Simple Summary
Long-finned gudgeon fish, Rhinogobio ventralis, is an economically important cyprinid species with a native distribution in the upper tributaries of the Yangtze River, China. Its natural population has significantly declined due to overfishing and habitat destruction in recent decades, and it is therefore classified as endangered in China. Despite its ecological and economic importance, the lack of genomic resources has restricted comprehensive studies in various areas including population conservation, ecological adaptation, and aquaculture development. To resolve this limitation, we established a high-quality chromosome-level genome assembly of Rhinogobio ventralis in this study. Furthermore, genome-wide prediction and localization of antimicrobial peptides (AMPs) containing genes were performed using some reference sequences from public databases. Our findings hence provide a valuable genetic resource for understanding innate immunity and developing novel bioactive compounds.
Abstract
As an economically important species endemic to the upper tributaries of Yangtze River in China, long-finned gudgeon fish (Rhinogobio ventralis) has been classified as endangered due to habitat destruction and population decline. In this study, we constructed a chromosome-level genome assembly of R. ventralis by integration of MGI, PacBio and Hi-C sequencing technologies. The final genome assembly was 1015.9 Mb in length (contig N50: 25.91 Mb; scaffold N50: 39.99 Mb), and 97.19% of the haplotypic genome sequences were anchored onto 25 chromosomes. Repetitive elements accounted for 51.00% of the entire genome assembly. A total of 23,220 protein-coding genes were predicted for the assembled genome, of which 99.79% were functionally annotated. Genome evaluation revealed 99.72% completeness for the genome assembly. Through genome-wide prediction of antimicrobial peptides (AMPs), we identified and localized 561 putative AMP-containing genes in the R. ventralis genome. These genes were further classified into 185 distinct functional categories based on public databases, with the top ten components of Penetratin (21.74%), Histone (5.70%), E6AP (4.09%), Scolopendin 1 (2.67%), D38 (2.31%), WBp-1 (2.13%), Defensin (2.13%), Claudin 1 (1.96%), Azurocidin (AZU1, 1.78%), and Ubiquitin (1.60%). Our data presented here provide a potential genetic resource for promoting fundamental research and wild population conservation of this endangered fish species.
1. Introduction
The Yangtze River is the longest river in China, and it traverses varied geological terrains with over 40 tributaries, sustaining a high biodiversity of aquatic and riparian species. In the 1990s, China initiated a fundamental construction project of Three Gorges Reservoir (TGR) in the upper Yangtze River. After completion of the Three Gorges Dam, the subsequent impoundment of the TGR transformed upper region ecosystems from a lotic (flowing-water) into a lentic (still-water) environment. This alteration significantly affected aquatic animals inhabiting both upstream and downstream of the dam [1,2]. Prior to 2017, a total of 443 fish species (including 194 endemic taxa) were recorded in the Yangtze River, but this number declined to 323 (109 endemic taxa) by 2021 [3]. Scientific studies revealed that lotic-adapted and insectivore fish populations dramatically declined in many areas closed to the TGR [4].
Genus Rhinogobio, belonging to the family Cyprinidae and the order Cypriniformes, contains two endangered species (R. ventralis and R. cylindricus) that inhabit the upper basins of the Yangtze River [5]. R. ventralis (long-finned gudgeon fish) is a benthic insectivore fish and inhabits high-flow streams of the upper tributaries of Yangtze River [5,6]. Following the first and second impoundment phases of the TGR, R. ventralis and other lotic-dependent fish species were presumed to have migrated away from the reservoir’s vicinity [1,7]. However, after the final impoundment in 2011, the natural population of R. ventralis experienced a sharp decline process [7]. In 2016, it was designated as a second-class protected aquatic wildlife species in China. Beyond its traditional importance as a local economic fish, R. ventralis also represents a promising model species for studying lotic-adaptation and endemic fish conservation [8]. Captive breeding is becoming a vital conservation tool for this endangered fish species. To date, previous studies have demonstrated that R. ventralis is susceptible to infection of certain pathogens, such as Aeromonas veronii [9] and Ichthyophthirius multifiliis [10] that are two major disease-causing agents in aquaculture practices.
Antimicrobial peptides (AMPs) are a class of short cationic polypeptides induced by pathogens infection, ultraviolet radiation, temperature or other environmental stresses, exhibiting broad-spectrum antimicrobial activity and playing significant immunomodulatory roles [11]. Natural AMPs originate either from specialized AMP genes (including piscidins, defensins, and cathelicidins) or are produced through proteolytic cleavage of proteins encoded by AMP-containing genes (such as histone, NK-lysin, and chemokine genes) [12]. In practical aquaculture, development of resistance to conventional antibiotics had caused a major economic loss, which led to considerable attention to the potential applications of AMPs. The immunomodulatory functions of various AMPs have drawn much more attention, especially from AMP-containing genes in recent years. Some AMPs from teleost also show potential application for drug development. For example, grass carp IFN1 derived AMPs were reported to have antimicrobial and anti-inflammatory effects in mammals [13].
Genome-wide screening is an effective high-throughput method to identify putative AMPs. With the availability of high-quality genome assemblies, numerous putative AMP sequences were successfully predicted and validated in various fishes, such as giant grouper [14], lined seahorse [15], amphibious mudskippers [16], and black rockfish [17]. The genome-wide prediction enables systematic characterization of AMP-containing genes in target species, revealing their abundance, diversity, and genomic organization, which are essential for investigation of antibacterial mechanisms, immune modulation, host-pathogen coevolutionary adaptation, as well as drug development.
In recent years, massive amounts of genome assemblies are available in public databases because of the rapid development of sequencing technologies with low cost. As a result, we have previously reported several chromosome-level genome assemblies of endemic fish distributed in the upper basins of Yangtze River, including elongate loach (Leptobotia elongata) [18], Lixian plateau loach (Triplophysa lixianensis) [19] and wide-bodied sand loach (Sinibotia reevesae) [20]. R. ventralis possesses significantly ecological and economic significance, but it remains poorly studied due to lack of valuable genomic data. In our present study, we obtained a chromosome-level genome assembly for R. ventralis by integration of MGI, PacBio and Hi-C sequencing technologies. Meanwhile, we conducted genome-wide prediction and localization of putative AMP-containing genes in the assembled genome. Our data presented in this study provide a potentially valuable genetic resource for in-depth studies on ecological adaptation, evolution, and population conservation of this endemic fish species.
2. Materials and Methods
2.1. Sample Collection
An adult female R. ventralis (body length: 244.3 mm, body weight: 152.7 g) was collected from the upper Yangtze River mainstem in Dadukou District (28°44′20.141″ N, 105°14′20.040″ E; Figure 1a) of Luzhou City, Sichuan Province, China. Tissue samples were stored at −80 °C for subsequent DNA and RNA extraction.
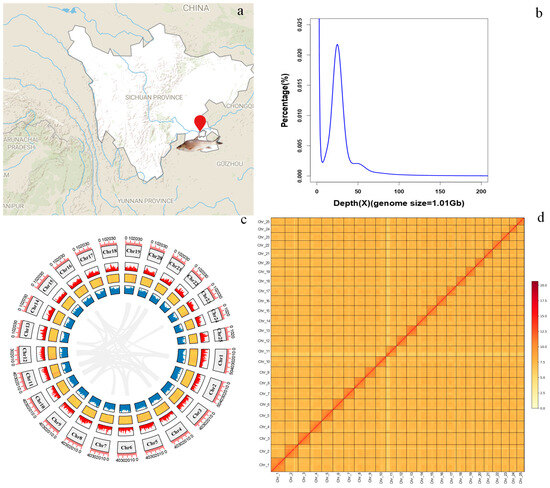
Figure 1.
Chromosome-level genome assembly of R. ventralis. (a) Geographic map of the sample collection site (indicated by a red symbol). (b) K_mer (17-mer) distribution for the genome-size estimation. (c) Genomic features of the 25 chromosomes. From the outer to the inner rings include chromosomes, gene distribution, GC content (a yellow-to-red color gradient represents increasing GC-content), repeat elements, and collinear blocks among each chromosome. (d) Genome-wide chromatin interactions at a 1000-kb resolution in the 25 chromosomes. Color blocks represent the interactions, with various strengths from yellow (low) to red (high).
Genomic DNA (gDNA) was extracted from muscle using QIAamp DNA Mini kit (Qiagen, Valencia, CA, USA), and then quality and quantity of the gDNA were assessed through agarose gel electrophoresis and an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). Total RNA samples were separately isolated from eight tissues (brain, eye, gill, muscle, heart, intestine, kidney, and liver) using TRIzol reagent (TIANGEN, Shanghai, China), and DNA contamination was eliminated with Qiagen RNeasy Mini Kits (Qiagen, Germantown, MD, USA).
2.2. Library Construction and Whole-Genome or Transcriptome Sequencing
MGI libraries with an insert size of 350 bp were constructed by using MGIEasy Universal DNA Library Preparation Kit (MGI, Shenzhen, China), and then they were sequenced on a DNBSEQ T7 platform (MGI, Shenzhen, China). To obtain PacBio HiFi long-reads, SMRT bell libraries (insert size of 15 kb) were generated and sequenced on a PacBio Sequel II platform in accordance with PacBio’s standard protocol (Pacific Biosciences, Menlo Park, CA, USA). For Hi-C sequencing, a Hi-C library was prepared under the manufacturer’s standard experimental guidance (GrandOmics, Wuhan, China) and then sequenced on a DNBSEQ T7 platform (MGI).
For the transcriptome sequencing of eight tissues (brain, eye, gill, muscle, heart, intestine, kidney, and liver), total RNA was used for construction of IIlumina cDNA libraries followed the manufacture’s protocol, which were subsequently sequenced on a HiSeq X Ten platform (Illumina, San Diego, CA, USA). Around 6~10 Gb of raw data were generated for assistance to gene annotation.
2.3. Genome Assembly and Evaluation
To estimate the genome size of R. ventralis, we performed a traditional 17-mer frequency distribution analysis using high-quality MGI short reads, according to the following formula: genome size = K_num/K_dept [21]. Genome-size estimation was conducted by utilizing KMC v3.0.3 and GCE v1.0.2 software [22].
De novo genome assembly of R. ventralis was constructed using Hifiasm (v0.16.0) [23] with default parameters and the high-quality HiFi long-read sequencing data. Subsequently, filtered Hi-C reads were aligned onto the initial assembly via Bowtie2 v2.2.5 [24] with default parameters. HiC-Pro v3 [25] pipeline with default parameters was employed to detect valid contact paired reads. Next, the chromosome-scale assembly was generated by using YaHS v1.2.2 [26] with default parameters, followed by manual curation with Juicebox v1.11.08 [27]. Finally, the Hi-C genome assembly was subjected to TGS-GapCloser v1.2.1 [28] with default parameters to fill N-gap with HiFi reads.
Genomic completeness was comprehensively evaluated using three complementary approaches with default parameters, including (1) CRAQ v1.0.9 [29] for systematic error assessment, (2) Merqury v1.3 [30] for k-mer-based analysis, and (3) Compleasm v0.2.6 [31] for alignment analysis with the actinopterygii_odb10 database as the reference.
2.4. Gene Prediction and Annotation
Homology-based and de novo prediction methods were employed to identify repeat elements (REs) in the R. ventralis genome assembly. For the homology-based prediction, RepeatMasker v4.0.6 [32] and RepeatProteinMask v4.0.6 [32] were applied to predict TEs. Meanwhile, LTR_FINDER v1.0.6 [33] and RepeatModeler v1.0.8 [34] were conducted to build a repeat library for de novo prediction, respectively. Then RepeatMasker v4.0.6 [32] were used to integrate the two libraries to identify TEs against the assembled genome.
To predict protein-coding genes, we implemented three strategies incorporating de novo prediction, homology-based method, and transcriptome-supported annotation. AUGUSTUS v3.2.1 [35] was applied for the ab initio gene prediction. Meanwhile, GeMoMa v1.6.4 [36] was conducted to perform homology prediction with genome annotation of five fish species as queries, including common carp (Cyprinus carpio), zebrafish (Danio rerio), medaka (Oryzias latipes), golden-line barbel (Sinocyclocheilus anshuiensi) and rohu (Labeo rohita). Additionally, transcriptome data from eight tissues (Table 1) were mapped on the genome assembly via Trinity v2.5.13 [37]. Finally, Evidence Modeler (EVM) pipeline v1.04 [38] and PASA v2.3.3 [38] software were employed for integrating the three sets of predicted genes.

Table 1.
Summary of genome and transcriptome sequencing data from an adult female R. ventralis. * For the PacBio HiFi sequencing, this number is read N50; for others, it denotes read length.
Finally, we conducted function assignments for all predicted genes by using BLASTP v 2.2.26 (e-value 1 × 10−5) against five public databases, including SwissProt, Kyoto Encyclopedia of Genes and Genomes (KEGG), EuKaryotic Orthologous Groups (KOG), Gene Ontology (GO) and NCBI Non-Redundant Protein Sequence (NR).
2.5. Genome Comparison and Genomic Synteny
To evaluate the quality of our R. ventralis genome assembly, we also performed a chromosomal collinearity analysis between R. ventralis and its relative R. nasutus [39] by using annotated protein-coding sequences and gene structures with JCVI v190213 [40]. MCscanX v0.8 [41] was employed to identify collinear regions of the 25 chromosomes of R. ventralis. TBtools-II v2.310 [42] was applied to visualize genomic features in a circos plot.
2.6. Genome-Wide Identification of AMP Sequences for Localization of AMP-Containing Genes
To predict putative AMPs, we downloaded 14,957 known AMP sequences from four public databases (accessed date: 13 January 2025), including Database of Antimicrobial Activity and Structure of Peptides (DBAASP, https://dbaasp.org/about, accessed date: 13 January 2025), Antimicrobial Peptide Database (APD3, https://aps.unmc.edu/AP/, accessed date: 13 January 2025), Data bank antimicrobial peptides (dbAMP, https://awi.cuhk.edu.cn/dbAMP/, accessed date: 13 January 2025) and Data repository of antimicrobial peptides (DRAMP, http://dramp.cpu-bioinfor.org/, accessed date: 13 January 2025). We constructed a reference database using the downloaded sequences with formatdb software v2.2.26. Subsequently, the protein sequences of R. ventralis and R. nasutus were aligned against this database using TBLASTN v 2.2.26 (-word_size 7-evalue 1 × 10−5-outfmt 6). Only BLAST hits exhibiting a query alignment ratio exceeding 0.8 and an query coverage of over 0.6 were retained [15]. Finally, putative AMPs were extracted from their corresponding peptide sequences using a custom script based on this refined database. MG2C v2.1 (http://mg2c.iask.in/mg2c_v2.1/, accessed on 25 July 2025) was employed to map putative AMPs onto the R. ventralis chromosomes, with the top ten AMP-containing gene types (Table 2) displayed in distinct color gradients. Phylogenetic analysis was performed on sequences from the major AMP-containing gene category. Multiple sequence alignment was first conducted using MUSCLE v3.8.31 [43], followed by the construction of a maximum likelihood phylogenetic tree with MEGA-X [44], both using default parameters. The phylogenetic tree was further refined for optimal visual presentation utilizing the Interactive Tree of Life (iTOL) v7 platform (https://itol.embl.de/).

Table 2.
Statistic of the assembled R. ventrali genome.
3. Results
3.1. Summary of the Genome Sequencing Data and Assembly
MGI, PacBio HiFi and Hi-C libraries were generated with 54.75, 25.76 and 106.45 Gb of sequencing reads, respectively; for the transcriptome sequencing, approximately 6~9 Gb of clean data from each tissue were obtained (see more details in Table 1). The genome size of R. ventralis was estimated to be 1.01 Gb using the 17-mer frequency distribution analysis (Table 2; Figure 1b).
The assembly of HiFi long reads covered a 1.03-Gb genome with a contig N50 of 25.91 Mb. Using Hi-C scaffolding, the final genome assembly of R. ventralis was 1015.9 Mb in length, within them 987.4 Mb (97.19%) were successfully anchored onto 25 chromosomes (Table 2; Figure 1d). Assessment of genome completeness revealed (1) CRAQ R-AQI (95.47) and S-AQI (99.70) scores, and (2) Merqury QV scores of 49.213 (MGI short reads) and 64.056 (HiFi long reads) and BUSCO completeness (99.72%), validating the high quality of this chromosome-level assembly. See detailed statistics of the assembly and the 25 chromosomes (Chr 1 to 25) in Table 2 and Table 3, respectively.

Table 3.
Length of the assembled chromosomes in the final genome assembly.
3.2. Genome Prediction and Annotation
Integrated homology-based and de novo analyses revealed that repetitive elements comprise 51.00% (545.4 Mb) of the assembled genome (Table 2, Figure 1c). Among the repeat elements, DNA transposons, long interspersed nuclear elements (LINEs), short interspersed nuclear elements (SINEs), and long terminal repeats (LTRs) accounted for 27.81%, 3.41%, 0.52%, and 8.04%, respectively. We employed three different methods (including de novo, homology and transcriptome-based) to predict protein-coding genes. In total, 23,220 genes were annotated in the R. ventralis genome assembly. Subsequent function assignment of these genes against five public databases was conducted. A total of 23,171 genes, accounting for 99.79% of the predicted genes were functionally annotated (see more details in Table 4). Notably, 94.07% completeness of the predicted protein-coding genes was exhibited in the genome assembly. Genomic features including chromosome numbers, gene distribution, GC content, repeat elements, and collinear blocks were illustrated in Figure 1c.

Table 4.
Function annotation of the total protein-coding genes. Total represents the number of annotated genes with at least one hit from the five searched public databases.
3.3. Genome Synteny
Chromosomal collinearity analysis revealed strong genomic synteny between the R. ventralis (RV) and R. nasutus (RN) genomes (Figure 2). All the 25 chromosomes displayed one-to-one synteny between the two relative species, validating the high quality and completeness of our R. ventralis assembly (Figure 1c) established in this study.
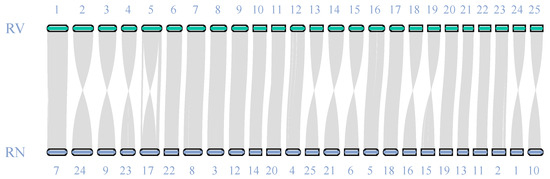
Figure 2.
Genome synteny between R. ventralis (RV) and its relative R. nasutus (RN) [39].
3.4. Identification and Localization of AMP-Containing Genes
Genome-wide screening revealed 561 putative AMP-containing genes in R. ventralis (Table S1); among them 524 (93.4%) were functionally annotated with classification into 185 distinct categories through database comparisons. The remaining 37 unannotated ones were mapped to R. ventralis genes with a prediction to encode 23 histone subunits, 4 complement system components, 2 lysozymes, and 8 other proteins (including RV_ACBP, RV_Ugt2a1, RV_KALM, RV_KPYM, RV_NLRP1 and RV_RNSL3).
Predicted AMPs were widely distributed across the 25 chromosomes of R. ventralis, with Chr 1–4 and 23 each containing ≥30 AMP-containing genes. Specifically, the Chr 24 exhibited the lowest AMP density with only 10 AMP-containing genes. These genes exhibited diverse classifications, with the ten most abundant categories (Table 5) including Penetratin (and deviated analogs, 21.74%,), Histone (5.70%), E6AP (4.09%), Scolopendin 1 (2.67%), D38 (2.31%), WBp-1 (2.13%), Defensin (2.13%), Claudin 1 (1.96%), Azurocidin (AZU1, 1.78%), and Ubiquitin (1.60%). Notably, penetratin genes formed distinct clusters on the Chr 4–5 and 16–17; histone and hemoglobin AMP-containing genes showed clustering on the Chr 5 and 23, respectively (see Figure 3). A phylogenetic analysis resolved the 122 penetratin-containing genes into several major clusters, with the most abundant one being identified as the Hox family (33 genes; see more details in Figure S1).

Table 5.
Number of the top ten categories of AMP-containning genes in R. ventralis and R. nasutus.
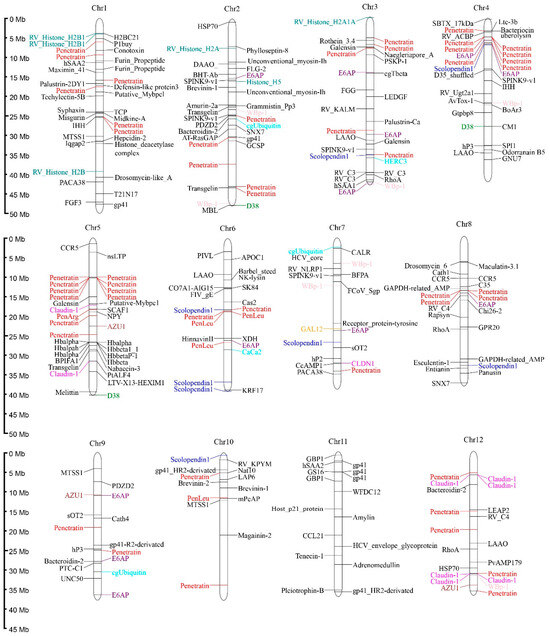
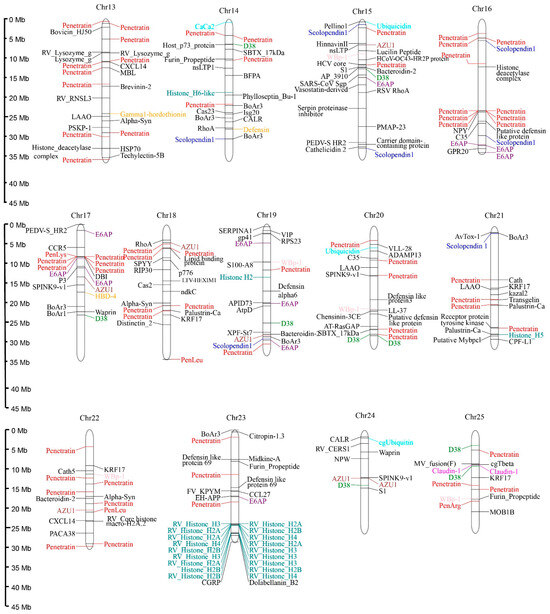
Figure 3.
Distribution of 561 AMP-containing genes on the 25 chromosomes of the R. ventralis genome. Various colors for different AMP categories: Penetratin, red; Histone, blue-green; E6AP, purple; Scolopendin 1, blue; D38, green; WBp-1, pink; Defensin, orange; Claudin 1, magenta; AZU1, brown; Ubiquitin, cyan; the others, black.
R. nasutus, a close relative of R. ventralis, is another endangered endemic fish restricted to the Yellow River. To compare their AMP profiles of these two phylogenetically related but geographically isolated species, we performed a genome-wide AMP-containing gene screening in R. nasutus [39] as well. Obviously, this species also possesses an extensive repertoire of putative AMP-containing genes (a total of 855; see Table S1). Among them, 24 annotated sequences were mapped to the histone-family members. Detailed numbers of the top ten categories of AMP-containing genes identified in both R. ventralis and R. nasutus are listed (in Table 5) for comparison.
4. Discussion
As an endangered and endemic cyprinid species of the Yangtze River, R. ventralis plays important roles with regard to biodiversity, adaptation, and population restoration. Despite its considerable ecological and economic importance, R. ventralis remains poorly studied. In this study, we produced a high-quality chromosome-level genome assembly for R. ventralis, with 97.19% (987.4 Mb) of the entire sequences anchored onto 25 chromosomes, consistent with the known karyotype for this species. To validate the quality of this genome assembly, we also performed chromosomal collinearity analysis with that of its close relative R. nasutus, which possesses a complete telomere-to-telomere reference genome assembly [39]. The high degree of synteny observed across the total 25 chromosome pairs (Figure 2) confirmed the high quality of our R. ventralis genome assembly (Figure 1c), which will be beneficial for further evolutionary and functional as well as conservative studies for this endangered species.
As part of the innate immune system, AMPs always play a critical role in immune defense and immune regulation. According to the genome-wide identification of AMPs, the entire number and category of AMPs are various in different species. The substantial difference in putative AMP-containing genes between R. ventralis (561) and R. nasutus (855) reveals different AMP profiles, which might be due to their distinct surrounding microbial environments in their respective riverine ecosystems. In contrast to prior studies that relied on a single AMP database, our present investigation integrated the most recent data from four public databases to ensure comprehensive coverage. Furthermore, we implemented strict e-value thresholds during sequence alignment to enhance detection reliability. In our current study, the Penetratin family (including its synthetic analogs) were identified as the predominant class of AMP-containing genes in both R. ventralis and R. nasutus genomes. Previous studies have demonstrated that thrombin are most abundant across 27 species, such as amphibious mudskippers [16], the lined seahorse (Hippocampus erectus) [15], the giant grouper (Epinephelus lanceolatus) [14], tilapias (Oreochromis niloticus and O. aureus), black rockfish (Sebastes schlegelii) [17] and golden pompano (Trachinotus ovatus) [45]. These differences may be caused by the distinct inhabited environments of these fish species, since the two species investigated in this study are living in freshwater ecosystems but the other examined fish species are commonly resident in seawater or euryhaline ecosystems.
Penetratin was initially discovered in the Antennapedia homeodomain of Drosophila, and belong to the cell-penetrating peptide (CPP) family [46]. Both penetratin and its derived analogs exhibit dual functional advantages, e.g., membrane-penetrating capability coupled with antimicrobial activity against bacteria and fungi [47,48,49,50]. Compared to native penetratin, a synthetic analog penArg was demonstrated to have enhanced antibacterial activity against Staphylococcus aureus and Escherichia coli, while PenLys and PenLeu were exhibited to possess significantly reduced cytotoxicity [48]. The high abundance of penetratin indicate its potential immune defense against both bacterial and fungi pathogens in Rhinogobio species. The second top abundant AMP-containing gene category in both Rhinogobio species was Histone. In addition to conventional antibacterial peptides, Histone-derived AMPs can synergize with other AMPs (e.g., magainin-2 and LL-37) to enhance antimicrobial activity, thus inducing the expression of major histocompatibility complex (MHC)-related genes [51]. We also identified several novel potential AMP-containing gene categories in both Rhinogobio fishes, such as antiviral CRISPR-associated endoribonuclease Cas2 family [52], intestinal microbiota-regulated bacteroidin-2 [53], and (3) immunomodulatory azurocidin 1 (enhancing cytokine release) [54], which are worthy of more investigations. In brief, our present study identified differential AMP-containing genes in a high through-put way between two Rhinogobio species that originated from different riverine systems.
5. Conclusions
In this study, we assembled a high-quality chromosome-level genome for the endangered endemic species R. ventralis through an integrative approach of MGI short-read, PacBio long-read, and Hi-C sequencing technologies. The final genome assembly was 1015.9 Mb with high continuity and completeness. Comparative synteny analysis of the 25 chromosomal pairs between R. ventralis (RV) and its relative R. nasutus (RN) revealed conserved one-to-one correspondence. To obtain comprehensive AMPs of Rhinogobio species from distinct riverine ecosystems, we performed genome-wide prediction in both Rhinogobio fishes to identify 561 and 855 putative AMP-containing genes, respectively. Among them, Penetratin and Histone genes represented the most abundant categories in both species. In R. ventralis, the identified AMP-containing genes were widely distributed across the total 25 chromosomes, with five chromosomes harboring ≥30 genes each. The high-quality genome assembly and comprehensive AMP-containing gene characterization of R. ventralis establish a potentially valuable genomic resource that could inform conservation strategies for this endangered species, elucidate mechanisms of lotic adaptation, and facilitate development of novel antimicrobial agents or drug delivery systems for both aquaculture and human medicine applications.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology14111486/s1. Figure S1: Phylogenetic analysis of penetratin-containing genes in R. ventralis. Table S1: A genome-wide comparison of AMP-containing genes between R. ventralis and its relative R. nasutus.
Author Contributions
Conceptualization, Z.W. and Q.S.; methodology, Z.W., X.Y. and Q.S.; software, J.C. and X.Z.; validation, Y.L. (Yunyun Lv); formal analysis, J.C., X.Z. and Y.L. (Yanping Li); investigation, J.C., X.Z., Y.L. (Yanping Li) and Z.W.; resources, X.Z. and Y.L. (Yunyun Lv); data curation, J.C. and X.Z.; writing—original draft preparation, J.C.; writing—review and editing, Z.W., X.Y. and Q.S.; visualization, Y.L. (Yanping Li); supervision, Z.W. and Q.S.; project administration, Z.W.; funding acquisition, Z.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Natural Science Fund of Sichuan Province of China (no. 2023NSFSC1221), the Project of Sichuan Provincial Department of Science and Technology (no. ZYZFSC22004), and the Research Fund from Key Laboratory of Sichuan Province for Fishes Conservation and Utilization in the Upper Reaches of Yangtze River (no. NJTCSC23-3).
Institutional Review Board Statement
The animal study protocol was approved by the Ethics Committee of Neijiang Normal University (protocol code SKXY2023008; 12 June 2023).
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article. Further inquiries can be directed to the corresponding authors.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of this study, data analysis, manuscript preparation, or the decision to publish the results.
References
- Gao, X.; Zeng, Y.; Wang, J.W.; Liu, H.Z. Immediate impacts of the second impoundment on fish communities in the Three Gorges Reservoir. Environ. Biol. Fish. 2010, 87, 163–173. [Google Scholar] [CrossRef]
- Li, Y.L.; Yang, J.J.; Wang, Y.H.; Wu, H.C.; Ma, Y.M.; Wu, F.X. Sediment eDNA reveals damming triggered changes in algal and fish communities at the Three Gorges Reservoir in China. Environ. Res. 2025, 276, 121474. [Google Scholar] [CrossRef]
- Yang, H.L.; Shen, L.; He, Y.F.; Tian, H.W.; Gao, L.; Wu, J.M.; Mei, Z.G.; Wei, N.; Lin, W.; Zhu, T.B.; et al. Status of aquatic organisms resources and their environments in Yangtze River system (2017–2021). Aquac. Fish. 2024, 9, 833–850. [Google Scholar] [CrossRef]
- Gao, X.; Masami, F.; Winemiller, K.O.; Lin, P.C.; Li, M.Z.; Liu, H.Z. Regime shift in fish assemblage structure in the Yangtze River following construction of the Three Gorges Dam. Sci. Rep. 2019, 9, 4212–4222. [Google Scholar] [CrossRef] [PubMed]
- Yue, P.Q. Gobioninae. In Fauna Sinica: Osteichthys, Cypriniformes, 2nd ed.; Chen, Y.Y., Ed.; Science Press: Beijing, China, 1998; Volume 2, pp. 232–389. (In Chinese) [Google Scholar]
- Wang, X.Z.; Liu, H.Z. Phylogenetic relationships of the Chinese cyprinid genus Rhinogobio Bleeker (Teleostei: Cyprinidae) based on sequences of the mitochondrial DNA control region, with comments on character adaptations. Hydrobiologia 2005, 532, 215–220. [Google Scholar] [CrossRef][Green Version]
- Liu, F.; Wang, J.; Cao, W. Long-term changes in fish assemblage following the impoundments of the Three Gorges Reservoir in Hejiang, a protected reach of the upper Yangtze River. Knowl. Manag. Aquat. Ecosyst. 2013, 407, 6–22. [Google Scholar] [CrossRef]
- Liu, F.; Wang, J.W.; Liu, H.Z. Seasonal variations in food resource partitioning among four sympatric gudgeon species in the upper Yangtze River. Ecol. Evol. 2019, 9, 7227–7236. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Cheng, B.; Xue, M.; Jiang, N.; Li, X.; Hu, X.; Li, X.; Zhu, T.; Zhu, Y.; Zhou, Y. Isolation, Characterization, and Pathogenicity of an Aeromonas veronii Strain Causing Disease in Rhinogobio ventralis. Fishes 2024, 9, 188–201. [Google Scholar] [CrossRef]
- Huang, K.; Wang, R.; Hu, G.; Zhou, W.; Li, W.; Zou, H.; Wang, G.; Li, M. Immune response of Rhinogobio ventralis to Ichthyophthirius multifiliis infection: Insights from histopathological and real-time gene expression analyses. Fish Shellfish Immunol. 2024, 153, 109801. [Google Scholar] [CrossRef]
- Li, H.; Niu, J.; Wang, X.; Niu, M.; Liao, C. The Contribution of Antimicrobial Peptides to Immune Cell Function: A Review of Recent Advances. Pharmaceutics 2023, 15, 2278–2315. [Google Scholar] [CrossRef]
- Masso-Silva, J.A.; Diamond, G. Antimicrobial peptides from fish. Pharmaceuticals 2014, 7, 265–310. [Google Scholar] [CrossRef]
- Xiao, X.; Lu, H.; Zhu, W.; Zhang, Y.; Huo, X.; Yang, C.; Xiao, S.; Zhang, Y.; Su, J.A. Novel Antimicrobial Peptide Derived from Bony Fish IFN1 Exerts Potent Antimicrobial and Anti-Inflammatory Activity in Mammals. Microbiol. Spectr. 2022, 10, e02013-21. [Google Scholar] [CrossRef]
- Wang, D.; Chen, X.; Zhang, X.; Li, J.; Yi, Y.; Bian, C.; Shi, Q.; Lin, H.; Li, S.; Zhang, Y.; et al. Whole Genome Sequencing of the Giant Grouper (Epinephelus lanceolatus) and High-Throughput Screening of Putative Antimicrobial Peptide Genes. Mar. Drugs 2019, 17, 503–518. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yi, Y.; You, X.; Liu, J.; Shi, Q. High-Throughput Identification of Putative Antimicrobial Peptides from Multi-Omics Data of the Lined Seahorse (Hippocampus erectus). Mar. Drugs 2019, 18, 30–48. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.; You, X.; Bian, C.; Chen, S.; Lv, Z.; Qiu, L.; Shi, Q. High-Throughput Identification of Antimicrobial Peptides from Amphibious Mudskippers. Mar. Drugs 2017, 15, 364–381. [Google Scholar] [CrossRef]
- Zhang, M.; Cao, M.; Xiu, Y.; Fu, Q.; Yang, N.; Su, B.; Li, C. Identification of Antimicrobial Peptide Genes in Black Rockfish Sebastes schlegelii and Their Responsive Mechanisms to Edwardsiella tarda Infection. Biology 2021, 10, 1015–1035. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Wei, X.; Chen, J.; Li, Y.; Zhou, B.; Zhang, C.; Fu, P.; Prathomya, P.; Li, R.; Lv, Y.; et al. Chromosome-level genome assemblies of vulnerable male and female elongate loach (Leptobotia elongata). Sci. Data 2024, 11, 924–931. [Google Scholar] [CrossRef]
- He, C.; Zhang, X.; Wen, Z.; Shi, Q.; Song, Z. A chromosome-scale reference genome assembly for Triplophysa lixianensis. Sci. Data 2024, 11, 1404–1410. [Google Scholar] [CrossRef]
- Lv, Y.; Li, Y.; Huang, Y.; Wang, J.; Tian, Z.; He, Y.; Shi, J.; Huang, Z.; Wen, Z.; Shi, Q.; et al. Deciphering genome-wide molecular pathways for exogenous Aeromonas hydrophila infection in wide-bodied sand loach (Sinibotia reevesae). Aquac. Rep. 2024, 35, 102033–102043. [Google Scholar] [CrossRef]
- Vurture, G.W.; Sedlazeck, F.J.; Nattestad, M.; Underwood, C.J.; Fang, H.; Gurtowski, J.; Schatz, M.C. GenomeScope: Fast reference-free genome profiling from short reads. Bioinformatics 2017, 33, 2202–2204. [Google Scholar] [CrossRef]
- Liu, B.; Shi, Y.; Yuan, J.; Hu, X.; Zhang, H.; Li, N.; Li, Z.; Chen, Y.; Mu, D.; Wei, F. Estimation of genomic characteristics by analyzing k-mer frequency in de novo genome projects. Quant. Biol. 2013, 35, 62–67. [Google Scholar]
- Cheng, H.; Concepcion, G.T.; Feng, X.; Zhang, H.; Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 2021, 18, 170–175. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Varoquaux, N.; Lajoie, B.R.; Viara, E.; Chen, C.; Vert, J.P.; Heard, E.; Job Dekker, J.; Barillot, E. HiC-Pro: An optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015, 16, 259–269. [Google Scholar]
- Zhou, C.; McCarthy, S.A.; Durbin, R. YaHS: Yet another Hi-C scaffolding tool. Bioinformatics 2023, 39, btac808. [Google Scholar] [CrossRef] [PubMed]
- Durand, N.C.; Robinson, J.T.; Shamim, M.S.; Machol, I.; Mesirov, J.P.; Lander, E.S.; Aiden, E.L. Juicebox Provides a Visualization System for Hi-C Contact Maps with Unlimited Zoom. Cell Syst. 2016, 3, 99–101. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Guo, L.; Gu, S.; Wang, O.; Zhang, R.; Peters, B.A.; Fan, G.; Liu, X.; Xu, X.; Deng, L.; et al. TGS-GapCloser: A fast and accurate gap closer for large genomes with low coverage of error-prone long reads. Gigascience 2020, 9, giaa094. [Google Scholar] [CrossRef]
- Li, K.; Xu, P.; Wang, J.; Yi, X.; Jiao, Y. Identification of errors in draft genome assemblies at single-nucleotide resolution for quality assessment and improvement. Nat. Commun. 2023, 14, 6556–6567. [Google Scholar] [CrossRef]
- Rhie, A.; Walenz, B.P.; Koren, S.; Phillippy, A.M. Merqury: Reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 2020, 21, 245–271. [Google Scholar] [CrossRef]
- Huang, N.; Li, H. compleasm: A faster and more accurate reimplementation of BUSCO. Bioinformatics 2023, 39, btad595. [Google Scholar] [CrossRef]
- Tarailo-Graovac, M.; Chen, N. Using Repeat Masker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. 2009, 5, 4–10. [Google Scholar]
- Xu, Z.; Wang, H. LTR_FINDER: An efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007, 35, W265–W268. [Google Scholar] [CrossRef]
- Flynn, J.M.; Hubley, R.; Goubert, C.; Rosen, J.; Clark, A.G.; Feschotte, C.; Smit, A.F. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. USA 2020, 117, 9451–9457. [Google Scholar] [CrossRef]
- Stanke, M.; Keller, O.; Gunduz, I.; Hayes, A.; Waack, S.; Morgenstern, B. AUGUSTUS: Ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006, 34, W435–W439. [Google Scholar] [CrossRef]
- Keilwagen, J.; Hartung, F.; Grau, J. GeMoMa: Homology-based gene prediction utilizing intron position conservation and RNA-seq data. In Gene Prediction; Springer: New York, NY, USA, 2019; pp. 161–177. [Google Scholar]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Salzberg, S.L.; Zhu, W.; Pertea, M.; Allen, J.E.; Orvis, J.; White, O.; Buell, C.R.; Wortman, J.R. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008, 9, R7. [Google Scholar] [CrossRef]
- Jiang, C.; Du, Y.; Lou, Z.; Zhang, Y.; Wang, T. Telomere-to-telomere reference genome of Rhinogobio nasutus, an endangered endemic fish from the Yellow River. Sci. Data 2025, 12, 462–471. [Google Scholar] [CrossRef]
- Tang, H.; Krishnakumar, V.; Bidwell, S.; Rosen, B.; Chan, A.; Zhou, S.; Gentzbittel, L.; Childs, K.L.; Yandell, M.; Gundlach, H.; et al. An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genom. 2014, 15, 312–325. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Quality measures for protein alignment benchmarks. Nucleic Acids Res. 2010, 38, 2145–2153. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Liang, Y.; Pan, J.M.; Zhu, K.C.; Xian, L.; Guo, H.Y.; Liu, B.S.; Zhang, N.; Yang, J.W.; Zhang, D.C. Genome-Wide Identification of Trachinotus ovatus Antimicrobial Peptides and Their Immune Response against Two Pathogen Challenges. Mar. Drugs 2023, 21, 505–532. [Google Scholar] [CrossRef]
- Derossi, D.; Joliot, A.H.; Chassaing, G.; Prochiantz, A. The third helix of the Antennapedia homeodomain translocates through biological membranes. J. Biol. Chem. 1994, 269, 10444–104450. [Google Scholar] [CrossRef] [PubMed]
- Garibotto, F.M.; Garro, A.D.; Rodríguez, A.M.; Raimondi, M.; Zacchino, S.A.; Perczel, A.; Somlai, C.; Penke, B.; Enriz, R.D. Penetratin analogues acting as antifungal agents. Eur. J. Med. Chem. 2011, 46, 370–377. [Google Scholar] [CrossRef]
- Bahnsen, J.S.; Franzyk, H.; Sandberg-Schaal, A.; Nielsen, H.M. Antimicrobial and cell-penetrating properties of penetratin analogs: Effect of sequence and secondary structure. Biochim. Biophys. Acta 2013, 1828, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.L.; Lan, H.; Park, I.S.; Kim, J.I.; Jin, H.Z.; Hahm, K.S.; Shin, S.Y. Design and mechanism of action of a novel bacteria-selective antimicrobial peptide from the cell-penetrating peptide Pep-1. Biochem. Biophys. Res. Commun. 2006, 349, 769–774. [Google Scholar] [CrossRef]
- Zhu, W.L.; Shin, S.Y. Antimicrobial and cytolytic activities and plausible mode of bactericidal action of the cell penetrating peptide penetratin and its lys-linked two-stranded peptide. Chem. Biol. Drug Des. 2009, 73, 209–215. [Google Scholar] [CrossRef]
- Duong, L.; Gross, S.P.; Siryaporn, A. A novel antibacterial strategy: Histone and antimicrobial peptide synergy. Microb. Cell 2020, 7, 309–311. [Google Scholar] [CrossRef] [PubMed]
- Beloglazova, N.; Brown, G.; Zimmerman, M.D.; Proudfoot, M.; Makarova, K.S.; Kudritska, M.; Kochinyan, S.; Wang, S.; Chruszcz, M.; Minor, W.; et al. A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. J. Biol. Chem. 2008, 283, 20361–20371. [Google Scholar] [CrossRef]
- Torres, M.D.T.; Brooks, E.F.; Cesaro, A.; Sberro, H.; Gill, M.O.; Nicolaou, C.; Bhatt, A.S.; de la Fuente-Nunez, C. Mining human microbiomes reveals an untapped source of peptide antibiotics. Cell 2024, 187, 5453–5467.e15. [Google Scholar] [CrossRef] [PubMed]
- Soehnlein, O.; Lindbom, L. Neutrophil-derived azurocidin alarms the immune system. J. Leukoc. Biol. 2009, 85, 344–351. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).