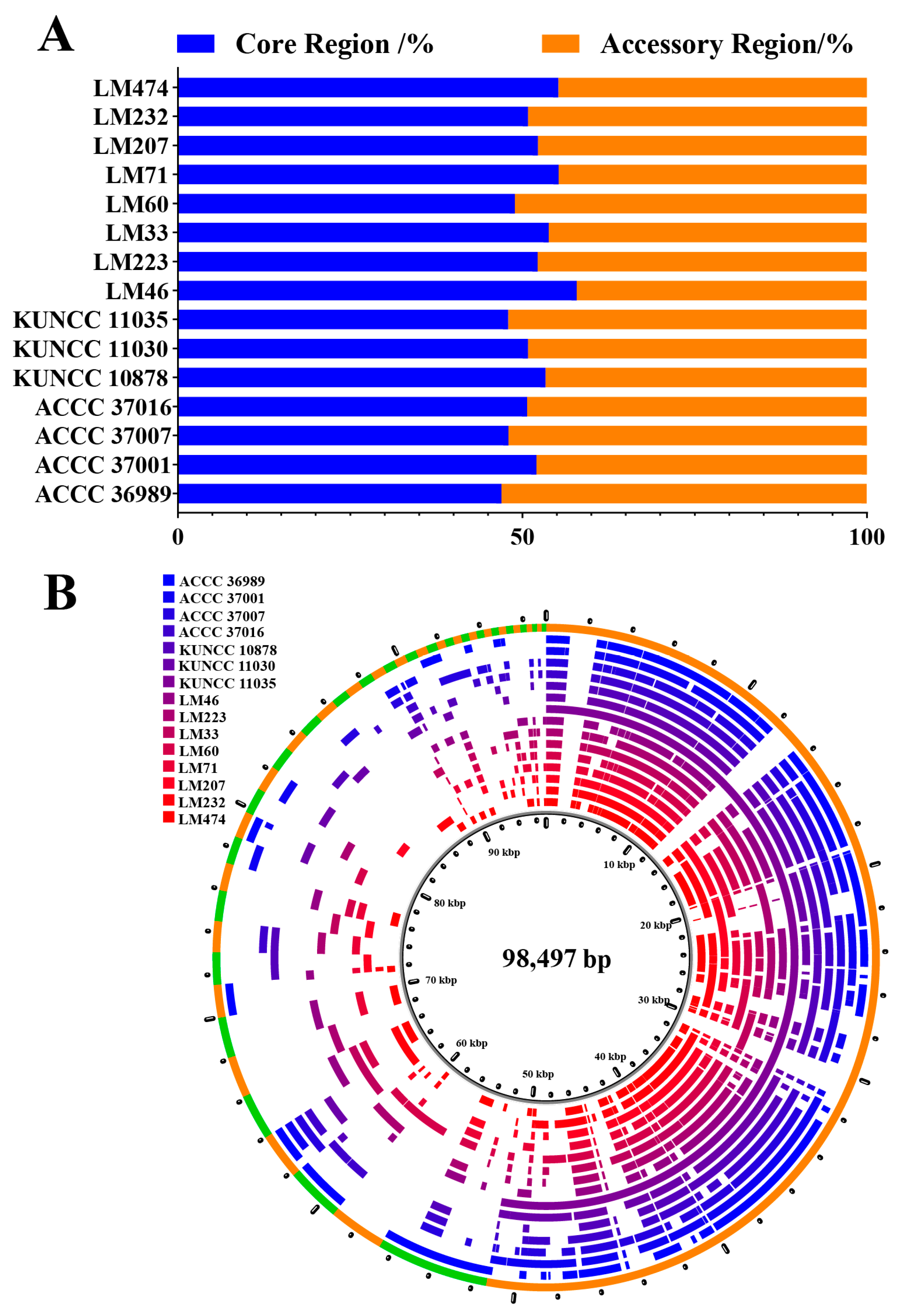
3.8. Pan-Mitogenome
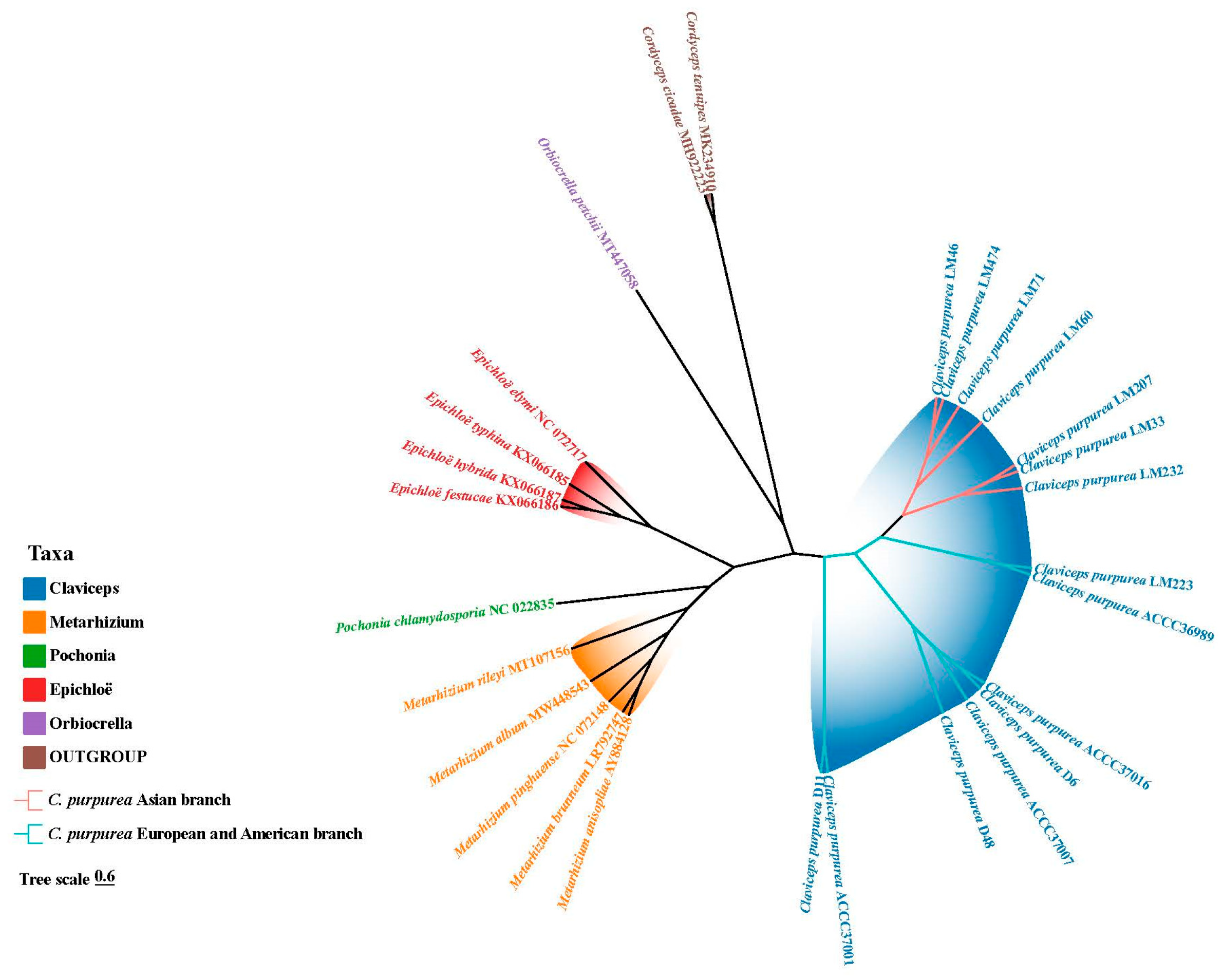
3.9. Phylogenetic Analysis of the Clavicipitaceae
4. Discussion
4.1. Structural Characteristics of the C. purpurea Mitogenome
4.2. Intraspecific Variation and Evolutionary Dynamics
4.3. Molecular Phylogeny and Biogeographic Inference
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PCG | protein-coding gene |
| ORF | Open Reading Frame |
| RSCU | Relative synonymous codon usage |
References
- Píchová, K.; Pažoutová, S.; Kostovčík, M.; Chudíčková, M.; Stodůlková, E.; Novák, P.; Flieger, M.; van der Linde, E.; Kolařík, M. Evolutionary History of Ergot with a New Infrageneric Classification (Hypocreales: Clavicipitaceae: Claviceps). Mol. Phylogenet. Evol. 2018, 123, 73–87. [Google Scholar] [CrossRef] [PubMed]
- Shoukouhi, P.; Hicks, C.; Menzies, J.G.; Popovic, Z.; Chen, W.; Seifert, K.A.; Assabgui, R.; Liu, M. Phylogeny of Canadian Ergot Fungi and a Detection Assay by Real-Time Polymerase Chain Reaction. Mycologia 2019, 111, 493–505. [Google Scholar] [CrossRef]
- Menzies, J.G.; Turkington, T.K. An overview of the ergot (Claviceps purpurea) issue in western Canada: Challenges and solutions. Can. J. Plant. Pathol. 2015, 37, 40–51. [Google Scholar] [CrossRef]
- Berraies, S.; Walkowiak, S.; Buchwaldt, L.; Menzies, J.G. Ergot (Claviceps spp.) of Cereals in Western Canada. Plant Health Cases 2023, 73, 1301–1316. [Google Scholar] [CrossRef]
- Belser-Ehrlich, S.; Harper, A.; Hussey, J.; Hallock, R. Human and Cattle Ergotism since 1900: Symptoms, Outbreaks, and Regulations. Toxicol. Ind. Health 2013, 29, 307–316. [Google Scholar] [CrossRef]
- Hersh, A.R.; Carroli, G.; Hofmeyr, G.J.; Garg, B.; Gülmezoglu, M.; Lumbiganon, P.; De Mucio, B.; Saleem, S.; Festin, M.P.R.; Mittal, S.; et al. Third Stage of Labor: Evidence-Based Practice for Prevention of Adverse Maternal and Neonatal Outcomes. Am. J. Obstet. Gynecol. 2024, 230, S1046–S1060.e1. [Google Scholar] [CrossRef]
- Puledda, F.; Sacco, S.; Diener, H.C.; Ashina, M.; Al-Khazali, H.M.; Ashina, S.; Burstein, R.; Liebler, E.; Cipriani, A.; Chu, M.K.; et al. International Headache Society Global Practice Recommendations for the Acute Pharmacological Treatment of Migraine. Cephalalgia 2024, 44, 03331024241252666. [Google Scholar] [CrossRef]
- Kutikuppala, L.V.S.; Sharma, S.; Chavan, M.; Rangari, G.; Misra, A.K.; Innamuri, S.R.; Vijayakumar, T.; Varshitha, G. Bromocriptine: Does This Drug of Parkinson’s Disease Have a Role in Managing Cardiovascular Diseases? Ann. Med. Surg. 2024, 86, 926–929. [Google Scholar] [CrossRef]
- Dara, T.; Zabihi, M.; Hoseinzade, F.; Rohani, M.; Saghafi, F. Bromocriptine in Type 2 Diabetes: A Promising Alternative, a Systematic Review and Meta-Analysis. Cardiovasc. Diabetol. Endocrinol. Rep. 2025, 11, 10. [Google Scholar] [CrossRef] [PubMed]
- Petersenn, S.; Fleseriu, M.; Casanueva, F.F.; Giustina, A.; Biermasz, N.; Biller, B.M.K.; Bronstein, M.; Chanson, P.; Fukuoka, H.; Gadelha, M.; et al. Diagnosis and Management of Prolactin-Secreting Pituitary Adenomas: A Pituitary Society International Consensus Statement. Nat. Rev. Endocrinol. 2023, 19, 722–740. [Google Scholar] [CrossRef]
- Joodi, S.A.; Khattab, M.M.; Ibrahim, W.W. Repurposing of Cabergoline to Improve Cognitive Decline in D-Galactose-Injected Ovariectomized Rats: Modulation of AKT/mTOR, GLT-1/P38-MAPK, and ERK1/2 Signaling Pathways. Toxicol. Appl. Pharmacol. 2025, 500, 117391. [Google Scholar] [CrossRef]
- Omidian, H.; Omidian, A. Clinical Research on Lysergic Acid Diethylamide (LSD) in Psychiatry and Neuroscience. Pharmaceuticals 2025, 18, 499. [Google Scholar] [CrossRef]
- Butenko, A.; Lukeš, J.; Speijer, D.; Wideman, J.G. Mitochondrial Genomes Revisited: Why Do Different Lineages Retain Different Genes? BMC Biol. 2024, 22, 15. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Kanai, M.; Durham, T.J.; Tsuo, K.; McCoy, J.G.; Kotrys, A.V.; Zhou, W.; Chinnery, P.F.; Karczewski, K.J.; Calvo, S.E.; et al. Nuclear Genetic Control of mtDNA Copy Number and Heteroplasmy in Humans. Nature 2023, 620, 839–848. [Google Scholar] [CrossRef]
- Shan, Y.; Li, J.; Zhang, X.; Yu, J. The Complete Mitochondrial Genome of Amorphophallus Albus and Development of Molecular Markers for Five Amorphophallus Species Based on Mitochondrial DNA. Front. Plant Sci. 2023, 14, 1180417. [Google Scholar] [CrossRef]
- Alqahtani, T.; Deore, S.L.; Kide, A.A.; Shende, B.A.; Sharma, R.; Dadarao Chakole, R.; Nemade, L.S.; Kishor Kale, N.; Borah, S.; Shrikant Deokar, S.; et al. Mitochondrial Dysfunction and Oxidative Stress in Alzheimer’s Disease, and Parkinson’s Disease, Huntington’s Disease and Amyotrophic Lateral Sclerosis—An Updated Review. Mitochondrion 2023, 71, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Luo, T.; Zhang, Q.; Pang, S.; Qin, Y.; Zhang, B.; Bian, X. Comparative Mitochondrial Genomic and Phylogenetic Study of Eight Species of the Family Lonchodidae (Phasmatodea: Euphasmatodea). Genes 2025, 16, 565. [Google Scholar] [CrossRef]
- Jia, J.; Li, C.; Tang, M.; Liao, H.; Yang, Z.; Wang, Y.; Li, M.; Zeng, W.; Wang, Y. Mitochondrial genome characterization, evolution and intron dynamics of the entomopathogenic genus Cordyceps. Front. Microbiol. 2025, 16, 1605218. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Overy, D.P.; Cayouette, J.; Shoukouhi, P.; Hicks, C.; Bisson, K.; Sproule, A.; Wyka, S.A.; Broders, K.; Popovic, Z.; et al. Four Phylogenetic Species of Ergot from Canada and Their Characteristics in Morphology, Alkaloid Production, and Pathogenicity. Mycologia 2020, 112, 974–988. [Google Scholar] [CrossRef]
- Schardl, C.L.; Young, C.A.; Moore, N.; Krom, N.; Dupont, P.-Y.; Pan, J.; Florea, S.; Webb, J.S.; Jaromczyk, J.; Jaromczyk, J.W.; et al. Genomes of Plant-Associated Clavicipitaceae. In Advances in Botanical Research; Academic Press: Cambridge, MA, USA, 2014; Volume 70, pp. 291–327. ISBN 978-0-12-397940-7. [Google Scholar]
- Jin, J.-J.; Yu, W.-B.; Yang, J.-B.; Song, Y.; dePamphilis, C.W.; Yi, T.-S.; Li, D.-Z. GetOrganelle: A Fast and Versatile Toolkit for Accurate de Novo Assembly of Organelle Genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De Novo Assembly of Organelle Genomes from Whole Genome Data. Nucleic Acids Res. 2016, 45, e18. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de Novo Metazoan Mitochondrial Genome Annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved Detection and Functional Classification of Transfer RNA Genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Lohse, M.; Drechsel, O.; Kahlau, S.; Bock, R. OrganellarGenomeDRAW—A Suite of Tools for Generating Physical Maps of Plastid and Mitochondrial Genomes and Visualizing Expression Data Sets. Nucleic Acids Res. 2013, 41, W575–W581. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An Integrated and Scalable Desktop Platform for Streamlined Molecular Sequence Data Management and Evolutionary Phylogenetics Studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Baeza, M.; Sepulveda, D.; Cifuentes, V.; Alcaíno, J. Codon Usage Bias in Yeasts and Its Correlation with Gene Expression, Growth Temperature, and Protein Structure. Front. Microbiol. 2024, 15, 1414422. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-R.; Chen, Y.-X.; Yan, Z.; Wei, X.-Y.; Qiu, J.-Z.; Zhang, Y.-J. Complete Mitogenome of the Entomopathogenic Fungus Orbiocrella Petchii. Mitochondrial DNA Part B Resour. 2020, 5, 2695–2696. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Tamura, K.; Nei, M. MEGA: Molecular Evolutionary Genetics Analysis Software for Microcomputers. Comput. Appl. Biosci. 1994, 10, 189–191. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-Web: A Web Server for Microsatellite Prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Benson, G. Tandem Repeats Finder: A Program to Analyze DNA Sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S. REPuter: The Manifold Applications of Repeat Analysis on a Genomic Scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Katoh, K. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Xiang, C.; Gao, F.; Jakovlić, I.; Lei, H.; Hu, Y.; Zhang, H.; Zou, H.; Wang, G.; Zhang, D. Using PhyloSuite for Molecular Phylogeny and Tree-based Analyses. iMeta 2023, 2, e87. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple Sequence Alignment Using ClustalW and ClustalX. CP Bioinform. 2003, 2.3.1–2.3.22. [Google Scholar] [CrossRef]
- He, Y.; Liu, W.; Wang, J. Assembly and Comparative Analysis of the Complete Mitochondrial Genome of Trigonella Foenum-Graecum L. BMC Genom. 2023, 24, 756. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Ozer, E.A.; Allen, J.P.; Hauser, A.R. Characterization of the Core and Accessory Genomes of Pseudomonas Aeruginosa Using Bioinformatic Tools Spine and AGEnt. BMC Genom. 2014, 15, 737. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Bliss, C.M.; Bennett, J.S.; Bratcher, H.B.; Brehony, C.; Colles, F.M.; Wimalarathna, H.; Harrison, O.B.; Sheppard, S.K.; Cody, A.J.; et al. Ribosomal Multilocus Sequence Typing: Universal Characterization of Bacteria from Domain to Strain. Microbiology 2012, 158, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Ozer, E.A. ClustAGE: A Tool for Clustering and Distribution Analysis of Bacterial Accessory Genomic Elements. BMC Bioinform. 2018, 19, 150. [Google Scholar] [CrossRef]
- Yildiz, G.; Ozkilinc, H. Pan-Mitogenomics Approach Discovers Diversity and Dynamism in the Prominent Brown Rot Fungal Pathogens. Front. Microbiol. 2021, 12, 647989. [Google Scholar] [CrossRef]
- Lang, B.F.; Laforest, M.-J.; Burger, G. Mitochondrial Introns: A Critical View. Trends Genet. 2007, 23, 119–125. [Google Scholar] [CrossRef]
- Song, X.; Geng, Y.; Xu, C.; Li, J.; Guo, Y.; Shi, Y.; Ma, Q.; Li, Q.; Zhang, M. The Complete Mitochondrial Genomes of Five Critical Phytopathogenic Bipolaris Species: Features, Evolution, and Phylogeny. IMA Fungus 2024, 15, 15. [Google Scholar] [CrossRef]
- Wolters, J.F.; LaBella, A.L.; Opulente, D.A.; Rokas, A.; Hittinger, C.T. Mitochondrial Genome Diversity across the Subphylum Saccharomycotina. bioRxiv 2023, 14, 1268944. [Google Scholar] [CrossRef]
- Ma, Q.; Wu, H.; Geng, Y.; Li, Q.; Zang, R.; Guo, Y.; Xu, C.; Zhang, M. Mitogenome-Wide Comparison and Phylogeny Reveal Group I Intron Dynamics and Intraspecific Diversification within the Phytopathogen Corynespora Cassiicola. Comput. Struct. Biotechnol. J. 2021, 19, 5987–5999. [Google Scholar] [CrossRef]
- Joardar, V.; Abrams, N.F.; Hostetler, J.; Paukstelis, P.J.; Pakala, S.; Pakala, S.B.; Zafar, N.; Abolude, O.O.; Payne, G.; Andrianopoulos, A.; et al. Sequencing of Mitochondrial Genomes of Nine Aspergillus and Penicillium Species Identifies Mobile Introns and Accessory Genes as Main Sources of Genome Size Variability. BMC Genom. 2012, 13, 698. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Liu, C.; Shen, B.; Bai, M.; Ling, J.; Chen, G.; Mao, Z.; Cheng, X.; Xie, B. Analysis of the Complete Mitochondrial Genome of Pochonia Chlamydosporia Suggests a Close Relationship to the Invertebrate-Pathogenic Fungi in Hypocreales. BMC Microbiol. 2015, 15, 5. [Google Scholar] [CrossRef]
- Campbell, M.A.; Tapper, B.A.; Simpson, W.R.; Johnson, R.D.; Mace, W.; Ram, A.; Lukito, Y.; Dupont, P.-Y.; Johnson, L.J.; Scott, D.B.; et al. Epichloë hybrida, sp. nov., an emerging model system for investigating fungal allopolyploidy. Mycologia 2017, 109, 715–729. [Google Scholar] [CrossRef]
- Ghikas, D.V.; Kouvelis, V.N.; Typas, M.A. The Complete Mitochondrial Genome of the Entomopathogenic Fungus Metarhizium Anisopliae Var. Anisopliae: Gene Order and Trn Gene Clusters Reveal a Common Evolutionary Course for All Sordariomycetes, While Intergenic Regions Show Variation. Arch. Microbiol. 2006, 185, 393–401. [Google Scholar] [CrossRef]
- Clark-Walker, G.D.; Francois, F.; Chen, X.J.; Silva, M.R.V.D.; Claisse, M.L. Mitochondrial ATP Synthase Subunit 9 Is Not Required for Viability of the Petite-Negative Yeast Kluyveromyces Lactis. Curr. Genet. 1997, 31, 488–493. [Google Scholar] [CrossRef]
- Fijarczyk, A.; Hessenauer, P.; Hamelin, R.C.; Landry, C.R. Lifestyles Shape Genome Size and Gene Content in Fungal Pathogens. eLife 2025, 14, RP104975. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, H.; Huo, S.; Lu, J.; Norvienyeku, J.; Miao, W.; Qin, C.; Liu, W. Comparative Mitogenomics Analysis Revealed Evolutionary Divergence among Neopestalotiopsis Species Complex (Fungi: Xylariales). Int. J. Mol. Sci. 2024, 25, 3093. [Google Scholar] [CrossRef]
- Xiong, C.; Lin, Y.; Keyhani, N.O.; Shang, J.; Mao, Y.; Yang, J.; Zheng, M.; Yang, L.; Pu, H.; Lin, L.; et al. Mitochondrial Genomes from Fungal the Entomopathogenic Moelleriella Genus Reveals Evolutionary History, Intron Dynamics and Phylogeny. J. Fungi 2025, 11, 94. [Google Scholar] [CrossRef]
- Kurata, S.; Mano, S.; Nakahama, N.; Hirota, S.; Suyama, Y.; Ito, M. Development of Mitochondrial DNA Cytochrome c Oxidase Subunit I Primer Sets to Construct DNA Barcoding Library Using Next-Generation Sequencing. Biodivers. Data J. 2024, 12, e117014. [Google Scholar] [CrossRef] [PubMed]
- Cheung, K.; Amos, T.; Shine, R.; DeVore, J.; Ducatez, S.; Edwards, R.; Rollins, L. Mitochondrial Genome Analyses Uncover Intriguing Population Structure across the Invasive Trajectory of an Iconic Invader. bioRxiv 2023, 9, 558141. [Google Scholar] [CrossRef]
- Li, H.; Liang, T.; Liu, Y.; Wang, P.; Wang, S.; Zhao, M.; Zhang, Y. Exploring Mitochondrial Heterogeneity and Evolutionary Dynamics in Thelephora Ganbajun through Population Genomics. Int. J. Mol. Sci. 2024, 25, 9013. [Google Scholar] [CrossRef]
- Wei, W.; Wang, H.; Liu, G. Transcriptional regulation of 10 mitochondrial genes in different tissues of NCa CMS system in Brassica napus L. and their relationship with sterility. J. Genet. Genom. 2007, 34, 72–80. [Google Scholar] [CrossRef]
- Hanson, M.R.; Wilson, R.K.; Bentolila, S.; Köhler, R.H.; Chen, H.C. Mitochondrial gene organization and expression in petunia male fertile and sterile plants. J. Hered. 1999, 90, 362–368. [Google Scholar] [CrossRef]
- Zhong, L.; Wang, M.; Li, D.; Tang, S.; Zhang, T.; Bian, W.; Chen, X. Complete Mitochondrial Genome of Odontobutis Haifengensis (Perciformes, Odontobutiae): A Unique Rearrangement of tRNAs and Additional Non-Coding Regions Identified in the Genus Odontobutis. Genomics 2018, 110, 382–388. [Google Scholar] [CrossRef]
- Kong, J.; Wang, J.; Nie, L.; Tembrock, L.R.; Zou, C.; Kan, S.; Ma, X.; Wendel, J.F.; Wu, Z. Evolutionary Dynamics of Mitochondrial Genomes and Intracellular Transfers among Diploid and Allopolyploid Cotton Species. BMC Biol. 2025, 23, 9. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, K.; Peng, Y.; Zhou, J.; Liu, Y.; Liu, B. Novel Gene Rearrangement in the Mitochondrial Genome of Three Garra and Insights into the Phylogenetic Relationships of Labeoninae. Front. Genet. 2022, 13, 922634. [Google Scholar] [CrossRef] [PubMed]
- Minhas, B.F.; Beck, E.A.; Cheng, C.-H.C.; Catchen, J. Novel Mitochondrial Genome Rearrangements Including Duplications and Extensive Heteroplasmy Could Underlie Temperature Adaptations in Antarctic Notothenioid Fishes. Sci. Rep. 2023, 13, 6939. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Z.; He, D.; Guang, X.; Li, Q. Novel tRNA Gene Rearrangements in the Mitochondrial Genomes of Poneroid Ants and Phylogenetic Implication of Paraponerinae (Hymenoptera: Formicidae). Life 2023, 13, 2068. [Google Scholar] [CrossRef] [PubMed]
- Aguileta, G.; De Vienne, D.M.; Ross, O.N.; Hood, M.E.; Giraud, T.; Petit, E.; Gabaldón, T. High Variability of Mitochondrial Gene Order among Fungi. Genome Biol. Evol. 2014, 6, 451–465. [Google Scholar] [CrossRef]
- Li, S.; Hu, X.; Song, Q. Comparative Analysis of the Mitochondrial Genome Sequences of Diaporthe Longicolla (Syn. Phomopsis Longicolla) Isolates Causing Phomopsis Seed Decay in Soybean. J. Fungi 2024, 10, 570. [Google Scholar] [CrossRef]
- Liu, C.; Li, W.-Y.; Zheng, L.-X.; Dao, M.; Chen, H.-H.; Han, L.-H. Comparative Mitogenomic Analysis Reveals Variations and Evolution of Ectomycorrhizal Fungal Strobilomyces. IMA Fungus 2025, 16, e141848. [Google Scholar] [CrossRef] [PubMed]
- Medina, R.; Franco, M.E.E.; Bartel, L.C.; Martinez Alcántara, V.; Saparrat, M.C.N.; Balatti, P.A. Fungal Mitogenomes: Relevant Features to Planning Plant Disease Management. Front. Microbiol. 2020, 11, 978. [Google Scholar] [CrossRef] [PubMed]
- Merheb, M.; Matar, R.; Hodeify, R.; Siddiqui, S.S.; Vazhappilly, C.G.; Marton, J.; Azharuddin, S.; Al Zouabi, H. Mitochondrial DNA, a Powerful Tool to Decipher Ancient Human Civilization from Domestication to Music, and to Uncover Historical Murder Cases. Cells 2019, 8, 433. [Google Scholar] [CrossRef]
- Dallagnol, L.C.; Cônsoli, F.L. Evolutionary and Phylogenetic Insights from the Mitochondrial Genomic Analysis of Diceraeus Melacanthus and D. Furcatus (Hemiptera: Pentatomidae). Sci. Rep. 2024, 14, 12861. [Google Scholar] [CrossRef]
- Thorn, V.; Xu, J. Mitogenome Variations in a Global Population of Aspergillus Fumigatus. J. Fungi 2023, 9, 995. [Google Scholar] [CrossRef]
- Zhang, M.-Z.; Xu, J.-P.; Callac, P.; Chen, M.-Y.; Wu, Q.; Wach, M.; Mata, G.; Zhao, R.-L. Insight into the Evolutionary and Domesticated History of the Most Widely Cultivated Mushroom Agaricus Bisporus via Mitogenome Sequences of 361 Global Strains. BMC Genom. 2023, 24, 182. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, M.; Hu, R.; Jia, J.; Wei, C.; Cui, Y.; Liao, H.; Yang, Z.; Guo, J.; Ma, Z.; Wang, Y. Pan-Mitogenome Construction, Intraspecific Variation, and Adaptive Evolution of the Plant Pathogenic Fungus Claviceps purpurea. Biology 2025, 14, 1548. https://doi.org/10.3390/biology14111548
Ding M, Hu R, Jia J, Wei C, Cui Y, Liao H, Yang Z, Guo J, Ma Z, Wang Y. Pan-Mitogenome Construction, Intraspecific Variation, and Adaptive Evolution of the Plant Pathogenic Fungus Claviceps purpurea. Biology. 2025; 14(11):1548. https://doi.org/10.3390/biology14111548
Chicago/Turabian StyleDing, Mingliang, Rui Hu, Jinlong Jia, Cuiyuan Wei, Yongzhen Cui, Hefa Liao, Zhuliang Yang, Jianwei Guo, Zhanhong Ma, and Yuanbing Wang. 2025. "Pan-Mitogenome Construction, Intraspecific Variation, and Adaptive Evolution of the Plant Pathogenic Fungus Claviceps purpurea" Biology 14, no. 11: 1548. https://doi.org/10.3390/biology14111548
APA StyleDing, M., Hu, R., Jia, J., Wei, C., Cui, Y., Liao, H., Yang, Z., Guo, J., Ma, Z., & Wang, Y. (2025). Pan-Mitogenome Construction, Intraspecific Variation, and Adaptive Evolution of the Plant Pathogenic Fungus Claviceps purpurea. Biology, 14(11), 1548. https://doi.org/10.3390/biology14111548






