Systematic Analysis of the CCoAOMT Gene Family in Isatis indigotica and the Molecular Mechanism of CCoAOMT8-Mediated Flavonoid Synthesis Under Alkaline Stress Treatment
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Identification and Characterization of IiCCoAOMT Genes
2.3. Analysis of the Gene Structure of I. indigotica CCoAOMT and the Conserved Motifs
2.4. Phylogenetic Analysis of the Isatis indigotica CCoAOMT Gene
2.5. Analysis of Gene Cis-Acting Elements
2.6. Gene Chromosome Location and Collinearity Analysis
2.7. RNA Extraction from Leaves at Different Treatment Times and Real-Time Fluorescence Quantitative PCR Analysis
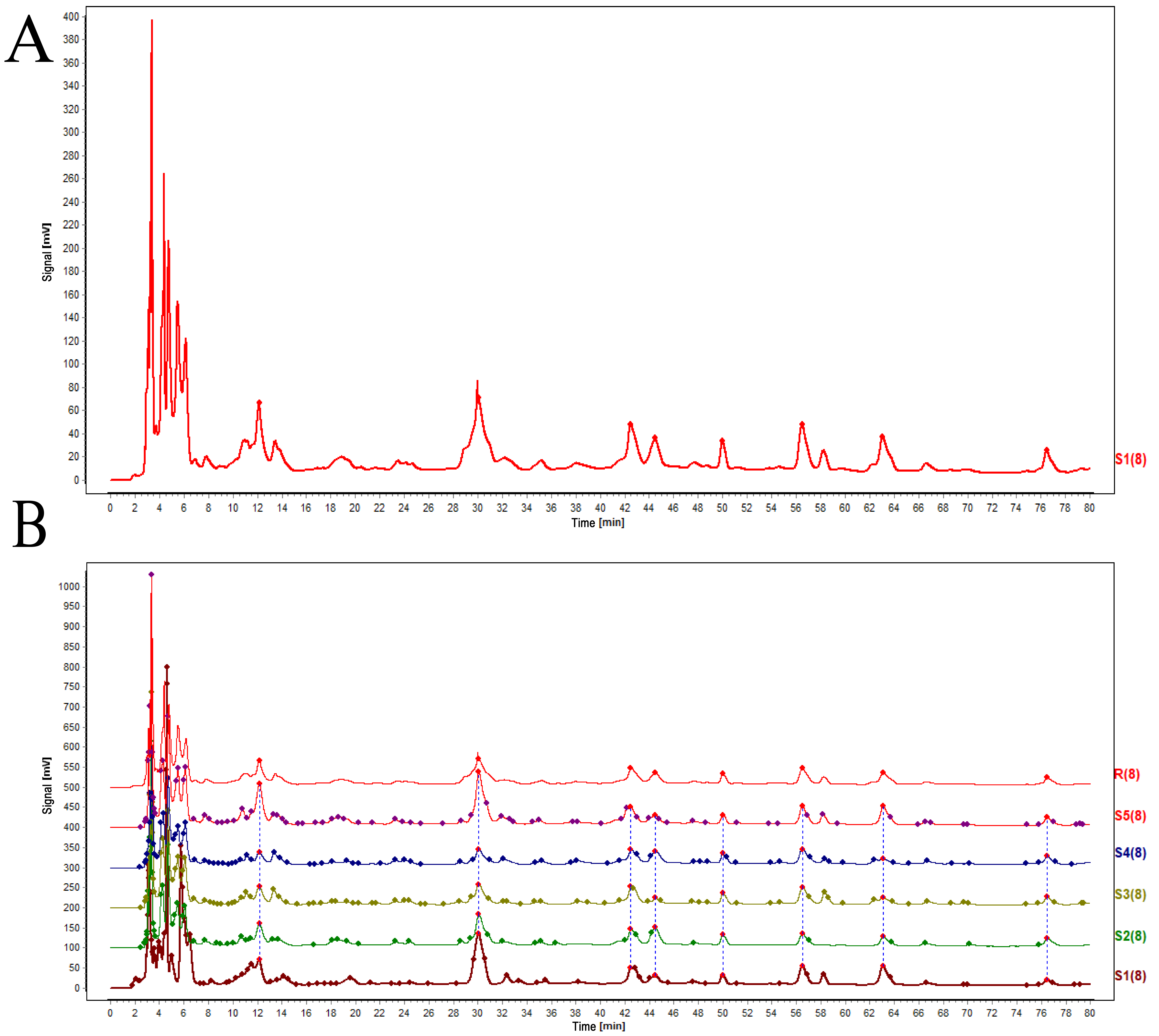
2.8. Determination of Flavonoid Chemical Constituents in I. indigotica Tissue Samples
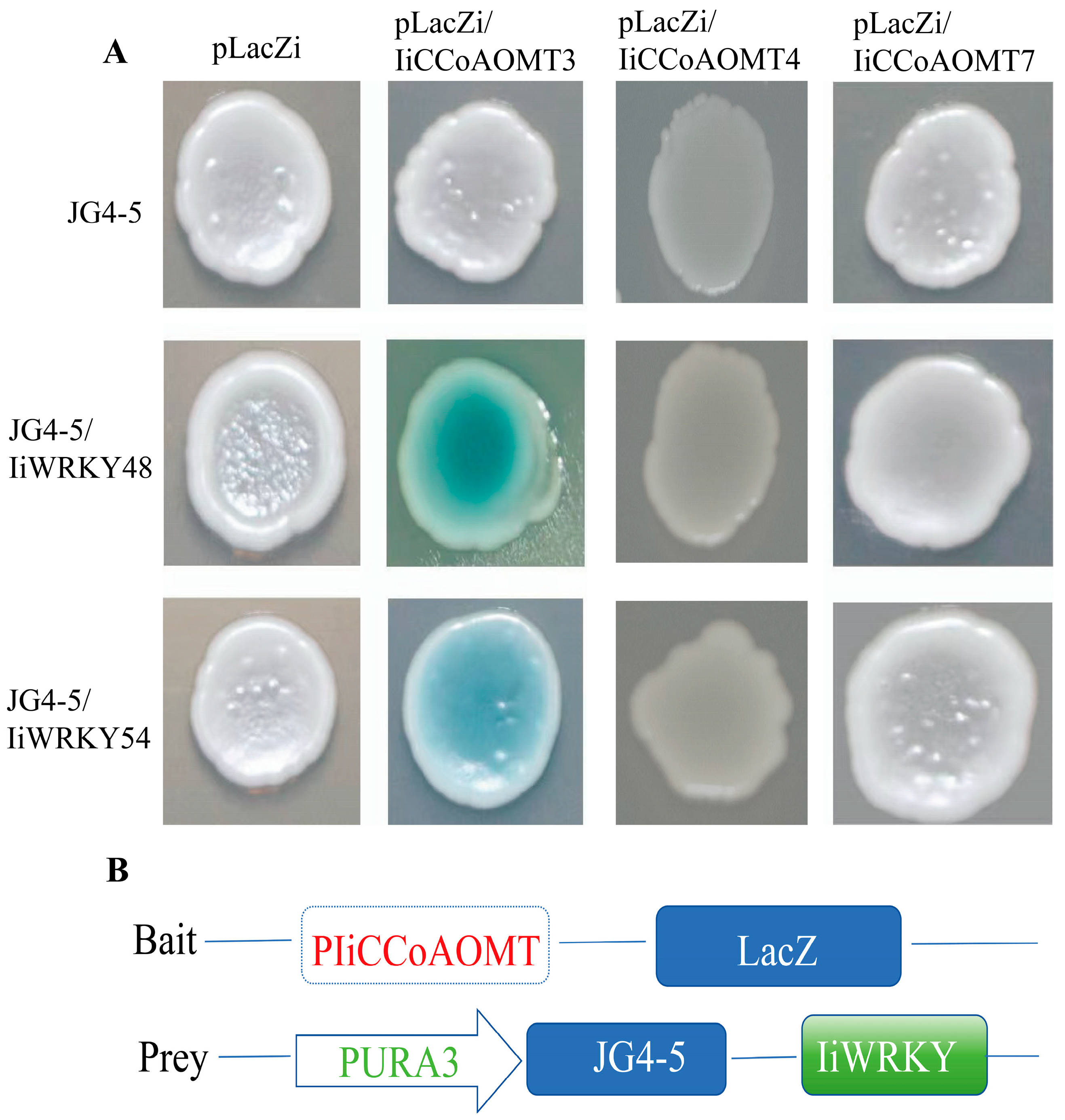
2.9. Yeast One-Hybrid Assay
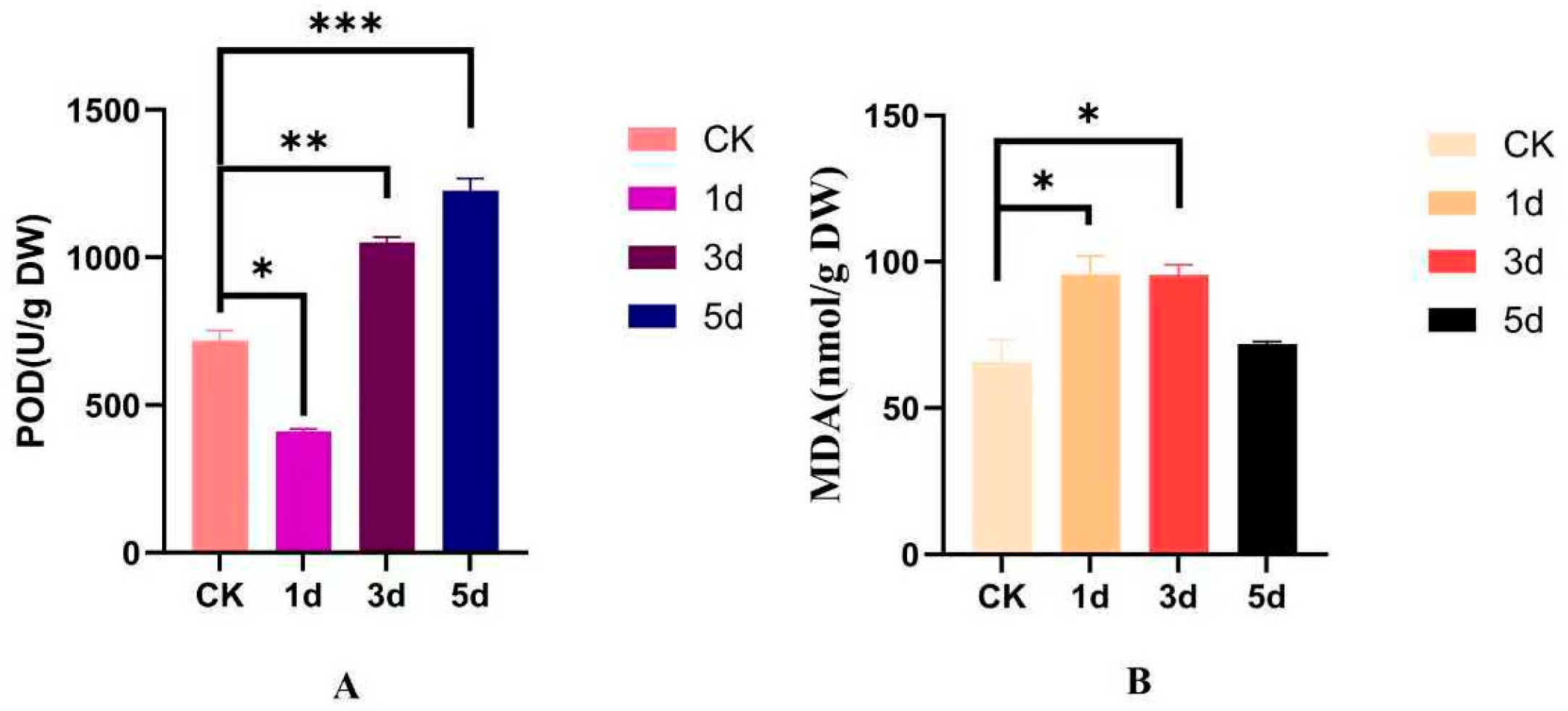
2.10. Determination of Antioxidant Content in I. indigotica
2.11. Statistical Analysis
3. Results
3.1. Identification and Physicochemical Property Analysis
3.2. Analysis of the Gene Structure of I. indigotica CCoAOMT and the Conserved Motifs and Sequence Alignment of Its Encoded Protein
3.3. Phylogenetic Analysis of IiCCoAOMT Family Members
3.4. Cis-Acting Element Analysis of IiCCoAOMT Gene Promoter
3.5. Chromosome Localization and Homology Analysis of IiCCoAOMT Genes
3.6. Analysis of IiCCoAOMT Gene Expression Patterns Under Different Time Periods of Alkali Treatment
3.7. Analysis of Flavonoid Metabolite Content of IiCCoAOMT Gene
3.8. The Effect of Alkaline Stress Treatment on POD Activity and MDA Content in I. indigotica Tissue Samples
3.9. IiWrky48 and IiWrky54 Can Bind to the Promoter CCoAOMT Gene
4. Discussion
4.1. Expansion of CCoAOMT Gene Family
4.2. Bioinformatics Analysis of the IiCCoAOMT Gene Family
4.3. Response of the IiCCoAOMT Family to Alkaline Treatment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Manzoor, M.A.; Manzoor, M.M.; Li, G.; Abdullah, M.; Han, W.; Wenlong, H.; Shakoor, A.; Riaz, M.W.; Rehman, S.; Cai, Y. Genome-wide identification and characterization of bZIP transcription factors and their expression profile under abiotic stress treatmentes in Chinese pear (Pyrus bretschneideri). BMC Plant Biol. 2021, 21, 413. [Google Scholar] [CrossRef]
- Huazhen, Q.; Bo, S.; Shiyang, L. Comparing Research in Antiviral Action of Rhizoma et Radix Baphicacanthis cusiae and Radix isatidis against Influenza A Virus. Chin. Arch. Tradit. Chin. Med. 2009, 27, 168–169. [Google Scholar] [CrossRef]
- Chang, S.-J.; Chang, Y.-C.; Lu, K.-Z.; Tsou, Y.-Y.; Lin, C.-W. Antiviral Activity of Isatis indigotica Extract and Its Derived Indirubin against Japanese Encephalitis Virus. Evid.-Based Complement. Altern. Med. 2012, 2012, 925830. [Google Scholar] [CrossRef]
- Zhou, W.; Zhang, X.-Y. Research Progress of Chinese Herbal Medicine Radix isatidis (Banlangen). Am. J. Chin. Med. 2013, 41, 743–764. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.H.; Du, T.T.; Zhang, Z.H.; Ji, M.; Hu, H.Y.; Chen, X.G. Research progress in pharmacological action and mechanism of chlorogenic acid, Yaoxue Xuebao. J. Pharm. 2020, 55, 2273–2280. [Google Scholar] [CrossRef]
- Akula, R.; Ravishankar, G.A. Influence of abiotic stress treatment signals on secondary metabolites in plants. Plant Signal. Behav. 2014, 6, 1720–1731. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, Z.; Wang, P.; Qin, C.; He, L.; Kong, L.; Ren, W.; Liu, X.; Ma, W. Genome-wide identification of the NAC transcription factors family and regulation of metabolites under salt stress treatment in Isatis indigotica. Int. J. Biol. Macromol. 2023, 240, 124436. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.N.; Wen, L.Y.; Liu, Z.W.; Li, X.X.; Gao, J.P.; Geng, R.M.; Jiang, C.H.; Sun, M.M.; Pu, W.X.; Yang, A.G. Identification and Functional Analysis of tobacco NtCCoAOMT Gene Family. Mol. Plant Breed. 2025, 1–11. Available online: https://link.cnki.net/urlid/46.1068.S.20230522.1128.006 (accessed on 18 July 2025).
- Golldack, D.; Li, C.; Mohan, H.; Probst, N. Tolerance to drought and salt stress treatment in plants: Unraveling the signaling networks. Front. Plant Sci. 2014, 5, 151. [Google Scholar] [CrossRef] [PubMed]
- Pitre, F.E.; Pollet, B.; Lafarguette, F.; Cooke, J.E.; MacKay, J.J.; Lapierre, C. Effects of increased nitrogen supply on the lignification of poplar wood. J. Agric. Food Chem. 2007, 55, 10306–10314. [Google Scholar] [CrossRef]
- Do, C.-T.; Pollet, B.; Thévenin, J.; Sibout, R.; Denoue, D.; Barrière, Y.; Lapierre, C.; Jouanin, L. Both caffeoyl Coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta 2007, 226, 1117–1129. [Google Scholar] [CrossRef] [PubMed]
- Day, A.; Dehorter, B.; Neutelings, G.; Czeszak, X.; Chabbert, B.; Belingheri, L.; David, H. Caffeoyl-coenzyme A 3-O-methyltransferase enzyme activity, protein and transcript accumulation in flax (Linum usitatissimum) stem during development. Physiol. Plant. 2001, 113, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Liu, Y.; Chen, J.J.; Zhang, H.P.; Zeng, J.W.; Huang, H.J.; Tian, J.; Peng, S.; Xu, J. The effect of Huanglong disease pathogen infection on flavonoids and volatile compounds in tea branches and citrus fruits. J. Huazhong Agric. Univ. 2020, 39, 24–33. [Google Scholar] [CrossRef]
- Zhang, C.L.; Chen, P.; Zhong, Y.M.; Zhou, C.Y.; Liang, G.H.; Shen, D.H.; Wu, Q.Y. Cloning and Expression of Flavonoid O-methyltransferase Gene in Ginkgo biloba, Yuanyi Xuebao. J. Hortic. 2012, 39, 355–362. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, T.; Qi, S.; Zhao, J.; Kong, J.; Xue, Z.; Sun, W.; Zeng, W. Genome-wide identification, expression profiling, and protein interaction analysis of the CCoAOMT gene family in the tea plant (Camellia sinensis). BMC Genom. 2024, 25, 238. [Google Scholar] [CrossRef]
- Kim, B.-G.; Sung, S.H.; Chong, Y.; Lim, Y.; Ahn, J.-H. Plant Flavonoid O-Methyltransferases: Substrate Specificity and Application. J. Plant Biol. 2010, 53, 321–329. [Google Scholar] [CrossRef]
- Liu, X.M. Cloning and Functional Validation of Flavonoids O-Methyltransferase Genes in Ponkan. Master’s Thesis, Southwest University, Chongqing, China, 2021. [Google Scholar] [CrossRef]
- Zhang, X.Y. Study on the Response of Fengdan to Drought Stress Treatment and the Function of CCoAOMT Gene. Master’s Thesis, Yangzhou University, Yangzhou, China, 2020. [Google Scholar] [CrossRef]
- Chun, H.J.; Lim, L.H.; Cheong, M.S.; Baek, D.; Park, M.S.; Cho, H.M.; Lee, S.H.; Jin, B.J.; No, D.H.; Cha, Y.J.; et al. Arabidopsis CCoAOMT1 Plays a Role in Drought Stress treatment Response via ROS- and ABA-Dependent Manners. Plants 2021, 10, 831. [Google Scholar] [CrossRef]
- Kai, K.; Mizutani, M.; Kawamura, N.; Yamamoto, R.; Tamai, M.; Yamaguchi, H.; Sakata, K.; Shimizu, B. Scopoletin is biosynthesized viaortho-hydroxylation of feruloyl CoA by a 2-oxoglutarate-dependent dioxygenase in Arabidopsis thaliana. Plant J. 2008, 55, 989–999. [Google Scholar] [CrossRef]
- Fellenberg, C.; van Ohlen, M.; Handrick, V.; Vogt, T. The role of CCoAOMT1 and COMT1 in Arabidopsis anthers. Planta 2012, 236, 51–61. [Google Scholar] [CrossRef]
- Chun, H.J.; Baek, D.; Cho, H.M.; Lee, S.H.; Jin, B.J.; Yun, D.-J.; Hong, Y.-S.; Kim, M.C. Lignin biosynthesis genes play critical roles in the adaptation of Arabidopsis plants to high-salt stress treatment. Plant Signal. Behav. 2019, 14, 1625697. [Google Scholar] [CrossRef]
- Gong, S.L. Study on the Function of Caffeoyl Coenzyme A-O-Methyltransferase Gene in Sugarbeet M14 Strain. Master’s Thesis, Heilongjiang University, Harbin, China, 2013. [Google Scholar]
- Huang, Y.Z.; Qiao, Z.Q.; Liu, S.S.; Chen, K.X.; Huang, X.Z.; He, G. Cloning and Functional Analysis of LmCCoAOMT Gene from Loniceramacranthoides Hand-Mazz. Genom. Appl. Biol. 2021, 40, 325–333. [Google Scholar]
- Miao, L.X. Cloning of AFLP Molecular Markers and Related Genes for Tomato Resistance to Bacterial Wilt Disease. Ph.D. Thesis, Zhejiang University, Hangzhou, China, 2008. [Google Scholar]
- Yokoyama, R.; Nishitani, K. Genomic basis for cell-wall diversity in plants. A comparative approach to gene families in rice and Arabidopsis. Plant Cell Physiol. 2004, 45, 1111–1121. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Sun, Y.-H.; Li, Q.; Heber, S.; Sederoff, R.; Chiang, V.L. Towards a Systems Approach for Lignin Biosynthesis in Populus trichocarpa: Transcript Abundance and Specificity of the Monolignol Biosynthetic Genes. Plant Cell Physiol. 2010, 51, 144–163. [Google Scholar] [CrossRef]
- Joshi, C.P.; Chiang, V.L. Conserved sequence motifs in plant S-adenosyl-L-methionine-dependent methyltransferases. Plant Mol. Biol. 1998, 37, 663–674. [Google Scholar] [CrossRef]
- Kang, M.; Wu, H.; Yang, Q.; Huang, L.; Hu, Q.; Ma, T.; Li, Z.; Liu, J. A chromosome-scale genome assembly of Isatis indigotica, an important medicinal plant used in traditional Chinese medicine. Hortic. Res. 2020, 7, 18. [Google Scholar] [CrossRef]
- Ma, L.; Kong, L.; Jiang, S.; Ma, J.; He, L.; Wu, J.; Zhang, X.; Wu, W.; Ma, W.; Ren, W. Genome-Wide Expression Profile of SOD Gene Family in Isatis indigotica and the Key Role of IiSOD2 and IiSOD7 in Alkaline Stress. Int. J. Mol. Sci. 2025, 26, 8131. [Google Scholar] [CrossRef]
- Jiang, M.; Wang, Z.; Ren, W.; Yan, S.; Xing, N.; Zhang, Z.; Li, H.; Ma, W. Identification of the bZIP gene family and regulation of metabolites under salt stress treatment in Isatis indigotica. Front. Plant Sci. 2022, 13, 1011616. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [PubMed]
- Newbigin, E.; Chou, K.-C.; Shen, H.-B. Plant-mPLoc: A Top-Down Strategy to Augment the Power for Predicting Plant Protein Subcellular Localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S.; Battistuzzi, F.U. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Qu, R.; Miao, Y.; Cui, Y.; Cao, Y.; Zhou, Y.; Tang, X.; Yang, J.; Wang, F. Selection of reference genes for the quantitative real-time PCR normalization of gene expression in Isatis indigotica fortune. BMC Mol. Biol. 2019, 20, 9. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.Y.; Duan, H.X.; Liu, X.P.; Chen, X.; Zhang, Y.; Li, Z.Y. Simultaneous determination of six anti-tumor active ingredients in Da Qing Ye by HPLC multiple evaluation method. Chin. Pharm. 2018, 29, 2635–2639. [Google Scholar]
- Xiao, Y.; Feng, J.X.; Li, Q.; Zhou, Y.Y.; Bu, Q.T.; Zhou, J.H.; Tan, H.X.; Yang, Y.B.; Zhang, L.; Chen, W.S. IiWRKY34 positively regulates yield, lignan biosynthesis and stress tolerance in Isatis indigotica. Acta Pharm. Sin. B 2020, 10, 2417–2432. [Google Scholar] [CrossRef]
- Ma, Q.; Yan, Q.; Zhang, Z.S.; Wu, F.; Zhang, J.Y. Identification, Evolution, and Expression Analysis of CCoAOMT Gene Family in Alfalfa. J. Grassl. Ind. 2021, 30, 144–156. [Google Scholar]
- Shiu, S.-H.; Bleecker, A.B. Expansion of the Receptor-Like Kinase/Pelle Gene Family and Receptor-Like Proteins in Arabidopsis. Plant Physiol. 2003, 132, 530–543. [Google Scholar] [CrossRef]
- Wang, L.; Zhu, W.; Fang, L.; Sun, X.; Su, L.; Liang, Z.; Wang, N.; Londo, J.P.; Li, S.; Xin, H. Genome-wide identification of WRKY family genes and their response to cold stress treatment in Vitis vinifera. BMC Plant Biol. 2014, 14, 103. [Google Scholar] [CrossRef]
- Akhter, S.; Sami, A.A.; Toma, T.I.; Jahan, B.; Islam, T. Caffeoyl-CoA 3-O-methyltransferase gene family in jute: Genome-wide identification, evolutionary progression and transcript profiling under different quandaries. Front. Plant Sci. 2022, 13, 1035383. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Qu, C.; Zuo, Z.; Cao, L.; Zhang, S.; Xu, X.; Xu, Z.; Liu, G. Genome Identification and Expression Profiles in Response to Nitrogen Treatment Analysis of the Class I CCoAOMT Gene Family in Populus. Biochem. Genet. 2021, 60, 656–675. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The Evolutionary Fate and Consequences of Duplicate Genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef]
- Xu, R.-X.; Ni, R.; Gao, S.; Fu, J.; Xiong, R.-L.; Zhu, T.-T.; Lou, H.-X.; Cheng, A.-X. Molecular cloning and characterization of two distinct caffeoyl CoA O-methyltransferases (CCoAOMTs) from the liverwort Marchantia paleacea. Plant Sci. 2022, 314, 111102. [Google Scholar] [CrossRef]
- Zhang, L.; Li, Q.; Chen, J.-F.; Chen, W.-S. Computational identification and systematic classification of novel GRAS genes in Isatis indigotica. Chin. J. Nat. Med. 2016, 14, 161–176. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Ren, J.-Z.; Li, Q.; Yang, B.; Liu, Z.-J.; Chen, R.-B.; Zhang, L. Genome-wide analysis of AP2/ERF superfamily in Isatis indigotica. J. Integr. Med. 2023, 21, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Barakat, A.; Choi, A.; Yassin, N.B.M.; Park, J.S.; Sun, Z.; Carlson, J.E. Comparative genomics and evolutionary analyses of the O-methyltransferase gene family in Populus. Gene 2011, 479, 37–46. [Google Scholar] [CrossRef]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217–218, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Tamagnone, L.; Merida, A.; Parr, A.; Mackay, S.; Culianez-Macia, F.A.; Roberts, K.; Martin, C. The AmMYB308 and AmMYB330 transcription factors from antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell. 1998, 10, 135–154. [Google Scholar] [CrossRef]
- Lin, S.-J.; Yang, Y.-Z.; Teng, R.-M.; Liu, H.; Li, H.; Zhuang, J. Identification and expression analysis of caffeoyl-coenzyme A O-methyltransferase family genes related to lignin biosynthesis in tea plant (Camellia sinensis). Protoplasma 2020, 258, 115–127. [Google Scholar] [CrossRef]
- Sehr, E.M.; Agusti, J.; Lehner, R.; Farmer, E.E.; Schwarz, M.; Greb, T. Analysis of secondary growth in the Arabidopsis shoot reveals a positive role of jasmonate signalling in cambium formation. Plant J. 2010, 63, 811–822. [Google Scholar] [CrossRef]
- Yang, K.; Li, L.; Lou, Y.; Zhu, C.; Li, X.; Gao, Z. A regulatory network driving shoot lignification in rapidly growing bamboo. Plant Physiol. 2021, 187, 900–916. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, C.; Gong, Q.; Wang, Y.; Cao, J.; Li, X.; Grierson, D.; Sun, C.; Costa, F. Characterization of a caffeoyl-CoA O-methyltransferase-like enzyme involved in biosynthesis of polymethoxylated flavones in Citrus reticulata. J. Exp. Bot. 2020, 71, 3066–3079. [Google Scholar] [CrossRef] [PubMed]
- Koirala, N.; Thuan, N.H.; Ghimire, G.P.; Thang, D.V.; Sohng, J.K. Methylation of flavonoids: Chemical structures, bioactivities, progress and perspectives for biotechnological production. Enzyme Microb. Technol. 2016, 86, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Zhou, Y.; Yang, Y.; Chi, Y.J.; Zhou, J.; Chen, J.Y.; Wang, F.; Fan, B.; Shi, K.; Zhou, Y.H.; et al. Structural and functional analysis of VQ motif-containing proteins in Arabidopsis as interacting proteins of WRKY transcription factors. Plant Physiol. 2012, 159, 810–825. [Google Scholar] [CrossRef] [PubMed]
- Widiez, T.; Hartman, T.G.; Dudai, N.; Yan, Q.; Lawton, M.; Havkin-Frenkel, D.; Belanger, F.C. Functional characterization of two new members of the caffeoyl CoA O-methyltransferase-like gene family from Vanilla planifolia reveals a new class of plastid-localized O-methyltransferases. Plant Mol. Biol. 2011, 76, 475–488. [Google Scholar] [CrossRef]
- Zhu, Z.; Sun, B.; Cai, W.; Zhou, X.; Mao, Y.; Chen, C.; Wei, J.; Cao, B.; Chen, C.; Chen, G.; et al. Natural variations in the MYB transcription factor MYB31 determine the evolution of extremely pungent peppers. New Phytol. 2019, 223, 922–938. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, D.; Zhang, W.; Shu, H.; Sun, P.; Huang, C.; Deng, Q.; Wang, Z.; Cheng, S. Genome-Wide Identification of WRKY Gene Family and Functional Characterization of CcWRKY25 in Capsicum chinense. Int. J. Mol. Sci. 2023, 24, 11389. [Google Scholar] [CrossRef]
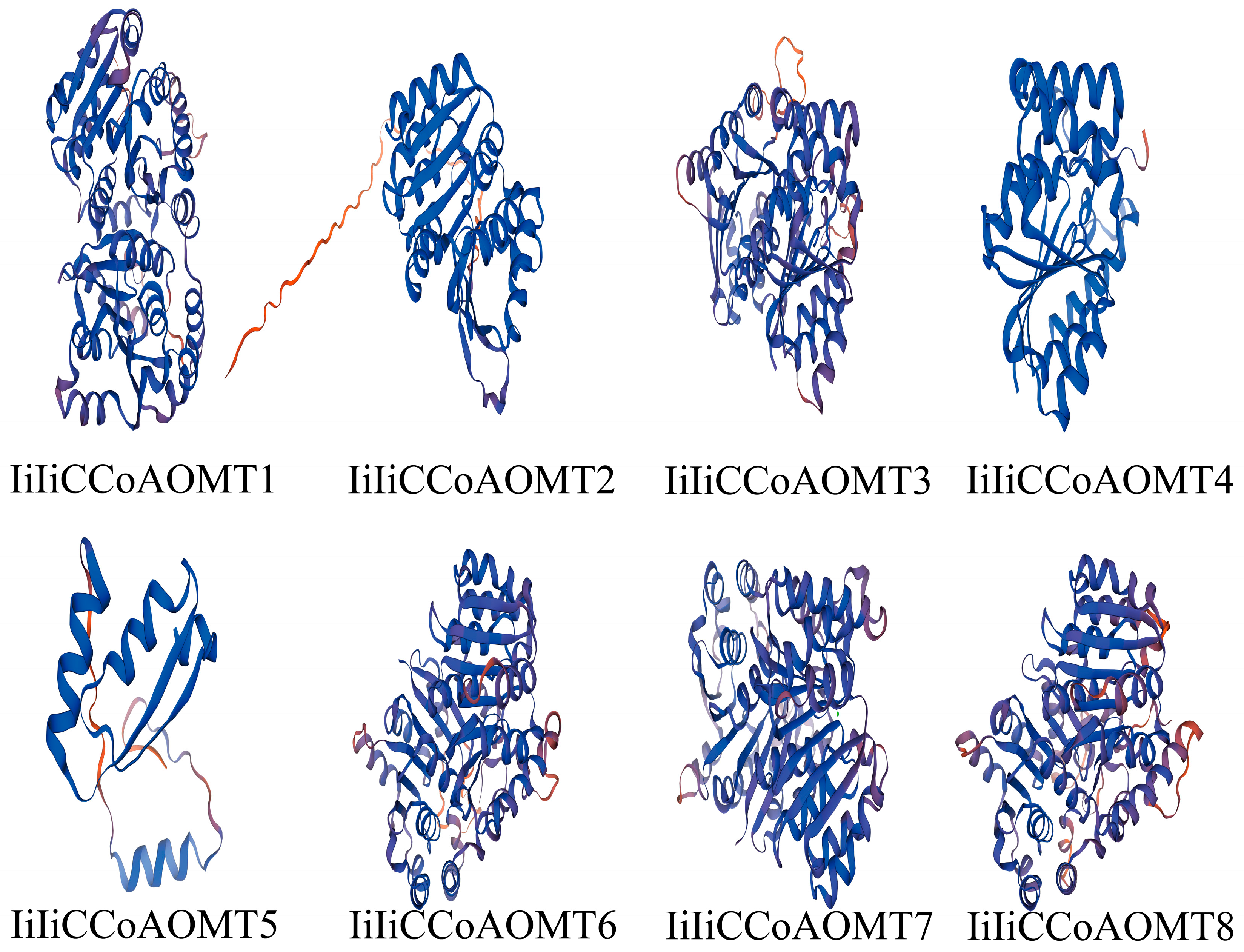

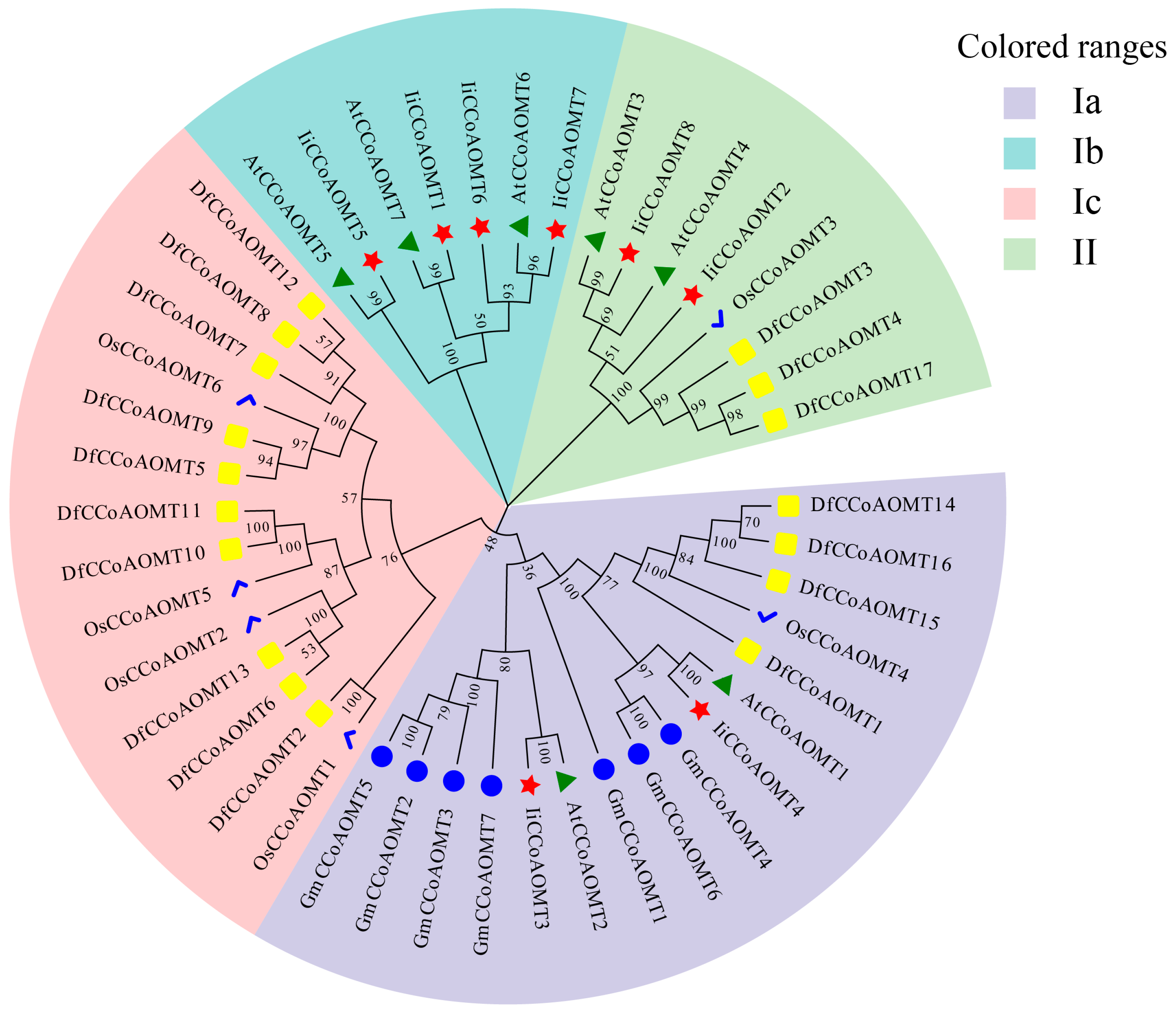
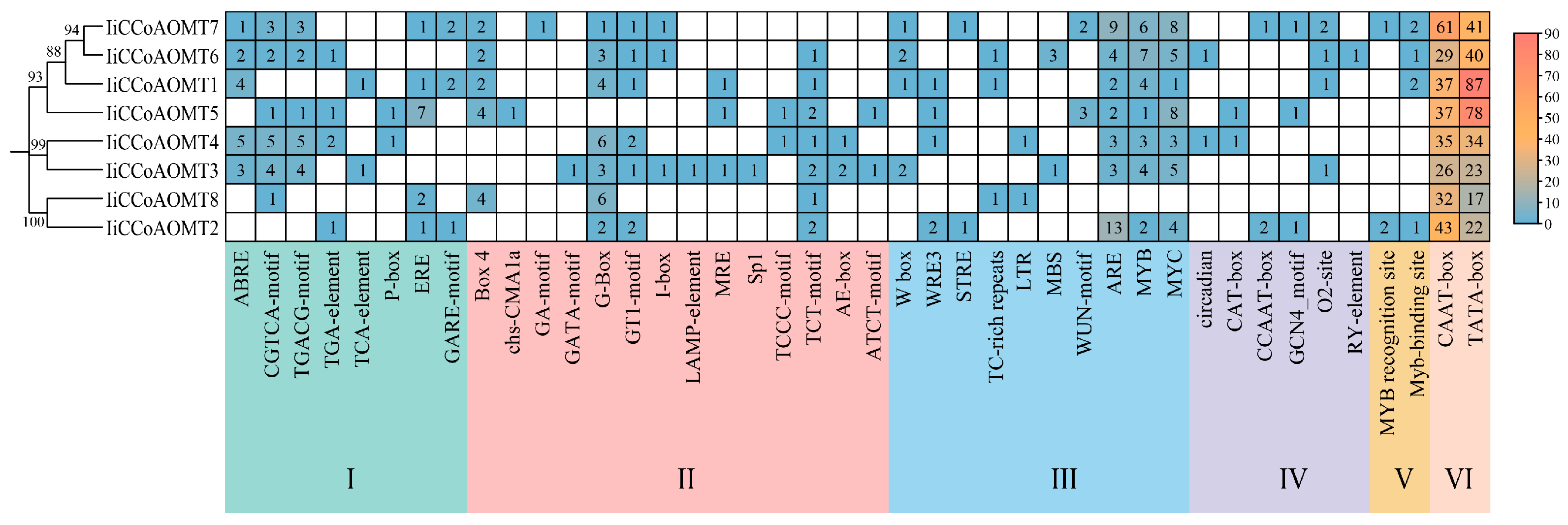



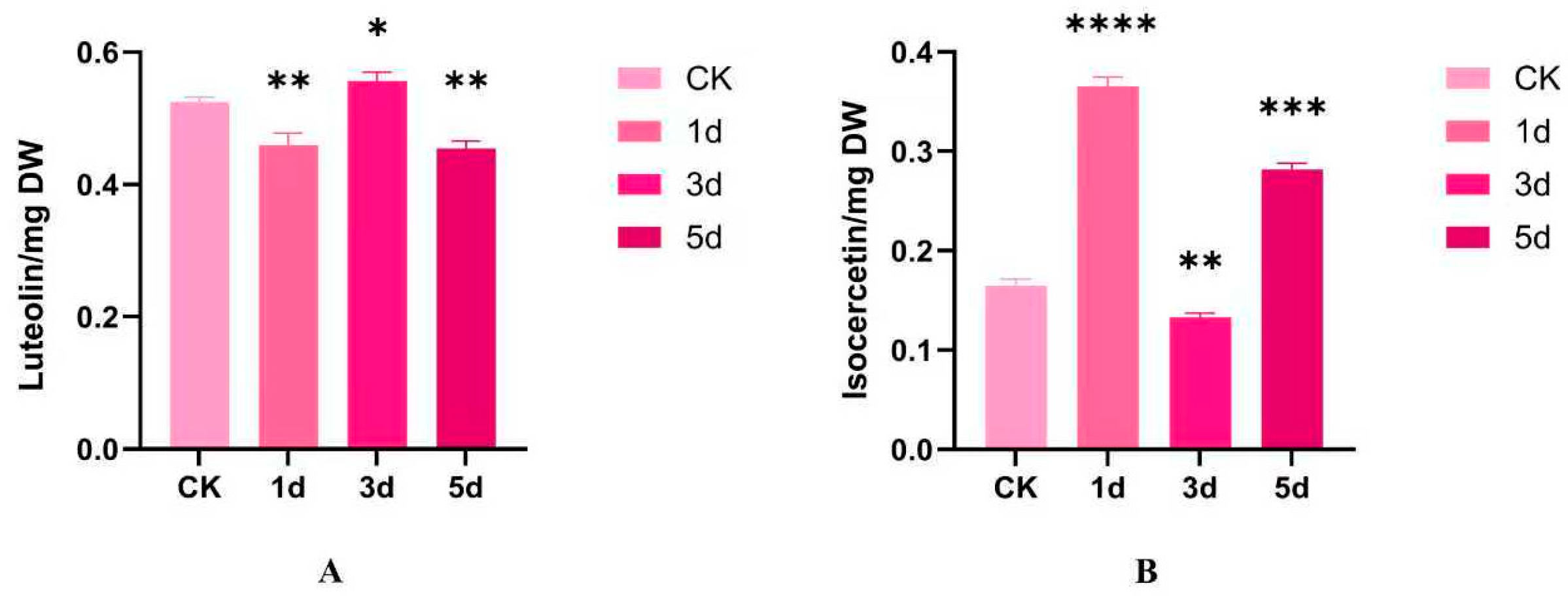
| Gene Name | Accession Number | Chromosome | Number of Amino Acids | Protein Length (aa) | Molecular Weight (KDa) | Isoelectric Point | Subcellular Localization Predicted |
|---|---|---|---|---|---|---|---|
| IiCCoAOMT1 | Iin23419.t1 | Chr3 | 774 | 257 | 28.94 | 5.21 | Specific location not predicted |
| IiCCoAOMT2 | Iin27867.t1 | Chr3 | 834 | 277 | 30.77 | 8.84 | Chloroplast. Cytoplasm |
| IiCCoAOMT3 | Iin25371.t1 | Chr6 | 699 | 232 | 26.19 | 4.96 | Chloroplast |
| IiCCoAOMT4 | Iin25370.t1 | Chr6 | 699 | 232 | 26.31 | 5.17 | Chloroplast |
| IiCCoAOMT5 | Iin11784.t1 | Chr7 | 444 | 147 | 16.67 | 9.37 | Nucleus |
| IiCCoAOMT6 | Iin10582.t1 | Chr7 | 705 | 234 | 26.45 | 4.97 | Chloroplast |
| IiCCoAOMT7 | Iin22645.t1 | Chr7 | 717 | 238 | 26.70 | 5.27 | Peroxisome |
| IiCCoAOMT8 | Iin25369.t1 | Chr7 | 801 | 266 | 30.24 | 5.48 | Cytoplasm |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, B.; Kong, L.; Ma, J.; Jiang, S.; Ma, L.; Xu, J.; Ren, W.; Ma, W. Systematic Analysis of the CCoAOMT Gene Family in Isatis indigotica and the Molecular Mechanism of CCoAOMT8-Mediated Flavonoid Synthesis Under Alkaline Stress Treatment. Biology 2025, 14, 1518. https://doi.org/10.3390/biology14111518
Liu B, Kong L, Ma J, Jiang S, Ma L, Xu J, Ren W, Ma W. Systematic Analysis of the CCoAOMT Gene Family in Isatis indigotica and the Molecular Mechanism of CCoAOMT8-Mediated Flavonoid Synthesis Under Alkaline Stress Treatment. Biology. 2025; 14(11):1518. https://doi.org/10.3390/biology14111518
Chicago/Turabian StyleLiu, Bo, Lingyang Kong, Junbai Ma, Shan Jiang, Lengleng Ma, Jiao Xu, Weichao Ren, and Wei Ma. 2025. "Systematic Analysis of the CCoAOMT Gene Family in Isatis indigotica and the Molecular Mechanism of CCoAOMT8-Mediated Flavonoid Synthesis Under Alkaline Stress Treatment" Biology 14, no. 11: 1518. https://doi.org/10.3390/biology14111518
APA StyleLiu, B., Kong, L., Ma, J., Jiang, S., Ma, L., Xu, J., Ren, W., & Ma, W. (2025). Systematic Analysis of the CCoAOMT Gene Family in Isatis indigotica and the Molecular Mechanism of CCoAOMT8-Mediated Flavonoid Synthesis Under Alkaline Stress Treatment. Biology, 14(11), 1518. https://doi.org/10.3390/biology14111518





