Dynamic Regulation of Gonadal Transposons and Pseudogenes via PIWI/piRNA Pathway in Gynogenetic Japanese Flounder (Paralichthys olivaceus)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples Used in This Study
2.2. Transcriptome Data Analysis
2.3. Small RNA Sequencing Data Analysis
2.4. Annotation and Ping-Pong Signature of Small RNAs
2.5. Prediction and Annotation of piRNA Cluster
2.6. Identification and Characterization of piRNA-Associated TEs and Genes
2.7. Identification and Characterization of Tc1/Mariner Transposon in P. olivaceus
2.8. Identification and Characterization of Pim Genes and Associated piRNAs in P. olivaceus
2.9. RNA Extraction and qRT-PCR
2.10. In Vitro Overexpression of piRC
3. Results
3.1. Expression of PIWI/piRNA Pathway Genes in P. olivaceus
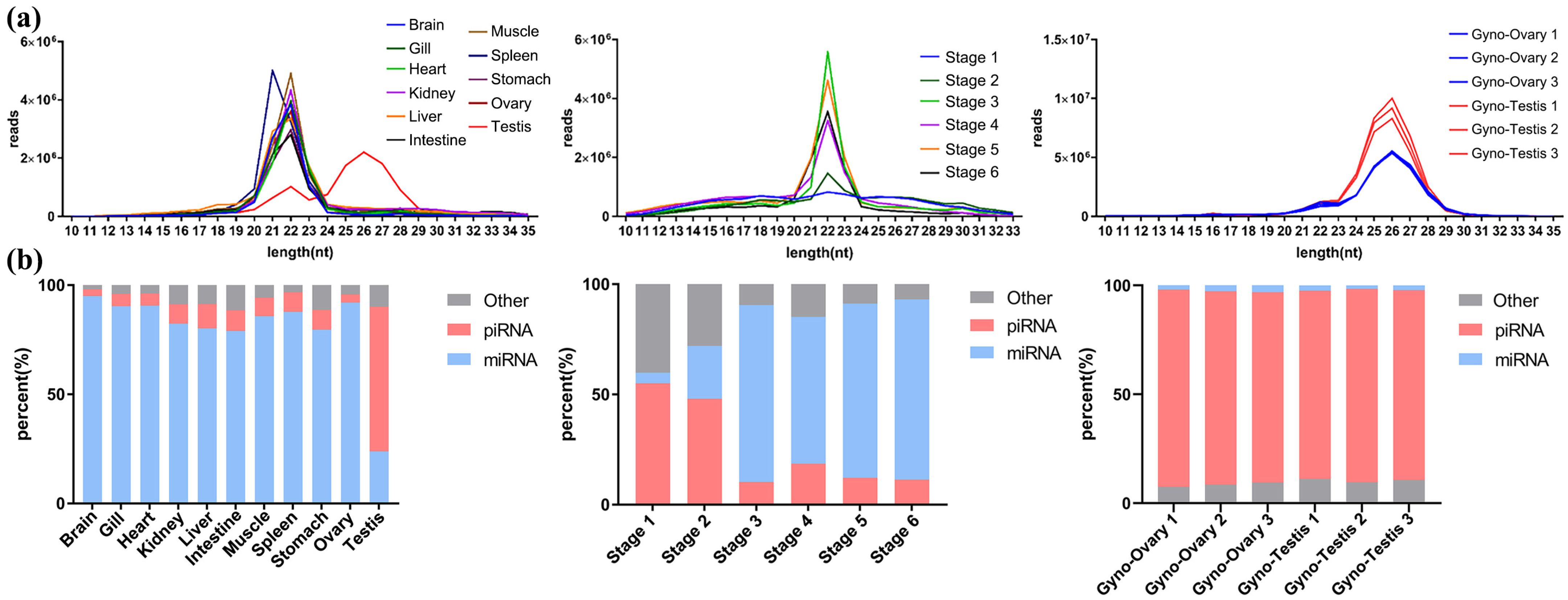
3.2. Small RNA Annotation and Spatio-Temporal Distribution in P. olivaceus
3.3. Ping-Pong Signature of P. olivaceus piRNAs
3.4. Characterization of piRNA Clusters in P. olivaceus
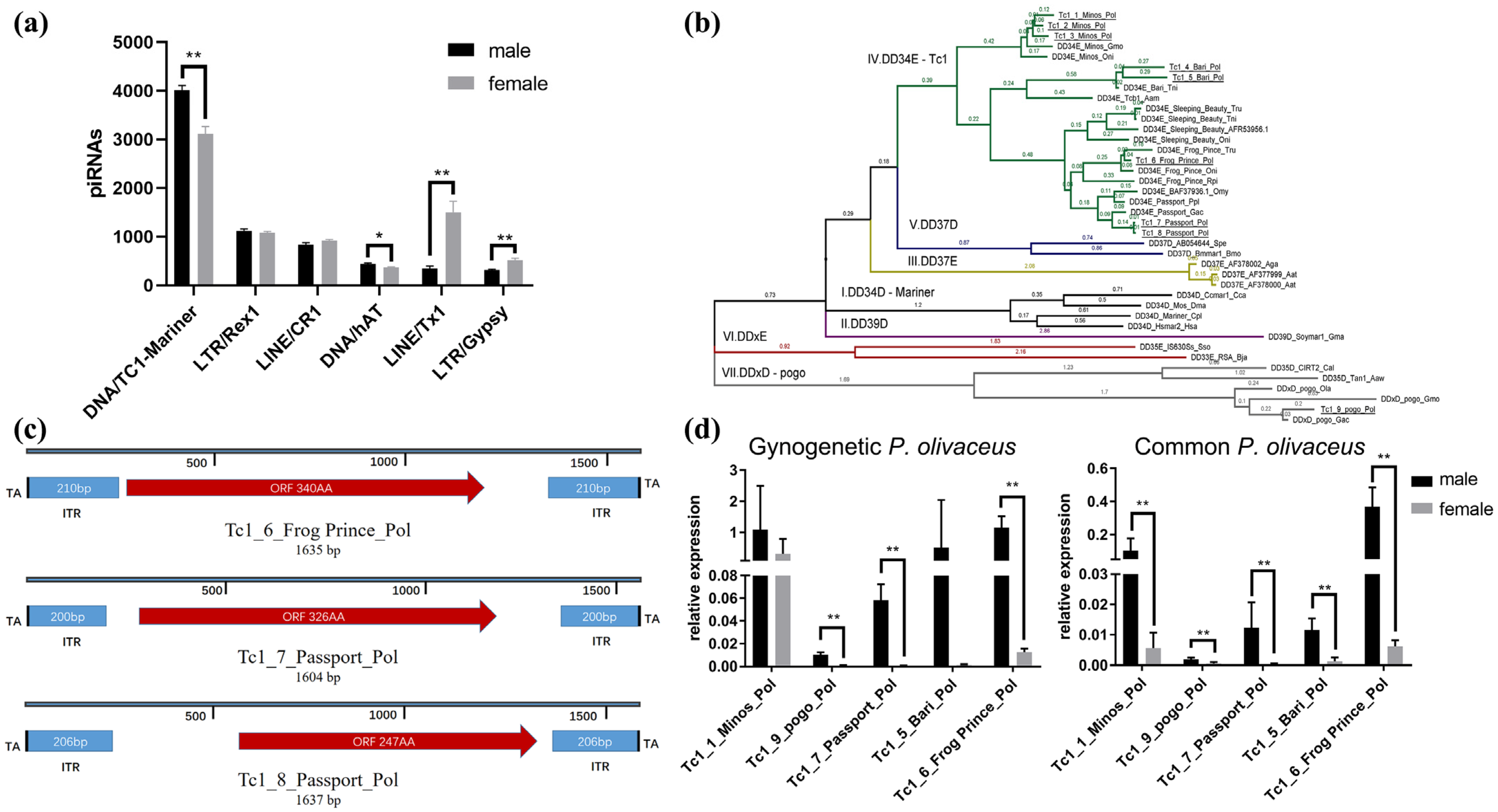
3.5. The piRNA-Targeted Tc1/Mariner Transposons in Gynogenetic P. olivaceus
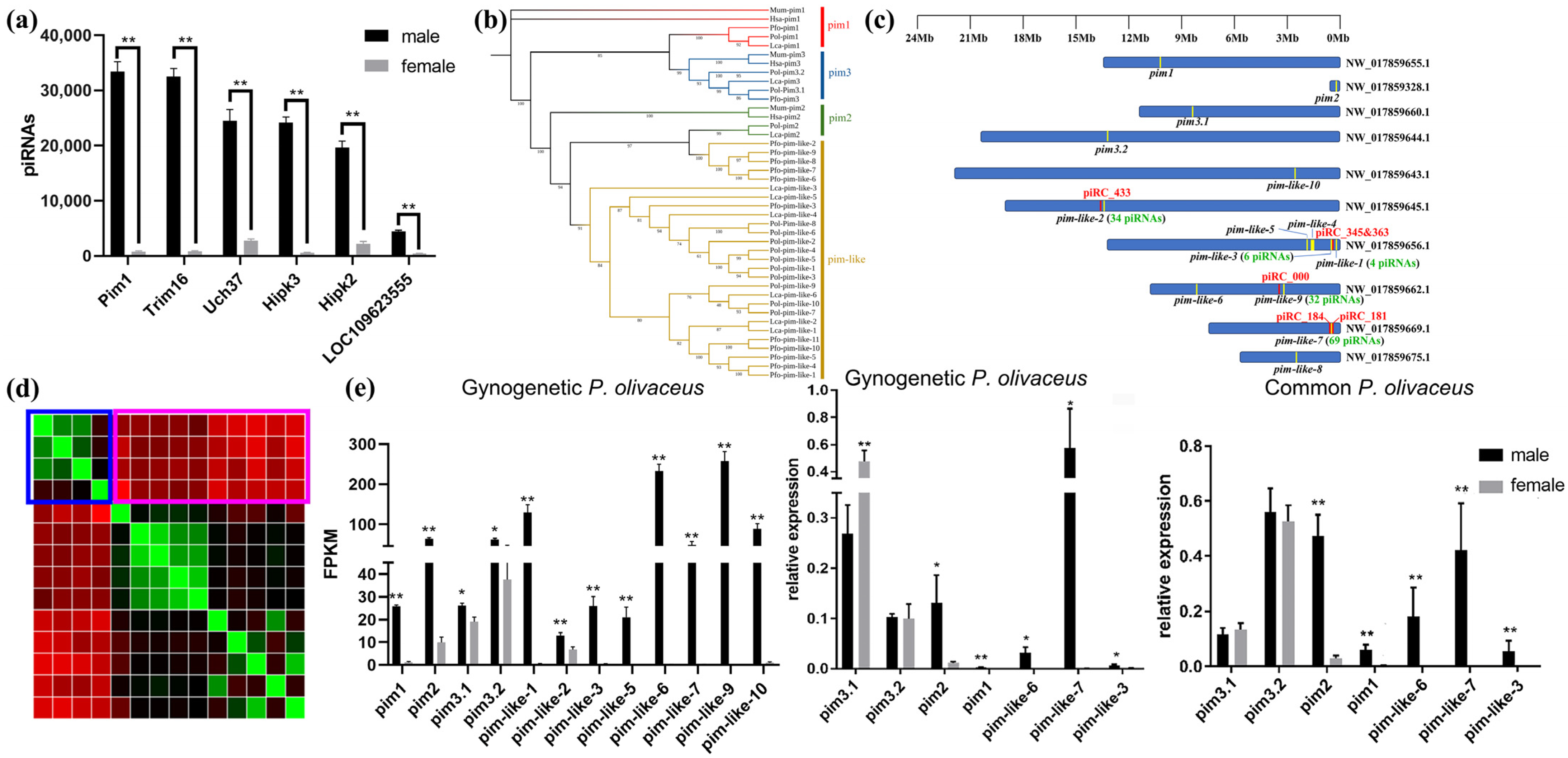
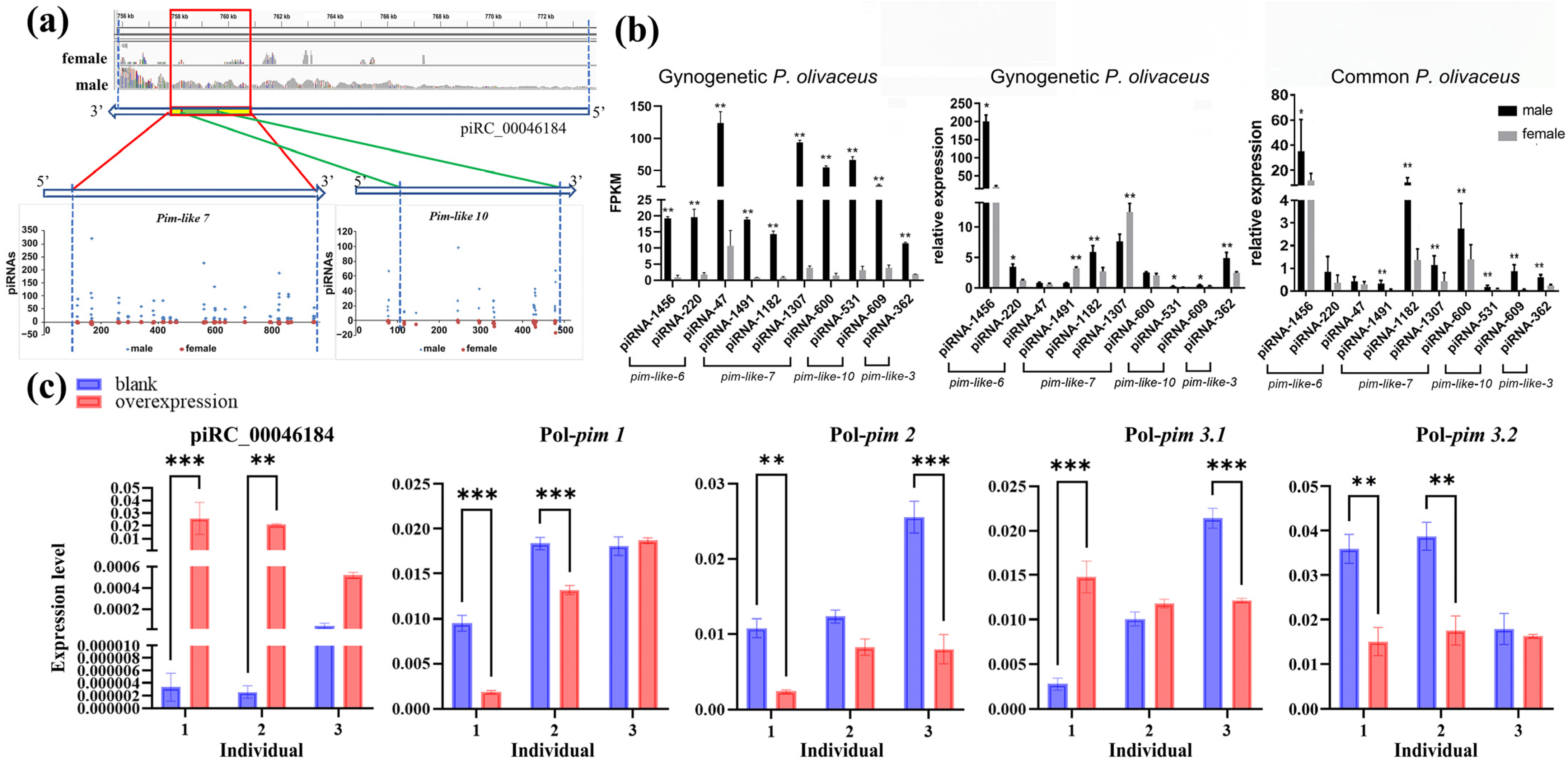
3.6. The piRNA-Targeted Pim Genes in Gynogenetic P. olivaceus
4. Discussion
4.1. Specific Characteristics of PIWI/piRNA Pathway in Common and Gynogenetic P. olivaceus
4.2. piRNA Targeting TEs in P. olivaceus
4.3. The Expanded Pim Family and piRNAs in P. olivaceus
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lau, N.C.; Seto, A.G.; Kim, J.; Kuramochi-Miyagawa, S.; Nakano, T.; Bartel, D.P.; Kingston, R.E. Characterization of the piRNA complex from rat testes. Science 2006, 313, 363–367. [Google Scholar] [CrossRef]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, N.; Nishimasu, H.; Sakakibara, K.; Nishida, K.M.; Hirano, T.; Ishitani, R.; Siomi, H.; Siomi, M.C.; Nureki, O. Crystal structure of silkworm PIWI-clade Argonaute Siwi bound to piRNA. Cell 2016, 167, 484–497. [Google Scholar] [CrossRef]
- Mohn, F.; Handler, D.; Brennecke, J. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science 2015, 348, 812–817. [Google Scholar] [CrossRef]
- Ding, D.; Liu, J.; Dong, K.; Midic, U.; Hess, R.A.; Xie, H.; Demireva, E.Y.; Chen, C. PNLDC1 is essential for piRNA 3′ end trimming and transposon silencing during spermatogenesis in mice. Nat. Commun. 2017, 8, 819. [Google Scholar] [CrossRef]
- Izumi, N.; Shoji, K.; Sakaguchi, Y.; Honda, S.; Kirino, Y.; Suzuki, T.; Katsuma, S.; Tomari, Y. Identification and functional analysis of the pre-piRNA 3′ trimmer in silkworms. Cell 2016, 164, 962–973. [Google Scholar] [CrossRef]
- Wang, W.; Yoshikawa, M.; Han, B.W.; Izumi, N.; Tomari, Y.; Weng, Z.; Zamore, P.D. The initial uridine of primary piRNAs does not create the tenth adenine that is the hallmark of secondary piRNAs. Mol. Cell 2014, 56, 708–716. [Google Scholar] [CrossRef] [PubMed]
- Gainetdinov, I.; Colpan, C.; Arif, A.; Cecchini, K.; Zamore, P.D. A single mechanism of biogenesis, initiated and directed by PIWI proteins, explains piRNA production in most animals. Mol. Cell 2018, 71, 775–790. [Google Scholar] [CrossRef]
- Han, B.W.; Zamore, P.D. PiRNAs. Curr. Biol. 2014, 24, R730–R733. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Wang, P.J. Mammalian piRNAs: Biogenesis, function, and mysteries. Spermatogenesis 2014, 4, e27889. [Google Scholar] [CrossRef]
- Shiromoto, Y.; Kuramochi-Miyagawa, S.; Daiba, A.; Chuma, S.; Katanaya, A.; Katsumata, A.; Nishimura, K.; Ohtaka, M.; Nakanishi, M.; Nakamura, T. GPAT2, a mitochondrial outer membrane protein, in piRNA biogenesis in germline stem cells. RNA 2013, 19, 803–810. [Google Scholar] [CrossRef]
- Xiol, J.; Spinelli, P.; Laussmann, M.A.; Homolka, D.; Yang, Z.; Cora, E.; Couté, Y.; Conn, S.; Kadlec, J.; Sachidanandam, R. RNA clamping by Vasa assembles a piRNA amplifier complex on transposon transcripts. Cell 2014, 157, 1698–1711. [Google Scholar] [CrossRef]
- Girard, A.; Sachidanandam, R.; Hannon, G.J.; Carmell, M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 2006, 442, 199–202. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Bogoch, Y.; Shvaizer, G.; Guler, N.; Levy, K.; Elkouby, Y.M. The piRNA protein Asz1 is essential for germ cell and gonad development in zebrafish and exhibits differential necessities in distinct types of germ granules. PLoS Genet. 2025, 21, e1010868. [Google Scholar] [CrossRef]
- Almeida, M.V.; Blumer, M.; Yuan, C.U.; Sierra, P.; Price, J.L.; Quah, F.X.; Friman, A.; Dallaire, A.; Vernaz, G.; Putman, A.L. Dynamic co-evolution of transposable elements and the piRNA pathway in African cichlid fishes. Genome Biol. 2025, 26, 14. [Google Scholar] [CrossRef]
- Ji, Q.; Xie, Z.; Gan, W.; Wang, L.; Song, W. Identification and Characterization of PIWI-Interacting RNAs in Spinyhead Croakers (Collichthys lucidus) by Small RNA Sequencing. Fishes 2022, 7, 297. [Google Scholar] [CrossRef]
- Akkouche, A.; Mugat, B.; Barckmann, B.; Varela-Chavez, C.; Li, B.; Raffel, R.; Pélisson, A.; Chambeyron, S. Piwi is required during Drosophila embryogenesis to license dual-strand piRNA clusters for transposon repression in adult ovaries. Mol. Cell 2017, 66, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Li, M.; Wei, L.; Yin, S.; Xiong, B.; Li, S.; Sheng-Li, L.; Schatten, H.; Sun, Q.-Y. Astrin regulates meiotic spindle organization, spindle pole tethering and cell cycle progression in mouse oocytes. Cell Cycle 2009, 8, 3384–3395. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhong, H.; Liu, S.; Yu, F.; Hu, J.; Zhang, C.; Tao, M.; Liu, Y. Elevated expression of Piwi and piRNAs in ovaries of triploid crucian carp. Mol. Cell. Endocrinol. 2014, 383, 1–9. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, G.; San, L.; Zhang, Y.; He, Z.; Liu, Y.; Ren, Y.; Wang, Y.; Hou, J. Establishment of recombinant doubled haploid lines of Japanese flounder and evaluation of their reproductive capacity. Aquaculture 2024, 587, 740899. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, S.; Guo, S.; Lu, W.; Zhang, Q.; Cheng, J. Evolutionary dynamics and conserved function of the Tudor domain-containing (TDRD) proteins in teleost fish. Mar. Life Sci. Technol. 2022, 4, 18–30. [Google Scholar] [CrossRef]
- Zong, W.; Wang, Y.; Zhang, L.; Lu, W.; Li, W.; Wang, F.; Cheng, J. DNA methylation mediates sperm quality via piwil1 and piwil2 regulation in Japanese flounder (Paralichthys olivaceus). Int. J. Mol. Sci. 2024, 25, 5935. [Google Scholar] [CrossRef]
- Song, H.; Xing, C.; Lu, W.; Liu, Z.; Wang, X.; Cheng, J.; Zhang, Q. Rapid evolution of piRNA pathway and its transposon targets in Japanese flounder (Paralichthys olivaceus). Comp. Biochem. Physiol. Part D Genom. Proteom. 2019, 31, 100609. [Google Scholar] [CrossRef]
- Yang, F.; Wang, Y.; Lu, W.; Zong, W.; Zhu, Q.; Cheng, J. The comparative survey of coordinated regulation of steroidogenic pathway in Japanese flounder (Paralichthys olivaceus) and Chinese tongue sole (Cynoglossus Semilaevis). Int. J. Mol. Sci. 2022, 23, 5520. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Yang, F.; Liu, S.; Zhao, H.; Lu, W.; Zhang, Q. Transcriptomic analysis reveals functional interaction of mRNA–lncRNA–miRNA in steroidogenesis and spermatogenesis of gynogenetic Japanese flounder (Paralichthys olivaceus). Biology 2022, 11, 213. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, F.; Liu, S.; Lu, W.; Zong, W.; Cheng, J. Coordinated regulatory feedback between estradiol and miR-17-92 cluster in ovaries of Japanese flounder Paralichthys olivaceus. Aquaculture 2024, 589, 740975. [Google Scholar] [CrossRef]
- Zhao, H.; Du, X.; Zhang, K.; Liu, Y.; Wang, Y.; Liu, J.; He, Y.; Wang, X.; Zhang, Q. Weighted correlation network analysis (WGCNA) of Japanese flounder (Paralichthys olivaceus) embryo transcriptome provides crucial gene sets for understanding haploid syndrome and rescue by diploidization. J. Ocean Univ. China 2018, 17, 1441–1450. [Google Scholar] [CrossRef]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Rosenkranz, D.; Han, C.-T.; Roovers, E.F.; Zischler, H.; Ketting, R.F. Piwi proteins and piRNAs in mammalian oocytes and early embryos: From sample to sequence. Genom. Data 2015, 5, 309–313. [Google Scholar] [CrossRef]
- Gebert, D.; Hewel, C.; Rosenkranz, D. unitas: The universal tool for annotation of small RNAs. BMC Genom. 2017, 18, 644. [Google Scholar] [CrossRef]
- Jehn, J.; Gebert, D.; Pipilescu, F.; Stern, S.; Kiefer, J.S.T.; Hewel, C.; Rosenkranz, D. PIWI genes and piRNAs are ubiquitously expressed in mollusks and show patterns of lineage-specific adaptation. Commun. Biol. 2018, 1, 137. [Google Scholar] [CrossRef]
- Rosenkranz, D.; Zischler, H. proTRAC-a software for probabilistic piRNA cluster detection, visualization and analysis. BMC Bioinform. 2012, 13, 5. [Google Scholar] [CrossRef]
- Chen, N. Using Repeat Masker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. 2004, 5, 4–10. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Mu, H.; Chen, J.; Huang, W.; Huang, G.; Deng, M.; Hong, S.; Ai, P.; Gao, C.; Zhou, H. OmicShare tools: A zero-code interactive online platform for biological data analysis and visualization. Imeta 2024, 3, e228. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Jurka, J. Repbase Update: A database and an electronic journal of repetitive elements. Trends Genet. 2000, 16, 418–420. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Chen, W.; Shen, D.; Wang, S.; Chen, C.; Zhang, L.; Wang, W.; Wang, X.; Song, C. Characterization of autonomous families of Tc1/mariner transposons in neoteleost genomes. Mar. Genom. 2017, 34, 67–77. [Google Scholar] [CrossRef]
- Rombel, I.T.; Sykes, K.F.; Rayner, S.; Johnston, S.A. ORF-FINDER: A vector for high-throughput gene identification. Gene 2002, 282, 33–41. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2017, 46, D493–D496. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.J.; Sun, L. Evaluation of housekeeping genes as references for quantitative real time RT-PCR analysis of gene expression in Japanese flounder (Paralichthys olivaceus). Fish. Shellfish Immunol. 2011, 30, 638–645. [Google Scholar] [CrossRef]
- Liu, S.; Song, H.; Liu, Z.; Lu, W.; Zhang, Q.; Cheng, J. Selection of references for microRNA quantification in Japanese flounder (Paralichthys olivaceus) normal tissues and Edwardsiella tarda-infected livers. Genes 2022, 13, 175. [Google Scholar] [CrossRef]
- Ivics, Z.; Izsvák, Z. Sleeping beauty transposition. In Mobile DNA III; Chandler, M., Gellert, M., Lambowitz, A.M., Rice, P.A., Sandmeyer, S.B., Eds.; John Wiley & Sons, Inc.: New York, NY, USA, 2015; pp. 851–872. [Google Scholar] [CrossRef]
- Carmell, M.A.; Girard, A.; Van De Kant, H.J.; Bourc’his, D.; Bestor, T.H.; De Rooij, D.G.; Hannon, G.J. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell 2007, 12, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.L.; Tsend-Ayush, E.; Kortschak, R.D.; Jacob, R.; Ricciardelli, C.; Oehler, M.K.; Grützner, F. Conservation and expression of PIWI-interacting RNA pathway genes in male and female adult gonad of amniotes. Biol. Reprod. 2013, 89, 136. [Google Scholar] [CrossRef]
- Wen, X.; Wang, D.; Li, X.; Zhao, C.; Wang, T.; Qian, X.; Yin, S. Differential expression of two Piwil orthologs during embryonic and gonadal development in pufferfish, Takifugu fasciatus. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2018, 219, 44–51. [Google Scholar] [CrossRef]
- Halbach, R.; Miesen, P.; Joosten, J.; Taşköprü, E.; Rondeel, I.; Pennings, B.; Vogels, C.B.; Merkling, S.H.; Koenraadt, C.J.; Lambrechts, L. A satellite repeat-derived piRNA controls embryonic development of Aedes. Nature 2020, 580, 274–277. [Google Scholar] [CrossRef]
- Zhang, B.; Zhao, N.; Jia, L.; Che, J.; He, X.; Liu, K.; Bao, B. Identification and application of piwi-interacting RNAs from seminal plasma exosomes in Cynoglossus semilaevis. BMC Genom. 2020, 21, 302. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Takeda, A.; Tsukiyama, T.; Mise, K.; Okuno, T.; Sasaki, H.; Minami, N.; Imai, H. Identification and characterization of two novel classes of small RNAs in the mouse germline: Retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006, 20, 1732–1743. [Google Scholar] [CrossRef] [PubMed]
- Reuter, M.; Berninger, P.; Chuma, S.; Shah, H.; Hosokawa, M.; Funaya, C.; Antony, C.; Sachidanandam, R.; Pillai, R.S. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 2011, 480, 264–267. [Google Scholar] [CrossRef]
- Wei, Z.; Liu, X.; Zhang, H. Identification and characterization of piRNA-like small RNAs in the gonad of sea urchin (Strongylocentrotus nudus). Mar. Biotechnol. 2012, 14, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Kneitz, S.; Mishra, R.R.; Chalopin, D.; Postlethwait, J.; Warren, W.C.; Walter, R.B.; Schartl, M. Germ cell and tumor associated piRNAs in the medaka and Xiphophorus melanoma models. BMC Genom. 2016, 17, 357. [Google Scholar] [CrossRef]
- Ivics, Z.; Kaufman, C.D.; Zayed, H.; Miskey, C.; Walisko, O.; Izsvák, Z. The Sleeping Beauty transposable element: Evolution, regulation and genetic applications. Curr. Issues Mol. Biol. 2004, 6, 43–56. [Google Scholar] [CrossRef]
- Kuraku, S.; Qiu, H.; Meyer, A. Horizontal transfers of Tc1 elements between teleost fishes and their vertebrate parasites, lampreys. Genome Biol. Evol. 2012, 4, 929–936. [Google Scholar] [CrossRef]
- Wallau, G.L.; Capy, P.; Loreto, E.; Hua-Van, A. Genomic landscape and evolutionary dynamics of mariner transposable elements within the Drosophila genus. BMC Genom. 2014, 15, 727. [Google Scholar] [CrossRef]
- Bachmann, M.; Möröy, T. The serine/threonine kinase Pim-1. Int. J. Biochem. Cell Biol. 2005, 37, 726–730. [Google Scholar] [CrossRef]
- Fox, C.J.; Hammerman, P.S.; Cinalli, R.M.; Master, S.R.; Chodosh, L.A.; Thompson, C.B. The serine/threonine kinase Pim-2 is a transcriptionally regulated apoptotic inhibitor. Genes Dev. 2003, 17, 1841–1854. [Google Scholar] [CrossRef]
- Asano, J.; Nakano, A.; Oda, A.; Amou, H.; Hiasa, M.; Takeuchi, K.; Miki, H.; Nakamura, S.; Harada, T.; Fujii, S. The serine/threonine kinase Pim-2 is a novel anti-apoptotic mediator in myeloma cells. Leukemia 2011, 25, 1182–1188. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhong, H.; Xiao, J.; Yan, J.; Luo, Y.; Gan, X.; Yu, F. Identification and comparative analysis of piRNAs in ovary and testis of Nile tilapia (Oreochromis niloticus). Genes Genom. 2016, 38, 519–527. [Google Scholar] [CrossRef]
- Glasauer, S.M.; Neuhauss, S.C. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol. Genet. Genom. 2014, 289, 1045–1060. [Google Scholar] [CrossRef] [PubMed]
- Pantano, L.; Jodar, M.; Bak, M.; Ballescà, J.L.; Tommerup, N.; Oliva, R.; Vavouri, T. The small RNA content of human sperm reveals pseudogene-derived piRNAs complementary to protein-coding genes. RNA 2015, 21, 1085–1095. [Google Scholar] [CrossRef] [PubMed]
- Gebert, D.; Zischler, H.; Rosenkranz, D. Primate piRNA cluster evolution suggests limited relevance of pseudogenes in piRNA-mediated gene regulation. Genome Biol. Evol. 2019, 11, 1088–1104. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Li, W.; Wang, F.; Lu, W.; Yang, F.; Zhang, Q.; Cheng, J. Dynamic Regulation of Gonadal Transposons and Pseudogenes via PIWI/piRNA Pathway in Gynogenetic Japanese Flounder (Paralichthys olivaceus). Biology 2025, 14, 1464. https://doi.org/10.3390/biology14101464
Liu Z, Li W, Wang F, Lu W, Yang F, Zhang Q, Cheng J. Dynamic Regulation of Gonadal Transposons and Pseudogenes via PIWI/piRNA Pathway in Gynogenetic Japanese Flounder (Paralichthys olivaceus). Biology. 2025; 14(10):1464. https://doi.org/10.3390/biology14101464
Chicago/Turabian StyleLiu, Zeyu, Weigang Li, Fengchi Wang, Wei Lu, Fan Yang, Qingke Zhang, and Jie Cheng. 2025. "Dynamic Regulation of Gonadal Transposons and Pseudogenes via PIWI/piRNA Pathway in Gynogenetic Japanese Flounder (Paralichthys olivaceus)" Biology 14, no. 10: 1464. https://doi.org/10.3390/biology14101464
APA StyleLiu, Z., Li, W., Wang, F., Lu, W., Yang, F., Zhang, Q., & Cheng, J. (2025). Dynamic Regulation of Gonadal Transposons and Pseudogenes via PIWI/piRNA Pathway in Gynogenetic Japanese Flounder (Paralichthys olivaceus). Biology, 14(10), 1464. https://doi.org/10.3390/biology14101464





