StomachDB: An Integrated Multi-Omics Database for Gastric Diseases
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Single-Cell and Spatial Transcriptome Data Collection and Reanalysis
2.2. Differential Expression Gene and Tissue-Specific Gene Analysis in Transcriptomics
2.3. Collection and Curation of Other Multi-Omics Data
2.4. Data Inclusion/Exclusion Criteria, Batch Effect Correction, and Quality Control
2.5. Data Integration, Standardization, and ID Mapping
2.6. Database Construction and Web Interface Implementation
3. Results
3.1. Construction Pipeline and Core Modules of StomachDB
3.2. Multifaceted Associations Between Gastric Diseases and Genes
3.3. Classification of Gastric Diseases, Treatment Strategies, and Multi-Omics Data Across Species
3.4. Spatial and Single-Cell Omics Data for Gastric Diseases
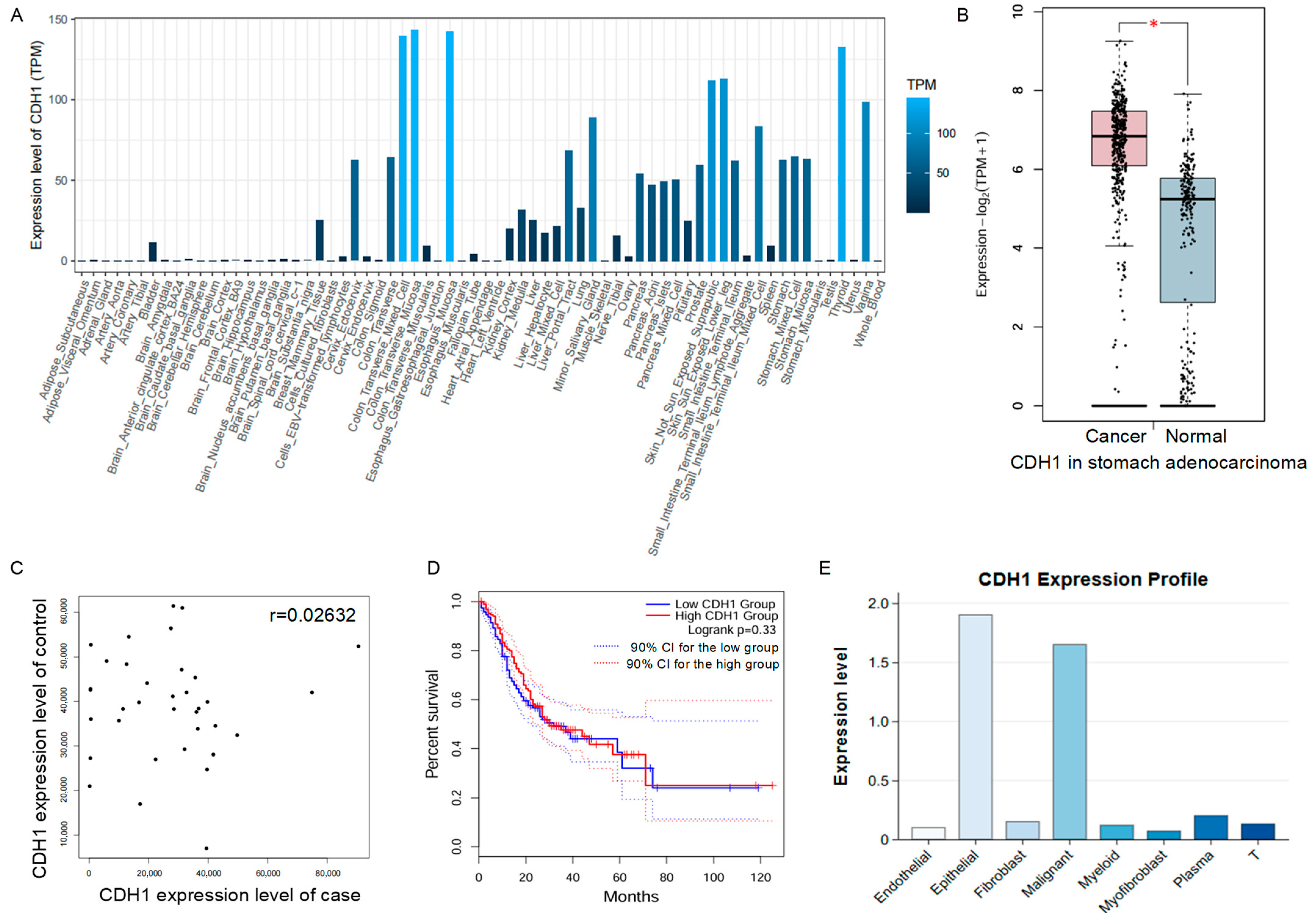
3.5. Interactive Visualization of Multi-Omics Profiles for Gastric Diseases
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Y.; Huang, Y.; Chase, R.C.; Li, T.; Ramai, D.; Li, S.; Huang, X.; Antwi, S.O.; Keaveny, A.P.; Pang, M. Global Burden of Digestive Diseases: A Systematic Analysis of the Global Burden of Diseases Study, 1990 to 2019. Gastroenterology 2023, 165, 773–783.e715. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Shi, Y.; Dai, S.; Deng, M.; Zhu, K.; Xu, Y.; Chen, Z.; Xu, Z.; Zhang, T.; Liang, S. The role of MAPK pathway in gastric cancer: Unveiling molecular crosstalk and therapeutic prospects. J. Transl. Med. 2024, 22, 1142. [Google Scholar] [CrossRef]
- Duan, Y.; Xu, Y.; Dou, Y.; Xu, D. Helicobacter pylori and gastric cancer: Mechanisms and new perspectives. J. Hematol. Oncol. 2025, 18, 10. [Google Scholar] [CrossRef]
- Ben-Baruch Morgenstern, N.; Shoda, T.; Rochman, Y.; Caldwell, J.M.; Collins, M.H.; Mukkada, V.; Putnam, P.E.; Bolton, S.M.; Felton, J.M.; Rochman, M.; et al. Local type 2 immunity in eosinophilic gastritis. J. Allergy Clin. Immunol. 2023, 152, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Tong, Q.Y.; Pang, M.J.; Hu, X.H.; Huang, X.Z.; Sun, J.X.; Wang, X.Y.; Burclaff, J.; Mills, J.C.; Wang, Z.N.; Miao, Z.F. Gastric intestinal metaplasia: Progress and remaining challenges. J. Gastroenterol. 2024, 59, 285–301. [Google Scholar] [CrossRef]
- Dong, L.; Zhu, J.; Deng, A.; Wei, J.; Li, J.; Mao, X.; Jia, Z. Relationship between histone demethylase LSD family and development and prognosis of gastric cancer. Front. Immunol. 2023, 14, 1170773. [Google Scholar] [CrossRef] [PubMed]
- Ni, T.; Wang, H.; Zhan, D.; Tao, L.; Lv, M.; Wang, W.; Chu, Z.; Zhou, Z.; Sunagawa, M.; Liu, Y. CD133+/CD166+ human gastric adenocarcinoma cells present the properties of neoplastic stem cells and emerge more malignant features. Life Sci. 2021, 269, 119021. [Google Scholar] [CrossRef]
- Wang, J.B.; Gao, Y.X.; Ye, Y.H.; Zheng, Q.L.; Luo, H.Y.; Wang, S.H.; Zhang, T.; Jin, Q.W.; Zheng, C.H.; Li, P.; et al. Comprehensive multi-omics analysis of pyroptosis for optimizing neoadjuvant immunotherapy in patients with gastric cancer. Theranostics 2024, 14, 2915–2933. [Google Scholar] [CrossRef]
- Sun, C.; Wang, A.; Zhou, Y.; Chen, P.; Wang, X.; Huang, J.; Gao, J.; Wang, X.; Shu, L.; Lu, J.; et al. Spatially resolved multi-omics highlights cell-specific metabolic remodeling and interactions in gastric cancer. Nat. Commun. 2023, 14, 2692. [Google Scholar] [CrossRef]
- Yang, Z.; Kotoge, R.; Piao, X.; Chen, Z.; Zhu, L.; Gao, P.; Matsubara, Y.; Sakurai, Y.; Sun, J. MLOmics: Cancer Multi-Omics Database for Machine Learning. Sci. Data. 2025, 12, 913. [Google Scholar] [CrossRef]
- Li, M.; Chen, Z.; Deng, S.; Wang, L.; Yu, X. MOSDNET: A multi-omics classification framework using simplified multi-view deep discriminant representation learning and dynamic edge GCN with multi-task learning. Comput Biol. Med. 2024, 181, 109040. [Google Scholar] [CrossRef]
- Yi, H.; Yang, Q.; Repaci, C.; Lee, C.M.; Heo, G.; Timsina, J.; Gorijala, P.; Yang, C.; Budde, J.; Wang, L.; et al. TOPMed imputed genomics enhances genomic atlas of the human proteome in brain, cerebrospinal fluid, and plasma. Sci. Data. 2024, 11, 387. [Google Scholar] [CrossRef]
- Zhou, K.; Yang, C.; Li, Y. Multi-omics in colorectal cancer liver metastasis: Applications and research advances. Cancer Biol. Med. 2025, 22, 618–638. [Google Scholar] [CrossRef]
- Zhou, K.; Yang, C.; Li, Y. Multi-Omics Graph Knowledge Representation for Pneumonia Prognostic Prediction. IEEE J. Biomed. Health Inform. 2025, 29, 3021–3034. [Google Scholar]
- Ren, Y.; Ren, F.; Yang, B. Multi-layer matrix factorization for cancer subtyping using full and partial multi-omics dataset. Brief. Bioinform. 2025, 26, bbaf448. [Google Scholar] [CrossRef] [PubMed]
- Tong, L.; Shi, W.; Isgut, M.; Zhong, Y.; Lais, P.; Gloster, L.; Sun, J.; Swain, A.; Giuste, F.; Wang, M.D. Integrating Multi-Omics Data With EHR for Precision Medicine Using Advanced Artificial Intelligence. IEEE Rev. Biomed. Eng. 2024, 17, 80–97. [Google Scholar] [CrossRef]
- Zhang, H.W.; Lv, C.; Zhang, L.J.; Guo, X.; Shen, Y.W.; Nagle, D.G.; Zhou, Y.D.; Liu, S.H.; Zhang, W.D.; Luan, X. Application of omics- and multi-omics-based techniques for natural product target discovery. Biomed. Pharmacother. 2021, 141, 111833. [Google Scholar] [CrossRef]
- Camps, J.; Noël, F.; Liechti, R.; Massenet-Regad, L.; Rigade, S.; Götz, L.; Hoffmann, C.; Amblard, E.; Saichi, M.; Ibrahim, M.M.; et al. Meta-Analysis of Human Cancer Single-Cell RNA-Seq Datasets Using the IMMUcan Database. Cancer Res. 2023, 83, 363–373. [Google Scholar] [CrossRef]
- Zhu, S.; Lian, Q.; Ye, W.; Qin, W.; Wu, Z.; Ji, G.; Wu, X. scAPAdb: A comprehensive database of alternative polyadenylation at single-cell resolution. Nucleic Acids Res. 2022, 50, D365–D370. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Pan, W.; Zhao, X.; Zhao, F.; Xu, Z.; Li, X.; Zhao, Y.; Zhang, M.Q.; Yao, J. SODB facilitates comprehensive exploration of spatial omics data. Nat. Methods 2023, 20, 387–399. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- Wang, Z.; Jensen, M.A.; Zenklusen, J.C. A Practical Guide to The Cancer Genome Atlas (TCGA). Methods Mol. Biol. 2016, 1418, 111–141. [Google Scholar]
- Whiteaker, J.R.; Halusa, G.N.; Hoofnagle, A.N.; Sharma, V.; MacLean, B.; Yan, P.; Wrobel, J.A.; Kennedy, J.; Mani, D.R.; Zimmerman, L.J.; et al. CPTAC Assay Portal: A repository of targeted proteomic assays. Nat. Methods 2014, 11, 703–704. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Bolstad, B.M. PreprocessCore: A Collection of Pre-Processing Functions. [Computer Software]. 2024. Available online: https://github.com/bmbolstad/preprocessCore (accessed on 16 April 2024).
- Wang, F.; Bai, X.; Wang, Y.; Jiang, Y.; Ai, B.; Zhang, Y.; Liu, Y.; Xu, M.; Wang, Q.; Han, X.; et al. ATACdb: A comprehensive human chromatin accessibility database. Nucleic Acids Res. 2021, 49, D55–D64. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zong, W.; Zou, D.; Wang, G.; Zhao, W.; Yang, F.; Wu, S.; Zhang, X.; Guo, X.; Ma, Y.; et al. MethBank 4.0: An updated database of DNA methylation across a variety of species. Nucleic Acids Res. 2023, 51, D208–D216. [Google Scholar] [CrossRef]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Amberger, J.S.; Hamosh, A. Searching Online Mendelian Inheritance in Man (OMIM): A Knowledgebase of Human Genes and Genetic Phenotypes. Curr. Protoc Bioinform. 2017, 58, 1.2.1–1.2.12. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef] [PubMed]
- UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Davis, A.P.; Wiegers, T.C.; Johnson, R.J.; Sciaky, D.; Wiegers, J.; Mattingly, C.J. Comparative Toxicogenomics Database (CTD): Update 2023. Nucleic Acids Res. 2023, 51, D1257–D1262. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.Y.; Zhang, Y.Q.; Liu, Z.M.; Chen, T.; Lv, C.Y.; Tang, S.H.; Zhang, X.B.; Zhang, W.; Li, Z.Y.; Zhou, R.R.; et al. ETCM: An encyclopaedia of traditional Chinese medicine. Nucleic Acids Res. 2019, 47, D976–D982. [Google Scholar] [CrossRef]
- Burset, M.; Seledtsov, I.A.; Solovyev, V.V. SpliceDB: Database of canonical and non-canonical mammalian splice sites. Nucleic Acids Res. 2001, 29, 255–259. [Google Scholar] [CrossRef]
- Lin, X.; Lu, Y.; Zhang, C.; Cui, Q.; Tang, Y.D.; Ji, X.; Cui, C. LncRNADisease v3.0: An updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 2024, 52, D1365–D1369. [Google Scholar] [CrossRef]
- Luo, X.; Li, H.; Liang, J.; Zhao, Q.; Xie, Y.; Ren, J.; Zuo, Z. RMVar: An updated database of functional variants involved in RNA modifications. Nucleic Acids Res. 2021, 49, D1405–D1412. [Google Scholar] [CrossRef]
- Luo, X.; Li, H.; Liang, J.; Zhao, Q.; Xie, Y.; Ren, J.; Zuo, Z. Genotype-first approach to identify associations between CDH1 germline variants and cancer phenotypes: A multicentre study by the European Reference Network on Genetic Tumour Risk Syndromes. Lancet Oncol. 2023, 24, 91–106. [Google Scholar]
- Zou, G.; Huang, Y.; Zhang, S.; Ko, K.P.; Kim, B.; Zhang, J.; Venkatesan, V.; Pizzi, M.P.; Fan, Y.; Jun, S.; et al. E-cadherin loss drives diffuse-type gastric tumorigenesis via EZH2-mediated reprogramming. J. Exp. Med. 2024, 221, e20230561. [Google Scholar] [CrossRef]
- Gilbert, M.A.; Schultz-Rogers, L.; Rajagopalan, R.; Grochowski, C.M.; Wilkins, B.J.; Biswas, S.; Conlin, L.K.; Fiorino, K.N.; Dhamija, R.; Pack, M.A.; et al. Protein-elongating mutations in MYH11 are implicated in a dominantly inherited smooth muscle dysmotility syndrome with severe esophageal, gastric, and intestinal disease. Hum. Mutat 2020, 41, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Tsai, R.X.; Fang, K.C.; Yang, P.C.; Hsieh, Y.H.; Chiang, I.T.; Chen, Y.; Lee, H.G.; Lee, J.T.; Chu, H.C. TERRA regulates DNA G-quadruplex formation and ATRX recruitment to chromatin. Nucleic Acids Res. 2022, 50, 12217–12234. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Jiang, T.; Shen, K.; Zhao, D.; Zhang, M.; Zhu, W.; Liu, Y.; Xu, C. GADD45B regulates the carcinogenesis process of chronic atrophic gastritis and the metabolic pathways of gastric cancer. Front. Endocrinol. 2023, 14, 1224832. [Google Scholar] [CrossRef]
- Dasari, S.; Tchounwou, P.B. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharmacol. 2014, 740, 364–378. [Google Scholar] [CrossRef] [PubMed]
- Dai, D.; Zhu, J.; Sun, C.; Li, M.; Liu, J.; Wu, S.; Ning, K.; He, L.J.; Zhao, X.M.; Chen, W.H. GMrepo v2: A curated human gut microbiome database with special focus on disease markers and cross-dataset comparison. Nucleic Acids Res. 2022, 50, D777–D784. [Google Scholar] [CrossRef]
- Qi, C.; Cai, Y.; Qian, K.; Li, X.; Ren, J.; Wang, P.; Fu, T.; Zhao, T.; Cheng, L.; Shi, L.; et al. gutMDisorder v2.0: A comprehensive database for dysbiosis of gut microbiota in phenotypes and interventions. Nucleic Acids Res. 2023, 51, D717–D722. [Google Scholar] [CrossRef]
- Yang, J.; Park, J.; Jung, Y.; Chun, J. AMDB: A database of animal gut microbial communities with manually curated metadata. Nucleic Acids Res. 2022, 50, D729–D735. [Google Scholar] [CrossRef]
- Chattopadhyay, A.; Lee, C.Y.; Lee, Y.C.; Liu, C.L.; Chen, H.K.; Li, Y.H.; Lai, L.C.; Tsai, M.H.; Ni, Y.H.; Chiu, H.M.; et al. Twnbiome: A public database of the healthy Taiwanese gut microbiome. BMC Bioinform. 2023, 24, 474. [Google Scholar] [CrossRef]
- Bao, Y.; Chen, Y.; Lin, L.; Li, J.; Liu, X.; Wang, G.; Li, Y.; Lin, Y.; Chen, Y.; Zhou, L.; et al. GutUDB: A comprehensive multiomics database for intestinal diseases. Imeta 2024, 3, e195. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, G.; Sun, Z.; Lee, S.Y.; Lai, M.; Wang, X.; An, S. StomachDB: An Integrated Multi-Omics Database for Gastric Diseases. Biology 2025, 14, 1484. https://doi.org/10.3390/biology14111484
Wang G, Sun Z, Lee SY, Lai M, Wang X, An S. StomachDB: An Integrated Multi-Omics Database for Gastric Diseases. Biology. 2025; 14(11):1484. https://doi.org/10.3390/biology14111484
Chicago/Turabian StyleWang, Gang, Zhe Sun, Shiou Yih Lee, Mingyu Lai, Xiaojuan Wang, and Sanqi An. 2025. "StomachDB: An Integrated Multi-Omics Database for Gastric Diseases" Biology 14, no. 11: 1484. https://doi.org/10.3390/biology14111484
APA StyleWang, G., Sun, Z., Lee, S. Y., Lai, M., Wang, X., & An, S. (2025). StomachDB: An Integrated Multi-Omics Database for Gastric Diseases. Biology, 14(11), 1484. https://doi.org/10.3390/biology14111484





