Genome-Wide Association Study Revealed Candidate Genes Associated with Litter Size, Weight, and Body Size Traits in Tianmu Polytocous Sheep (Ovis aries)
Simple Summary
Abstract
1. Background
2. Materials and Methods
2.1. Collection of Phenotypic Data and Blood Samples
2.2. Extraction of Genomic DNA, Library Construction, and Sequencing
2.3. Preprocessing of Whole-Genome Resequencing Data
- 1.
- Removed sequencing adapter/primer sequences from the reads;
- 2.
- Trimmed bases at the 3′ end with a quality score (Q) below 20 (i.e., base error rate < 0.01), where Q = −10log(error_ratio);
- 3.
- Discarded reads containing more than 5 Ns to filter out reads with excessive ambiguous bases.
2.4. GWAS
2.5. Bioinformatics Analysis After GWAS
3. Results
3.1. Overview of the Reproductive/Growth Trait Phenotypes of Tianmu Polytocous Sheep
3.2. Sequencing Data Quality Control
3.3. Detection and Localization of SNPs
3.4. GWAS Results of Litter Size
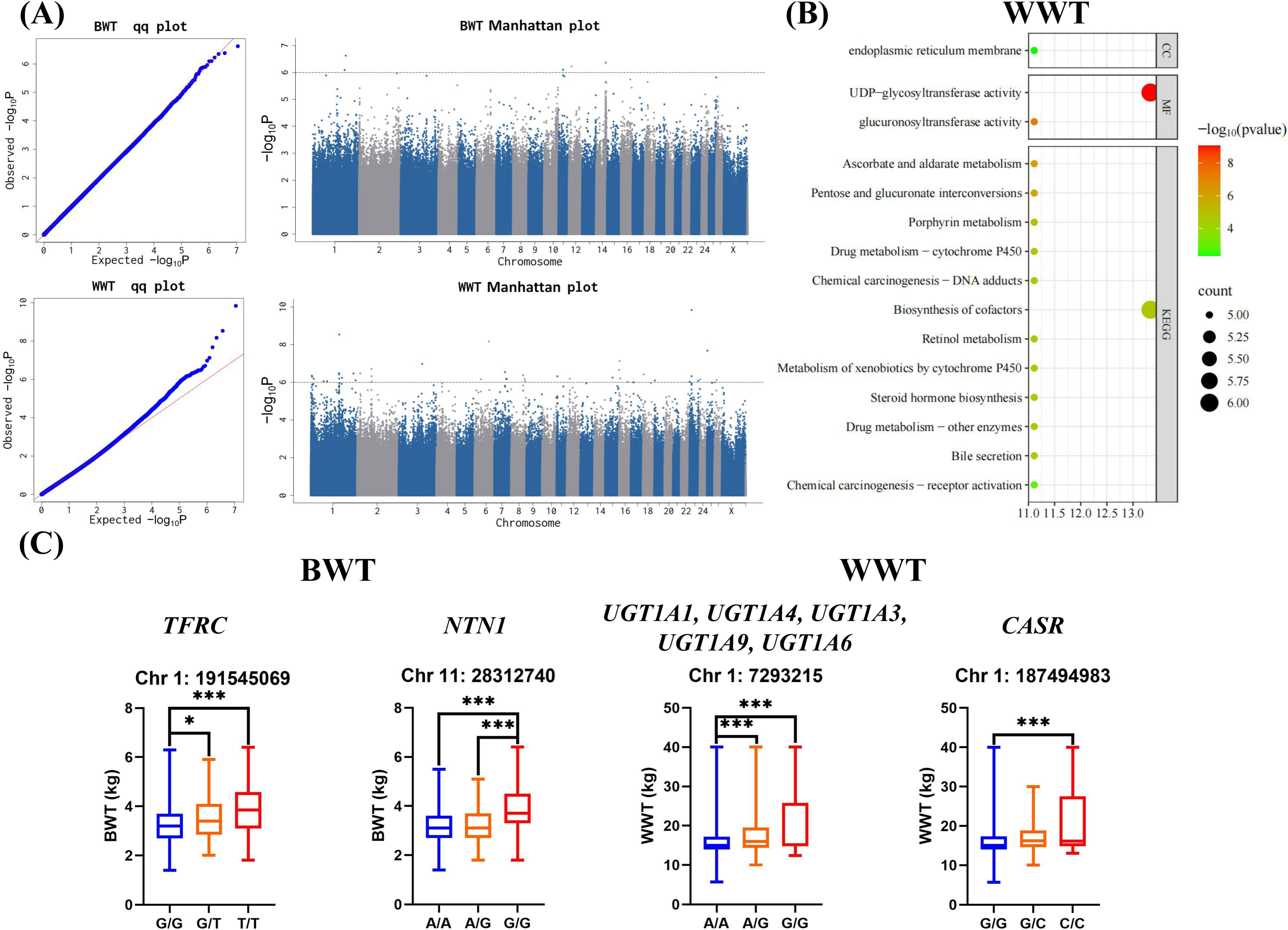
3.5. GWAS Results of Body Weight Traits
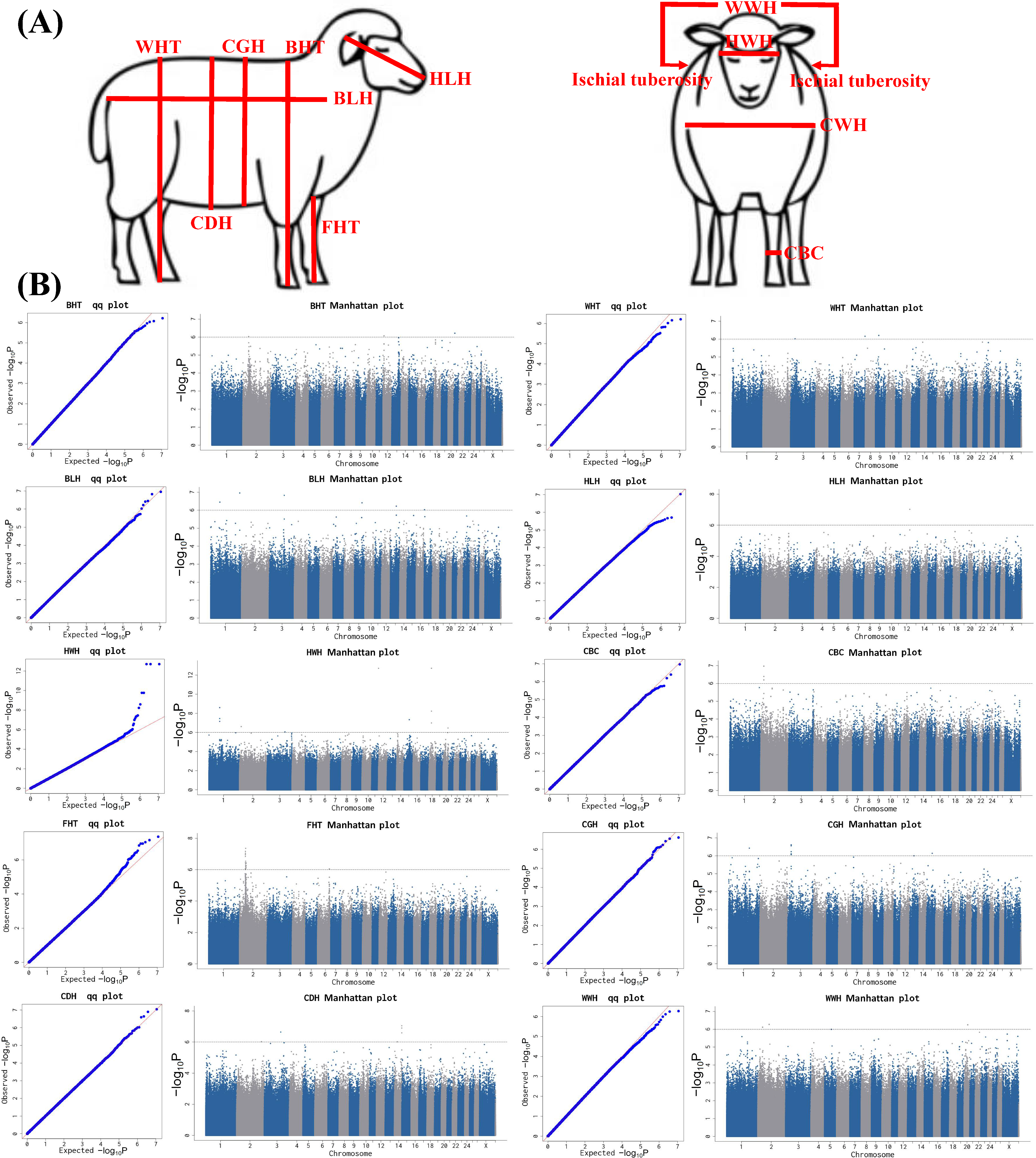
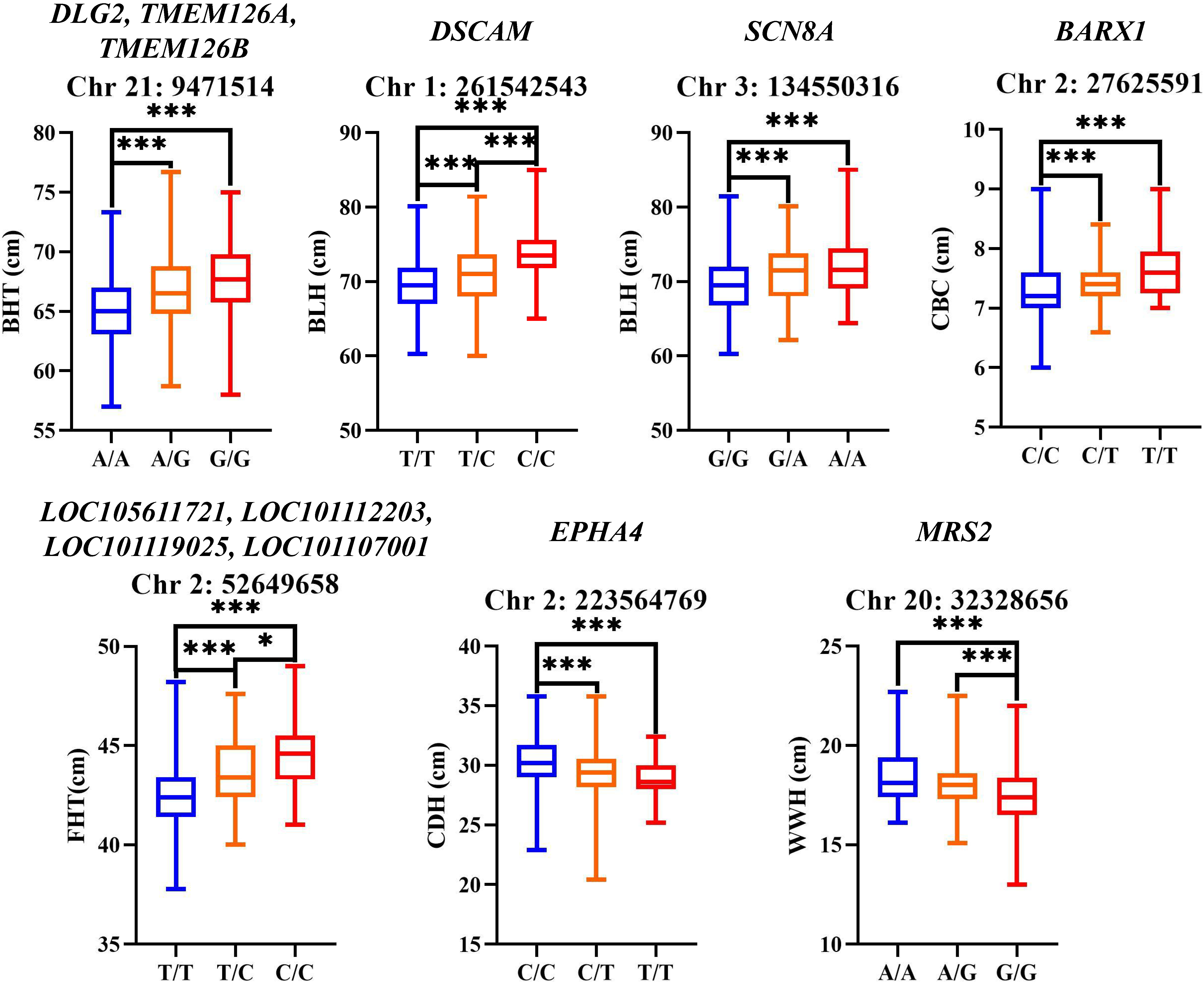
3.6. GWAS Results of Body Size Traits
4. Discussion
4.1. Identification of Candidate Genes Associated with Litter Size
4.2. Identification of Key Candidate Genes Associated with Body Weight Traits
4.3. Identification of Candidate Genes Associated with Body Size Traits
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, Y. Production situation of sheep industry in China in 2021 and trend prospect in 2022. Anim. Agric. 2022, 28–31. [Google Scholar]
- Jones, H.E.; Amer, P.R.; Lewis, R.M.; Emmans, G.C. Economic values for changes in carcass lean and fat weights at a fixed age for terminal sire breeds of sheep in the UK. Livest. Prod. Sci. 2004, 89, 1–17. [Google Scholar] [CrossRef]
- Murray, D.M.; Slezacek, O. Growth rate and its effect on empty body weight, carcass weight and dissected carcass composition of sheep. J. Agric. Sci. 1976, 87, 171–179. [Google Scholar] [CrossRef]
- Okeudo, N.J.; Moss, B.W. Interrelationships amongst carcass and meat quality characteristics of sheep. Meat Sci. 2005, 69, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Makarechian, M.; Whiteman, J.V.; Walters, L.E.; Munson, A.W. Relationships between growth rate, dressing percentage and carcass composition in lambs. J. Anim. Sci. 1978, 46, 1610–1617. [Google Scholar] [CrossRef]
- Camacho, A.; Torres, A.; Capote, J.; Mata, J.; Viera, J.; Bermejo, L.A.; Argüello, A. Meat quality of lambs (hair and wool) slaughtered at different live weights. J. Appl. Anim. Res. 2017, 45, 400–408. [Google Scholar] [CrossRef]
- Rasali, D.P.; Shrestha, J.N.B.; Crow, G.H. Development of composite sheep breeds in the world: A review. Can. J. Anim. Sci. 2006, 86, 1–24. [Google Scholar]
- Rauw, W.M.; Gomez-Raya, L. Genotype by environment interaction and breeding for robustness in livestock. Front. Genet. 2015, 6, 310. [Google Scholar] [CrossRef]
- Burrow, H.M. Importance of adaptation and genotype×environment interactions in tropical beef breeding systems. Animal 2012, 6, 729–740. [Google Scholar] [CrossRef]
- Ruan, J.; Xu, J.; Chen-Tsai, R.Y.; Li, K. Genome editing in livestock: Are we ready for a revolution in animal breeding industry? Transgenic Res. 2017, 26, 715–726. [Google Scholar] [CrossRef]
- Osinowo, O.A.; Abubakar, B.Y.; Trimnell, A.R. Genetic and phenotypic relationships between gestation length, litter size and litter birth weight in Yankasa sheep. Anim. Reprod. Sci. 1993, 34, 111–118. [Google Scholar] [CrossRef]
- Analla, M.; Munoz-Serrano, A.; Serradilla, J.M. Analysis of the genetic relationship between litter size and weight traits in Segurena sheep. Can. J. Anim. Sci. 1997, 77, 17–21. [Google Scholar] [CrossRef]
- Barbato, G.F. Genetic relationships between selection for growth and reproductive effectiveness. Poult. Sci. 1999, 78, 444–452. [Google Scholar] [CrossRef]
- Brito, L.F.; McEwan, J.C.; Miller, S.; Bain, W.; Lee, M.; Dodds, K.; Newman, S.-A.; Pickering, N.; Schenkel, F.S.; Clarke, S. Genetic parameters for various growth, carcass and meat quality traits in a New Zealand sheep population. Small Rumin. Res. 2017, 154, 81–91. [Google Scholar] [CrossRef]
- Meuwissen, T.; Hayes, B.; Goddard, M. Genomic selection: A paradigm shift in animal breeding. Anim. Front. 2016, 6, 6–14. [Google Scholar] [CrossRef]
- Wakchaure, R.; Ganguly, S.; Praveen, P.K.; Kumar, A.; Sharma, S.; Mahajan, T. Marker assisted selection (MAS) in animal breeding: A review. J. Drug Metab. Toxicol. 2015, 6, e127. [Google Scholar] [CrossRef]
- Zhang, Y.D.; Johnston, D.J.; Bolormaa, S.; Hawken, R.J.; Tier, B. Genomic selection for female reproduction in Australian tropically adapted beef cattle. Anim. Prod. Sci. 2013, 54, 16–24. [Google Scholar] [CrossRef]
- Tade, B.; Melesse, A. A review on the application of genomic selection in the improvement of dairy cattle productivity. Ecol. Genet. Genom. 2024, 31, 100257. [Google Scholar] [CrossRef]
- Goddard, M.E.; Hayes, B.J. Genomic selection. J. Anim. Breed Genet. 2007, 124, 323–330. [Google Scholar] [CrossRef]
- Ibtisham, F.; Zhang, L.; Xiao, M.; An, L.; Ramzan, M.B.; Nawab, A.; Zhao, Y.; Li, G.; Xu, Y. Genomic selection and its application in animal breeding. Thai J. Vet. Med. 2017, 47, 301–310. [Google Scholar] [CrossRef]
- Hidalgo, J.; Tsuruta, S.; Lourenco, D.; Masuda, Y.; Huang, Y.; Gray, K.A.; Misztal, I. Changes in genetic parameters for fitness and growth traits in pigs under genomic selection. J. Anim. Sci. 2020, 98, skaa032. [Google Scholar] [CrossRef]
- Xu, S.S.; Gao, L.; Xie, X.L.; Ren, Y.L.; Shen, Z.Q.; Wang, F.; Shen, M.; Eyϸórsdóttir, E.; Hallsson, J.H.; Kiseleva, T.; et al. Genome-wide association analyses highlight the potential for different genetic mechanisms for litter size among sheep breeds. Front. Genet. 2018, 9, 118. [Google Scholar] [CrossRef]
- Gholizadeh, M.; Esmaeili-Fard, S.M. Meta-analysis of genome-wide association studies for litter size in sheep. Theriogenology 2022, 180, 103–112. [Google Scholar] [CrossRef]
- Tuersuntuoheti, M.; Zhang, J.; Zhou, W.; Zhang, C.L.; Liu, C.; Chang, Q.; Liu, S. Exploring the growth trait molecular markers in two sheep breeds based on Genome-wide association analysis. PLoS ONE 2023, 18, e0283383. [Google Scholar] [CrossRef]
- Yang, H.; Li, T.; Zhang, N.; Chen, J.; Zhang, Y.; Peng, S.; Zhou, L.; Ma, R.; Zhang, Z.; Liu, Q.; et al. Identification of Candidate Genes and Functional Pathways Associated with Body Size Traits in Hulunbuir Sheep Through GWAS Analysis. Genes 2025, 16, 410. [Google Scholar] [CrossRef]
- Simon, A. FastQC: A Quality Control Tool for High Throughput Sequence Data; Version 0.10; Babraham Bioinformatics: Cambridge, UK, 2010. [Google Scholar]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Tarasov, A.; Vilella, A.J.; Cuppen, E.; Nijman, I.J.; Prins, P. Sambamba: Fast processing of NGS alignment formats. Bioinformatics 2015, 31, 2032–2034. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Stephens, M. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nat. Methods 2014, 11, 407–409. [Google Scholar] [CrossRef]
- Yin, L.; Zhang, H.; Tang, Z.; Xu, J.; Yin, D.; Zhang, Z.; Yuan, X.; Zhu, M.; Zhao, S.; Li, X.; et al. rMVP: A memory-efficient, visualization-enhanced, and parallel-accelerated tool for genome-wide association study. Genom. Proteom. Bioinf. 2021, 19, 619–628. [Google Scholar] [CrossRef]
- Turner, S.D. qqman: An R package for visualizing GWAS results using QQ and manhattan plots. Biorxiv 2014, 005165. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic. Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Melo, T.P.D.; De Camargo, G.M.F.; De Albuquerque, L.G.; Carvalheiro, R. Genome-wide association study provides strong evidence of genes affecting the reproductive performance of Nellore beef cows. PLoS ONE 2017, 12, e0178551. [Google Scholar] [CrossRef]
- Horodyska, J.; Hamill, R.M.; Varley, P.F.; Reyer, H.; Wimmers, K. Genome-wide association analysis and functional annotation of positional candidate genes for feed conversion efficiency and growth rate in pigs. PLoS ONE 2017, 12, e0173482. [Google Scholar] [CrossRef]
- Huang, J.; Lin, X. Advances in animal disease resistance research: Discoveries of genetic markers for disease resistance in cattle through GWAS. Biol. Evid. 2024, 14, 24–31. [Google Scholar] [CrossRef]
- Saleh, A.A.; Hammoud, M.H.; Dabour, N.A.; Hafez, E.E.; Sharaby, M.A. BMPR-1B, BMP-15 and GDF-9 genes structure and their relationship with litter size in six sheep breeds reared in Egypt. BMC Res. Notes 2020, 13, 215. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, Y.; Cao, J.M. G protein-coupled receptors: Extranuclear mediators for the non-genomic actions of steroids. Int. J. Mol. Sci. 2014, 15, 15412–15425. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhong, J.; Inuzuka, H.; Gao, D.; Shaik, S.; Sarkar, F.H.; Wei, W. An evolving role for DEPTOR in tumor development and progression. Neoplasia 2012, 14, 368–375. [Google Scholar] [CrossRef]
- Ran, Z.; Liu, R.; Shi, H.; Wang, X.; Wu, Z.; Zhou, S.; Liao, J.; Hu, L.; Hu, Y.; Zhou, J.; et al. mTOR signaling mediates energy metabolic equilibrium in bovine and mouse oocytes during the ovulatory phase. Biol. Reprod. 2025, 112, 474–484. [Google Scholar] [CrossRef] [PubMed]
- Panidis, D.; Balaris, C.; Farmakiotis, D.; Rousso, D.; Kourtis, A.; Balaris, V.; Katsikis, I.; Zournatzi, V.; Diamanti-Kandarakis, E. Serum parathyroid hormone concentrations are increased in women with polycystic ovary syndrome. Clin. Chem. 2005, 51, 1691–1697. [Google Scholar] [CrossRef]
- Wang, X.; Lu, T.; Qu, Y.; Fan, L. PTH2R is related to cell proliferation and migration in ovarian cancer: A multi-omics analysis of bioinformatics and experiments. Cancer Cell Int. 2022, 22, 148. [Google Scholar]
- Yan, H.; Tong, J.; Lin, X.; Han, Q.; Huang, H. Effect of the WWOX gene on the regulation of the cell cycle and apoptosis in human ovarian cancer stem cells. Mol. Med. Rep. 2015, 12, 1783–1788. [Google Scholar] [CrossRef]
- Zhang, D.; Sliwkowski, M.X.; Mark, M.; Frantz, G.; Akita, R.; Sun, Y.; Hillan, K.; Crowley, C.; Brush, J.; Godowski, P.J. Neuregulin-3 (NRG3): A novel neural tissue-enriched protein that binds and activates ErbB4. Proc. Natl. Acad. Sci. USA 1997, 94, 9562–9567. [Google Scholar] [CrossRef]
- Maihle, N.J.; Baron, A.T.; Barrette, B.A.; Boardman, C.H.; Christensen, T.A.; Cora, E.M.; Faupel-Badger, J.M.; Greenwood, T.; Juneja, S.C.; Lafky, J.M.; et al. EGF/ErbB receptor family in ovarian cancer. Cancer Treat Res. 2002, 107, 247–258. [Google Scholar]
- Welt, C.; Sidis, Y.; Keutmann, H.; Schneyer, A. Activins, inhibins, and follistatins: From endocrinology to signaling. A paradigm for the new millennium. Exp. Biol. Med. 2002, 227, 724–752. [Google Scholar] [CrossRef]
- Cui, Z.; Liu, L.; Zhao, X.; Ran, J.; Wang, Y.; Yin, H.; Li, D.; Zhu, Q. Analysis of expression and single nucleotide polymorphisms of INHA gene associated with reproductive traits in chickens. Biomed. Res. Int. 2019, 2019, 8572837. [Google Scholar] [CrossRef] [PubMed]
- McClain, D.A.; Sharma, N.K.; Jain, S.; Harrison, A.; Salaye, L.N.; Comeau, M.E.; Langefeld, C.D.; Lorenzo, F.R.; Das, S.K. Adipose tissue transferrin and insulin resistance. J. Clin. Endocrinol. Metab. 2018, 103, 4197–4208. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhao, L.; Zhang, L.; Liu, X.; Hou, X.; Gao, H.; Yan, H.; Zhao, F.; Wang, L. NTN1 affects porcine intramuscular fat content by affecting the expression of myogenic regulatory factors. Animals 2019, 9, 609. [Google Scholar] [CrossRef]
- Mentxaka, A.; Gómez-Ambrosi, J.; Ramírez, B.; Rodríguez, A.; Becerril, S.; Neira, G.; Valentí, V.; Moncada, R.; Silva, C.; Unamuno, X.; et al. Netrin-1 promotes visceral adipose tissue inflammation in obesity and is associated with insulin resistance. Nutrients 2022, 14, 4372. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zhang, H.; Teng, J.; Huang, L.; Li, Y.; Sun, C. Involvement of calcium-sensing receptor in inhibition of lipolysis through intracellular cAMP and calcium pathways in human adipocytes. Biochem. Biophys. Res. Commun. 2011, 404, 393–399. [Google Scholar] [CrossRef]
- Cifuentes, M.; Rojas, C.V. Antilipolytic effect of calcium-sensing receptor in human adipocytes. Mol. Cell. Biochem. 2008, 319, 17–21. [Google Scholar] [CrossRef]
- Zhang, N.; Jiang, H.; Bai, Y.; Lu, X.; Feng, M.; Guo, Y.; Zhang, S.; Luo, Q.; Wu, H.; Wang, L. The molecular mechanism study of insulin on proliferation and differentiation of osteoblasts under high glucose conditions. Cell Biochem. Funct. 2019, 37, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Aldiss, P.; Lewis, J.E.; Boocock, D.J.; Miles, A.K.; Bloor, I.; Ebling, F.J.; Budge, H.; Symonds, M.E. Interscapular and perivascular brown adipose tissue respond differently to a short-term high-fat diet. Nutrients 2019, 11, 1065. [Google Scholar] [CrossRef]
- Rauthan, K.; Joshi, S.; Kumar, L.; Kumar, S. Functional annotation and identification of novel drug targets from uncharacterized proteome of Trichuris trichiura. J. Infect. Dev. Ctries. 2025, 19, 948–961. [Google Scholar] [CrossRef]
- Wu, W.; Zhu, Z.; Mao, S.; Qiu, X.; Qian, B.; Liu, Z.; Qiu, Y. Lack of association between DSCAM gene polymorphisms and adolescent idiopathic scoliosis susceptibility in a Chinese Han population. J. Back Musculoskelet 2015, 28, 681–687. [Google Scholar] [CrossRef]
- Zhou, S.; Zhu, Z.; Qiu, X.; Wu, W.; Wang, W.; Liu, Z.; Lv, F.; Qiu, Y. Association study of IL-17RC, CHL1, DSCAM and CNTNAP2 genes polymorphisms with adolescent idiopathic scoliosis susceptibility in a Chinese Han population. Stud. Health Technol. Inform. 2012, 176, 47–51. [Google Scholar] [PubMed]
- Meisler, M.H.; Kearney, J.; Escayg, A.; Macdonald, B.T.; Sprunger, L.K. Sodium channels and neurological disease: Insights from Scn8a mutations in the mouse. Neurosci. 2001, 7, 136–145. [Google Scholar] [CrossRef]
- Liu, Y.; Koko, M.; Lerche, H. A SCN8A variant associated with severe early onset epilepsy and developmental delay: Loss-or gain-of-function? Epilepsy Res. 2021, 178, 106824. [Google Scholar] [CrossRef] [PubMed]
- Salsi, V.; Vigano, M.A.; Cocchiarella, F.; Mantovani, R.; Zappavigna, V. Hoxd13 binds in vivo and regulates the expression of genes acting in key pathways for early limb and skeletal patterning. Dev. Biol. 2008, 317, 497–507. [Google Scholar] [CrossRef]
- Nichols, J.T.; Pan, L.; Moens, C.B.; Kimmel, C.B. barx1 represses joints and promotes cartilage in the craniofacial skeleton. Development 2013, 140, 2765–2775. [Google Scholar] [CrossRef] [PubMed]
- Methner, A.; Hermey, G.; Schinke, B.; Hermans-Borgmeyer, I. A novel G protein-coupled receptor with homology to neuropeptide and chemoattractant receptors expressed during bone development. Biochem. Biophys. Res. Commun. 1997, 233, 336–342. [Google Scholar] [CrossRef]
- Bowler, W.B.; Gallagher, J.A.; Bilbe, G. G-protein coupled receptors in bone. Front. Biosci. 1998, 3, d769–d780. [Google Scholar] [CrossRef][Green Version]
- Stiffel, V.; Amoui, M.; Sheng, M.H.C.; Mohan, S.; Lau, K.H.W. EphA4 receptor is a novel negative regulator of osteoclast activity. J. Bone Miner. Res. 2014, 29, 804–819. [Google Scholar] [CrossRef]
- Ting, M.C.; Wu, N.L.; Roybal, P.G.; Sun, J.; Liu, L.; Yen, Y.; Maxson Jr, R.E. EphA4 as an effector of Twist1 in the guidance of osteogenic precursor cells during calvarial bone growth and in craniosynostosis. Development 2009, 136, 855–864. [Google Scholar] [CrossRef]
- Li, M.; Li, Y.; Lu, Y.; Li, J.; Lu, X.; Ren, Y.; Wen, T.; Wang, Y.; Chang, S.; Zhang, X.; et al. Molecular basis of Mg2+ permeation through the human mitochondrial Mrs2 channel. Nat. Commun. 2023, 14, 4713. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Leng, H.; Guo, L.; Huang, Y.; Zheng, T.; Zhao, Z.; Liu, X.; Zhang, X.; Cai, Q.; Yang, X. Calcium silicate scaffolds promoting bone regeneration via the doping of Mg2+ or Mn2+ ion. Compos. Part B Eng. 2020, 190, 107937. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Ma, S.; Lu, Q.; Tang, S.; Mamat, N.; Wang, Y.; Hong, W.; Hu, X.; Wu, C.; Fu, X. Genome-Wide Association Study Revealed Candidate Genes Associated with Litter Size, Weight, and Body Size Traits in Tianmu Polytocous Sheep (Ovis aries). Biology 2025, 14, 1446. https://doi.org/10.3390/biology14101446
Liu W, Ma S, Lu Q, Tang S, Mamat N, Wang Y, Hong W, Hu X, Wu C, Fu X. Genome-Wide Association Study Revealed Candidate Genes Associated with Litter Size, Weight, and Body Size Traits in Tianmu Polytocous Sheep (Ovis aries). Biology. 2025; 14(10):1446. https://doi.org/10.3390/biology14101446
Chicago/Turabian StyleLiu, Wenna, Shengchao Ma, Qingwei Lu, Sen Tang, Nuramina Mamat, Yaqian Wang, Wei Hong, Xiangrong Hu, Cuiling Wu, and Xuefeng Fu. 2025. "Genome-Wide Association Study Revealed Candidate Genes Associated with Litter Size, Weight, and Body Size Traits in Tianmu Polytocous Sheep (Ovis aries)" Biology 14, no. 10: 1446. https://doi.org/10.3390/biology14101446
APA StyleLiu, W., Ma, S., Lu, Q., Tang, S., Mamat, N., Wang, Y., Hong, W., Hu, X., Wu, C., & Fu, X. (2025). Genome-Wide Association Study Revealed Candidate Genes Associated with Litter Size, Weight, and Body Size Traits in Tianmu Polytocous Sheep (Ovis aries). Biology, 14(10), 1446. https://doi.org/10.3390/biology14101446





