First Identification of MORF Family in Ferns: Molecular Regulation of Organellar RNA Editing in Osmunda japonica and Plenasium vachellii
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material Collection and Sequencing
2.2. Chloroplast Genome Assembly and Annotation
2.3. Public Data Acquisition and Transcriptome Analysis
2.4. Analysis of Protein Structure and Physicochemical Properties
2.5. Prediction of RNA Editing Sites in Organelle Genomes Across Multiple Plant Groups
2.6. Identification of Tissue-Specific RNA Editing Sites in Osmundaceae Species
2.7. Statistical Analysis
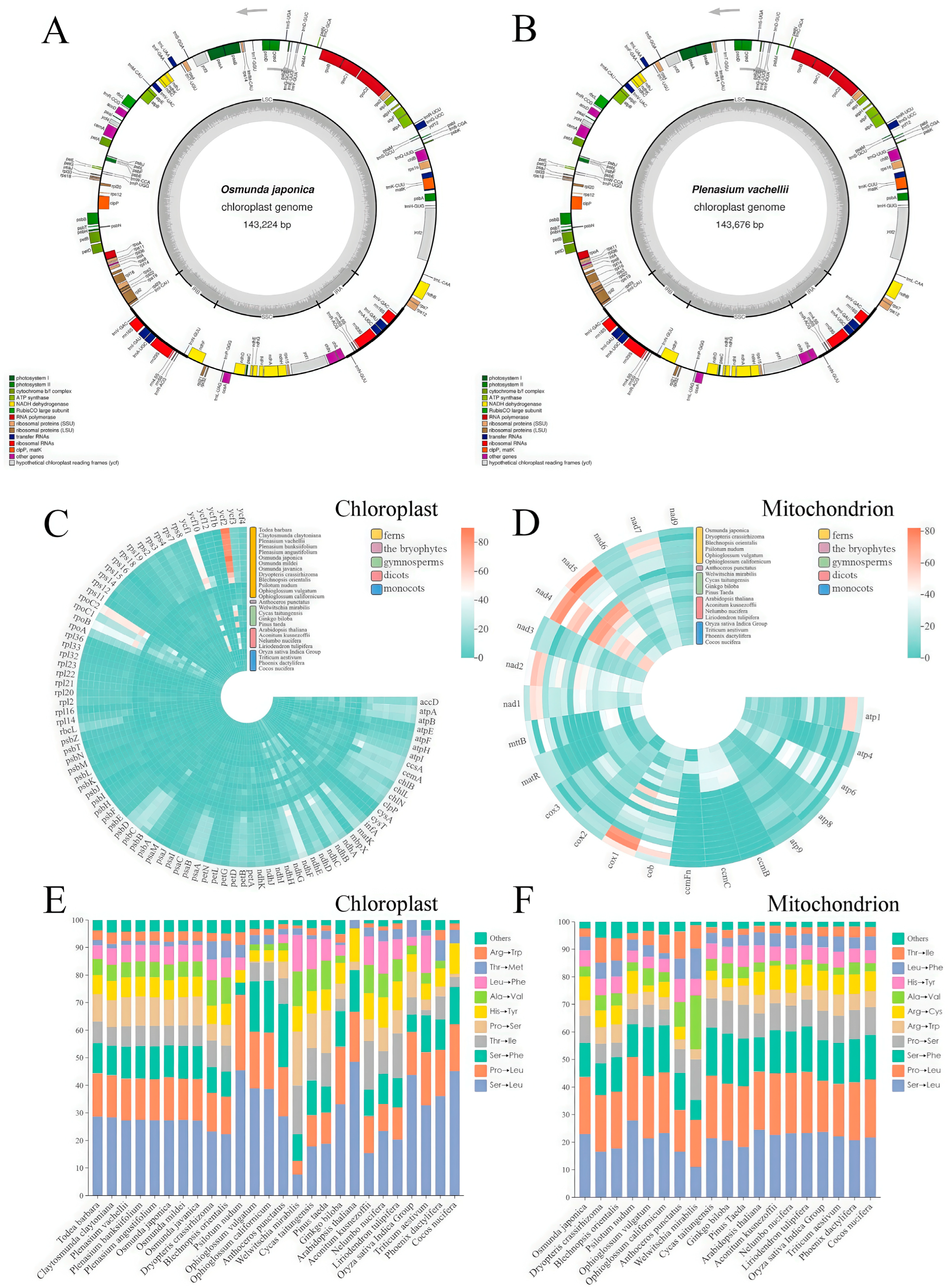
3. Results
3.1. Identification and Characterization of MORF Proteins in O. japonica and P. vachellii
3.2. Comparative Analysis of Organellar RNA Editing Across Plant Taxa
3.3. Interplay Among RNA Editing-Induced Amino Acid Changes Across Plant Taxa
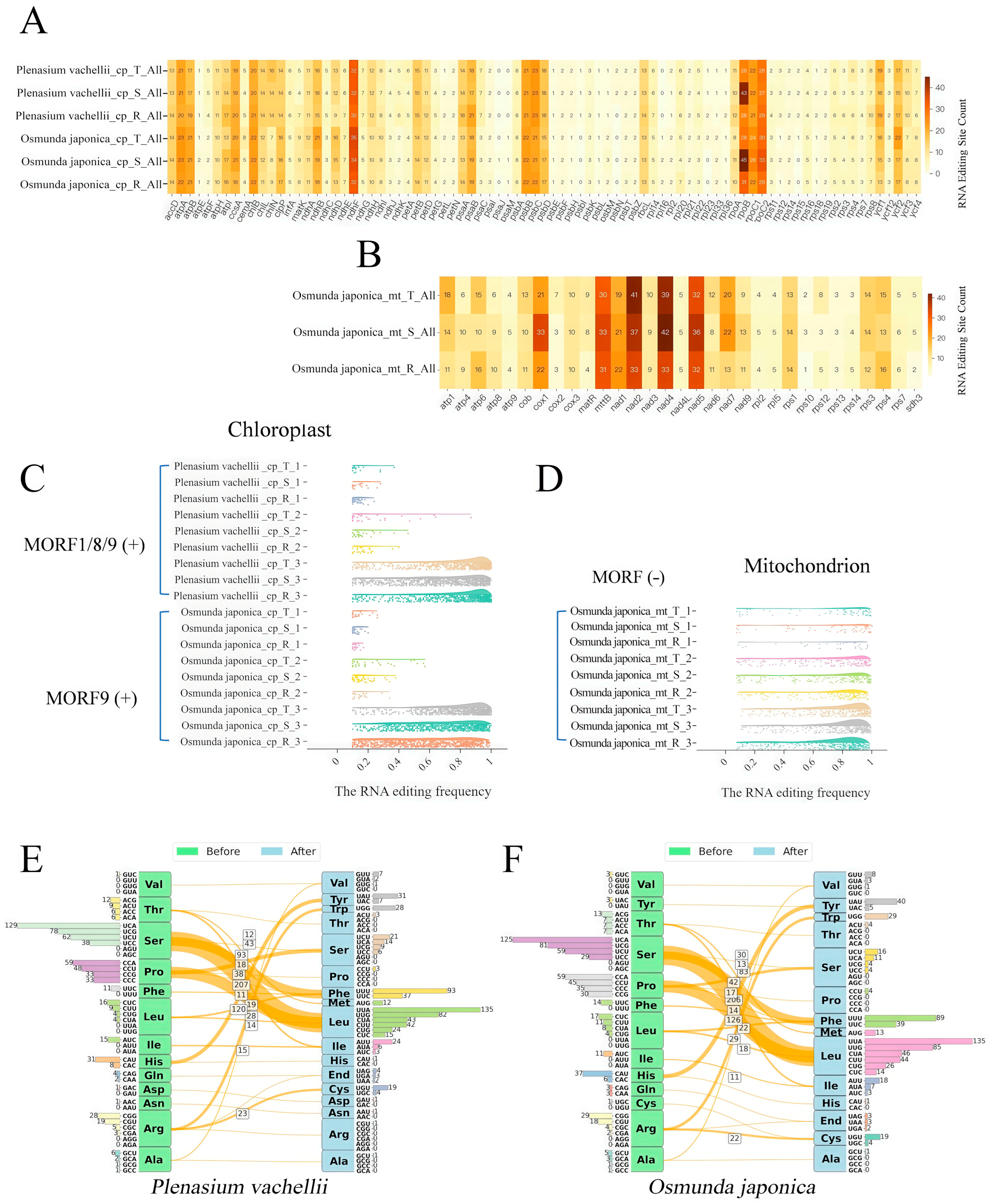
3.4. Tissue- and Organelle-Specific RNA Editing Sites in O. japonica and P. vachellii
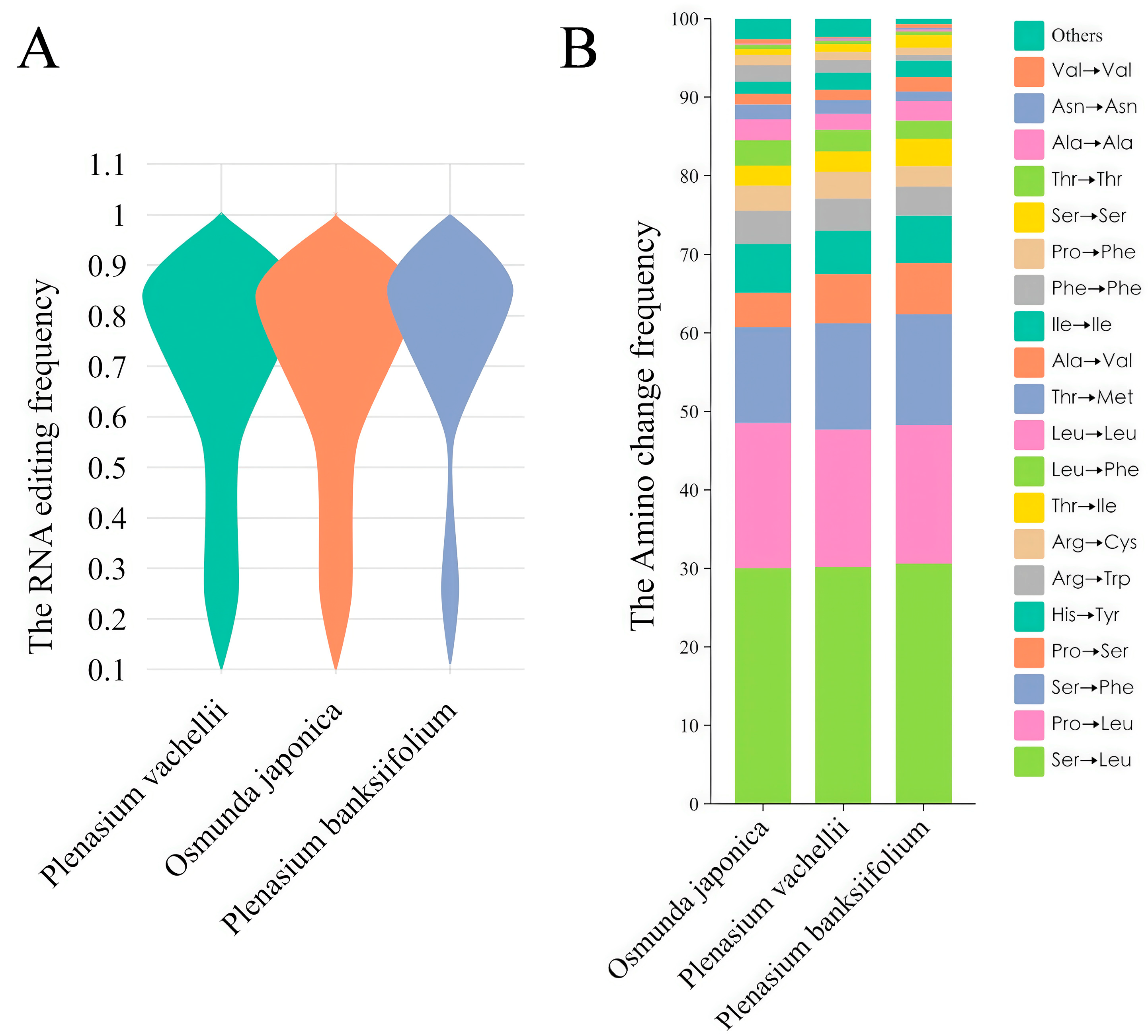
3.5. Highly Conserved Chloroplast RNA Editing Sites of O. japonica, P. vachellii and P. banksiifolium
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gott, J.M.; Emeson, R.B. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 2000, 34, 499–531. [Google Scholar] [CrossRef]
- Benne, R.; Van den Burg, J.; Brakenhoff, J.P.; Sloof, P.; Van Boom, J.H.; Tromp, M.C. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 1986, 46, 819–826. [Google Scholar] [CrossRef]
- Ichinose, M.; Sugita, M. RNA Editing and Its Molecular Mechanism in Plant Organelles. Genes 2016, 8, 5. [Google Scholar] [CrossRef]
- Hao, W.; Liu, G.; Wang, W.; Shen, W.; Zhao, Y.; Sun, J.; Yang, Q.; Zhang, Y.; Fan, W.; Pei, S.; et al. RNA Editing and Its Roles in Plant Organelles. Front. Genet. 2021, 12, 757109. [Google Scholar] [CrossRef]
- Covello, P.S.; Gray, M.W. RNA editing in plant mitochondria. Nature 1989, 341, 662–666. [Google Scholar] [CrossRef] [PubMed]
- Hoch, B.; Maier, R.M.; Appel, K.; Igloi, G.L.; Kossel, H. Editing of a chloroplast mRNA by creation of an initiation codon. Nature 1991, 353, 178–180. [Google Scholar] [CrossRef]
- Gao, C.; Li, T.; Zhao, X.; Wu, C.; Zhang, Q.; Zhao, X.; Wu, M.; Lian, Y.; Li, Z. Comparative analysis of the chloroplast genomes of Rosa species and RNA editing analysis. BMC Plant Biol. 2023, 23, 318. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tan, B.C. Pentatricopeptide repeat proteins in plants: Cellular functions, action mechanisms, and potential applications. Plant Commun. 2025, 6, 101203. [Google Scholar] [CrossRef]
- Shi, X.; Bentolila, S.; Hanson, M.R. Organelle RNA recognition motif-containing (ORRM) proteins are plastid and mitochondrial editing factors in Arabidopsis. Plant Signal. Behav. 2016, 11, e1167299. [Google Scholar] [CrossRef] [PubMed]
- Boyd, R.D.; Hayes, M.L. A ribonuclease activity linked to DYW1 in vitro is inhibited by RIP/MORF proteins. Sci. Rep. 2023, 13, 10723. [Google Scholar] [CrossRef]
- Barkan, A.; Small, I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014, 65, 415–442. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, H.; Huang, Z.Q.; Ma, B.; Yang, Y.Z.; Xiu, Z.H.; Wang, L.; Tan, B.C. Maize PPR-E proteins mediate RNA C-to-U editing in mitochondria by recruiting the trans deaminase PCW1. Plant Cell 2023, 35, 529–551. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Dong, J.; Fu, D.; Shi, M.; Zheng, Z.; Zhong, M.; Wang, H.B.; Duan, S.J.; Jin, H.L. The HPE1 RNA-binding protein modulates chloroplast RNA editing to promote photosynthesis under cold stress in Arabidopsis. FEBS Lett. 2024, 598, 1888–1898. [Google Scholar] [CrossRef]
- Takenaka, M.; Zehrmann, A.; Verbitskiy, D.; Kugelmann, M.; Hartel, B.; Brennicke, A. Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proc. Natl. Acad. Sci. USA 2012, 109, 5104–5109. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Shen, L.; Ren, D.; Hu, J.; Chen, G.; Zhu, L.; Gao, Z.; Zhang, G.; Guo, L.; Zeng, D.; et al. Characterization, Expression, and Interaction Analyses of OsMORF Gene Family in Rice. Genes 2019, 10, 694. [Google Scholar] [CrossRef]
- Wang, D.; Meng, S.; Su, W.; Bao, Y.; Lu, Y.; Yin, W.; Liu, C.; Xia, X. Genome-Wide Analysis of Multiple Organellar RNA Editing Factor Family in Poplar Reveals Evolution and Roles in Drought Stress. Int. J. Mol. Sci. 2019, 20, 1425. [Google Scholar] [CrossRef]
- Xiong, Y.; Fang, J.; Jiang, X.; Wang, T.; Liu, K.; Peng, H.; Zhang, X.; Zhang, A. Genome-Wide Analysis of Multiple Organellar RNA Editing Factor (MORF) Family in Kiwifruit (Actinidia chinensis) Reveals Its Roles in Chloroplast RNA Editing and Pathogens Stress. Plants 2022, 11, 146. [Google Scholar] [CrossRef]
- Xing, J.; Zhang, Y.; Song, W.; Ali, N.A.; Su, K.; Sun, X.; Sun, Y.; Jiang, Y.; Zhao, X. Comprehensive identification, characterization, and expression analysis of the MORF gene family in Brassica napus. BMC Plant Biol. 2024, 24, 475. [Google Scholar] [CrossRef]
- Yan, J.; Zhang, Q.; Guan, Z.; Wang, Q.; Li, L.; Ruan, F.; Lin, R.; Zou, T.; Yin, P. MORF9 increases the RNA-binding activity of PLS-type pentatricopeptide repeat protein in plastid RNA editing. Nat. Plants 2017, 3, 17037. [Google Scholar] [CrossRef]
- Wu, J.; Wang, Y.; Chen, H.; Xu, T.; Yang, W.; Fang, X. Solid-like condensation of MORF8 inhibits RNA editing under heat stress in Arabidopsis. Nat. Commun. 2025, 16, 2789. [Google Scholar] [CrossRef]
- Ali, N.A.; Song, W.; Huang, J.; Wu, D.; Zhao, X. Recent advances and biotechnological applications of RNA metabolism in plant chloroplasts and mitochondria. Crit. Rev. Biotechnol. 2024, 44, 1552–1573. [Google Scholar] [CrossRef]
- Li, J.; Yuan, J.; Jing, Y.; Lin, R. MORF proteins: A small family regulating organellar RNA editing and beyond. J. Integr. Plant Biol. 2025, 67, 2532–2544. [Google Scholar] [CrossRef]
- Shim, H.; Lee, H.J.; Lee, J.; Lee, H.O.; Kim, J.H.; Yang, T.J.; Kim, N.S. Plastid Genomes of the Early Vascular Plant Genus Selaginella Have Unusual Direct Repeat Structures and Drastically Reduced Gene Numbers. Int. J. Mol. Sci. 2021, 22, 641. [Google Scholar] [CrossRef]
- Huang, Y.; Xing, Y.; Men, W.; Xue, H.; Hou, W.; Huang, Y.; Dou, D.; Kang, T.; Yang, Y.; Xu, L. The first complete mitochondrial genome assembly and comparative analysis of the fern Blechnaceae family: Blechnopsis orientalis. Front. Plant Sci. 2025, 16, 1534171. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; dePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Chen, H.; Jiang, M.; Wang, L.; Wu, X.; Huang, L.; Liu, C. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 2019, 47, W65–W73. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Zhang, C.T. GC-Profile: A web-based tool for visualizing and analyzing the variation of GC content in genomic sequences. Nucleic Acids Res. 2006, 34, W686–W691. [Google Scholar] [CrossRef]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Chen, Y.; Ye, W.; Zhang, Y.; Xu, Y. High speed BLASTN: An accelerated MegaBLAST search tool. Nucleic Acids Res. 2015, 43, 7762–7768. [Google Scholar] [CrossRef]
- Lewis, S.E.; Searle, S.M.; Harris, N.; Gibson, M.; Lyer, V.; Richter, J.; Wiel, C.; Bayraktaroglu, L.; Birney, E.; Crosby, M.A.; et al. Apollo: A sequence annotation editor. Genome Biol. 2002, 3, RESEARCH0082.1. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Picardi, E.; Pesole, G. REDItools: High-throughput RNA editing detection made easy. Bioinformatics 2013, 29, 1813–1814. [Google Scholar] [CrossRef]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: https://www.R-project.org/ (accessed on 28 March 2025).
- Knoop, V. When you can’t trust the DNA: RNA editing changes transcript sequences. Cell. Mol. Life Sci. 2011, 68, 567–586. [Google Scholar] [CrossRef]
- Chen, M.; Xia, L.; Tan, X.; Gao, S.; Wang, S.; Li, M.; Zhang, Y.; Xu, T.; Cheng, Y.; Chu, Y.; et al. Seeing the unseen in characterizing RNA editome during rice endosperm development. Commun. Biol. 2024, 7, 1314. [Google Scholar] [CrossRef]
- Nie, Y.; Li, Y.; Yuan, P.; Wu, C.; Wang, X.; Wang, C.; Xu, X.; Shen, Z.; Hu, Z. Arabidopsis Pentatricopeptide Repeat Protein GEND2 Participates in Mitochondrial RNA Editing. Plant Cell Physiol. 2024, 65, 1849–1861. [Google Scholar] [CrossRef] [PubMed]
- Schuettpelz, E.; Schneider, H.; Smith, A.R.; Hovenkamp, P.; Prado, J.; Rouhan, G.; Salino, A.; Sundue, M.; Almeida, T.E.; Parris, B.; et al. A community-derived classification for extant lycophytes and ferns. J. Syst. Evol. 2016, 54, 563–603. [Google Scholar] [CrossRef]
- Fujii, S.; Small, I. The evolution of RNA editing and pentatricopeptide repeat genes. New Phytol. 2011, 191, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Fauskee, B.D.; Sigel, E.M.; Pryer, K.M.; Grusz, A.L. Variation in frequency of plastid RNA editing within Adiantum implies rapid evolution in fern plastomes. Am. J. Bot. 2021, 108, 820–827. [Google Scholar] [CrossRef]
- Small, I.D.; Schallenberg-Rudinger, M.; Takenaka, M.; Mireau, H.; Ostersetzer-Biran, O. Plant organellar RNA editing: What 30 years of research has revealed. Plant J. 2020, 101, 1040–1056. [Google Scholar] [CrossRef]
- Rivals, E.; Bruyere, C.; Toffano-Nioche, C.; Lecharny, A. Formation of the Arabidopsis pentatricopeptide repeat family. Plant Physiol. 2006, 141, 825–839. [Google Scholar] [CrossRef]
- Liu, R.; Cao, S.K.; Sayyed, A.; Yang, H.H.; Zhao, J.; Wang, X.; Jia, R.X.; Sun, F.; Tan, B.C. The DYW-subgroup pentatricopeptide repeat protein PPR27 interacts with ZmMORF1 to facilitate mitochondrial RNA editing and seed development in maize. J. Exp. Bot. 2020, 71, 5495–5505. [Google Scholar] [CrossRef]
- Yang, Y.; Fan, G.; Zhao, Y.; Wen, Q.; Wu, P.; Meng, Y.; Shan, W. Cytidine-to-Uridine RNA Editing Factor NbMORF8 Negatively Regulates Plant Immunity to Phytophthora Pathogens. Plant Physiol. 2020, 184, 2182–2198. [Google Scholar] [CrossRef]
- Liang, H.; Deng, J.; Wang, Y.; Gao, G.; Yang, R. The first complete mitochondrial genome of Curcuma amarissima (Zingiberaceae): Insights into multi-branch structure, codon usage, and phylogenetic evolution. BMC Genom. 2025, 26, 343. [Google Scholar] [CrossRef]
- Wagoner, J.A.; Sun, T.; Lin, L.; Hanson, M.R. Cytidine deaminase motifs within the DYW domain of two pentatricopeptide repeat-containing proteins are required for site-specific chloroplast RNA editing. J. Biol. Chem. 2015, 290, 2957–2968. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Qu, G.; Zhang, Y.; Liu, J. Assembly and comparative analysis of the first complete mitochondrial genome of Astragalus membranaceus (Fisch.) Bunge: An invaluable traditional Chinese medicine. BMC Plant Biol. 2024, 24, 1055. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Lu, P.; Long, X.; Cao, X.; Wu, M.; Xin, K.; Xue, T.; Gao, X.; Huang, Y.; Wang, Q.; et al. Adaptive advantages of restorative RNA editing in fungi for resolving survival-reproduction trade-offs. Sci. Adv. 2024, 10, eadk6130. [Google Scholar] [CrossRef] [PubMed]


| Protein ID | Molecular Weight | Theoretical pI | Number of Amino Acids | Instability Index | Aliphatic Index | Grand Average of Hydropathicity |
|---|---|---|---|---|---|---|
| MORF1 | 45,157.41 | 7.65 | 419 | 56.54 | 44.06 | −0.891 |
| MORF2 | 24,714.83 | 8.52 | 219 | 54.46 | 64.98 | −0.700 |
| MORF3 | 27,558.30 | 9.02 | 244 | 67.46 | 62.37 | −0.625 |
| MORF4 | 78,822.18 | 8.47 | 721 | 44.76 | 43.29 | −1.001 |
| MORF5 | 26,024.51 | 9.21 | 229 | 51.15 | 66.86 | −0.669 |
| MORF6 | 26,367.98 | 9.08 | 232 | 52.03 | 64.27 | −0.601 |
| MORF7 | 21,622.74 | 9.35 | 188 | 55.85 | 79.79 | −0.538 |
| MORF8 | 42,869.70 | 9.38 | 395 | 66.48 | 47.95 | −0.949 |
| MORF9 | 26,173.49 | 8.99 | 232 | 56.06 | 64.31 | −0.709 |
| OsMORF9 | 24,818.96 | 8.90 | 229 | 56.89 | 58.60 | −0.476 |
| OsMORF3 | 25,140.50 | 9.16 | 228 | 62.26 | 71.54 | −0.486 |
| OsMORF2a | 24,668.83 | 9.21 | 223 | 65.48 | 63.90 | −0.661 |
| OsMORF2b | 19,720.09 | 5.64 | 172 | 61.97 | 60.06 | −0.897 |
| OsMORF2c | 24,690.81 | 9.24 | 223 | 63.14 | 62.20 | −0.709 |
| OsMORF8a | 43,298.76 | 8.65 | 398 | 62.28 | 47.41 | −0.949 |
| OsMORF8b | 42,933.10 | 8.35 | 396 | 61.12 | 47.40 | −0.998 |
| OJ_27007 | 21,169.77 | 8.40 | 191 | 58.30 | 58.32 | −0.684 |
| PV_23919 | 53,930.48 | 6.87 | 495 | 60.53 | 39.11 | −1.079 |
| PV_35689 | 26,111.66 | 9.55 | 231 | 61.90 | 62.51 | −0.672 |
| PV_33438 | 43,009.38 | 9.31 | 393 | 63.51 | 48.96 | −0.898 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Gu, X.; Lu, C.; Liang, Y.; Ping, J.; Su, Y.; Wang, T. First Identification of MORF Family in Ferns: Molecular Regulation of Organellar RNA Editing in Osmunda japonica and Plenasium vachellii. Biology 2025, 14, 1463. https://doi.org/10.3390/biology14101463
Li L, Gu X, Lu C, Liang Y, Ping J, Su Y, Wang T. First Identification of MORF Family in Ferns: Molecular Regulation of Organellar RNA Editing in Osmunda japonica and Plenasium vachellii. Biology. 2025; 14(10):1463. https://doi.org/10.3390/biology14101463
Chicago/Turabian StyleLi, Lingling, Xiaolin Gu, Chuying Lu, Yingyi Liang, Jingyao Ping, Yingjuan Su, and Ting Wang. 2025. "First Identification of MORF Family in Ferns: Molecular Regulation of Organellar RNA Editing in Osmunda japonica and Plenasium vachellii" Biology 14, no. 10: 1463. https://doi.org/10.3390/biology14101463
APA StyleLi, L., Gu, X., Lu, C., Liang, Y., Ping, J., Su, Y., & Wang, T. (2025). First Identification of MORF Family in Ferns: Molecular Regulation of Organellar RNA Editing in Osmunda japonica and Plenasium vachellii. Biology, 14(10), 1463. https://doi.org/10.3390/biology14101463






