Comparative Proteomics Analysis Reveals Differential Immune Responses of Paralichthys olivaceus to Edwardsiella tarda Infection Under High and Low Temperature
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Japanese Flounder
2.3. In Vivo Infection Experiment of Flounder and Samples Collection
2.4. Sample Preparation for Proteomics and Data Analysis
2.5. Identification of Differentially Abundant Proteins (DAPs)
2.6. GO and KEGG Annotation Analysis
2.7. Construction of the Interaction Network and Key Proteins Selection
2.8. Key Proteins Analysis and Validation
3. Results
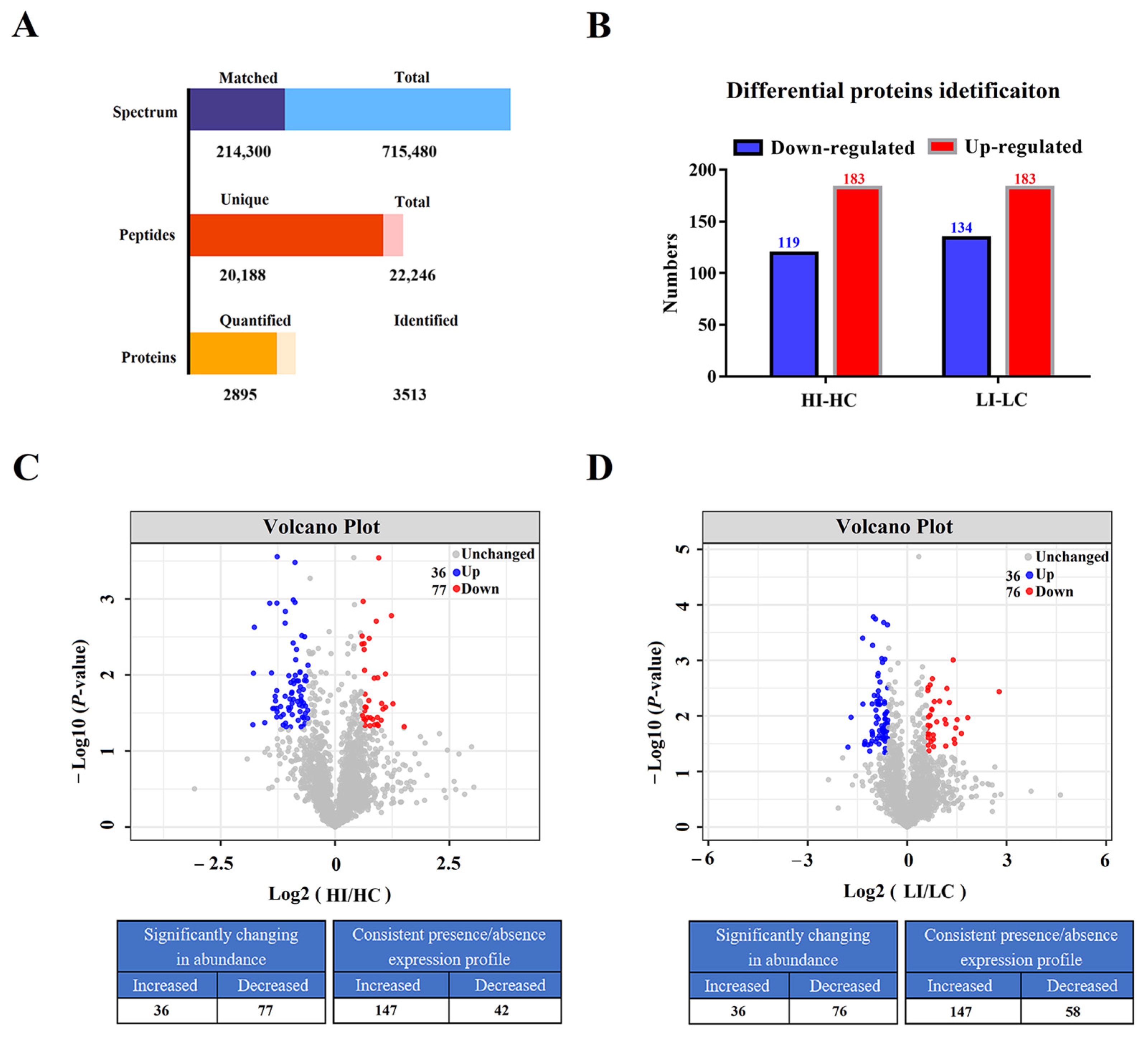
3.1. A Large Number of Proteins Exhibit Differential Expressions in Flounder Infected with E. tarda at Low and High Temperatures
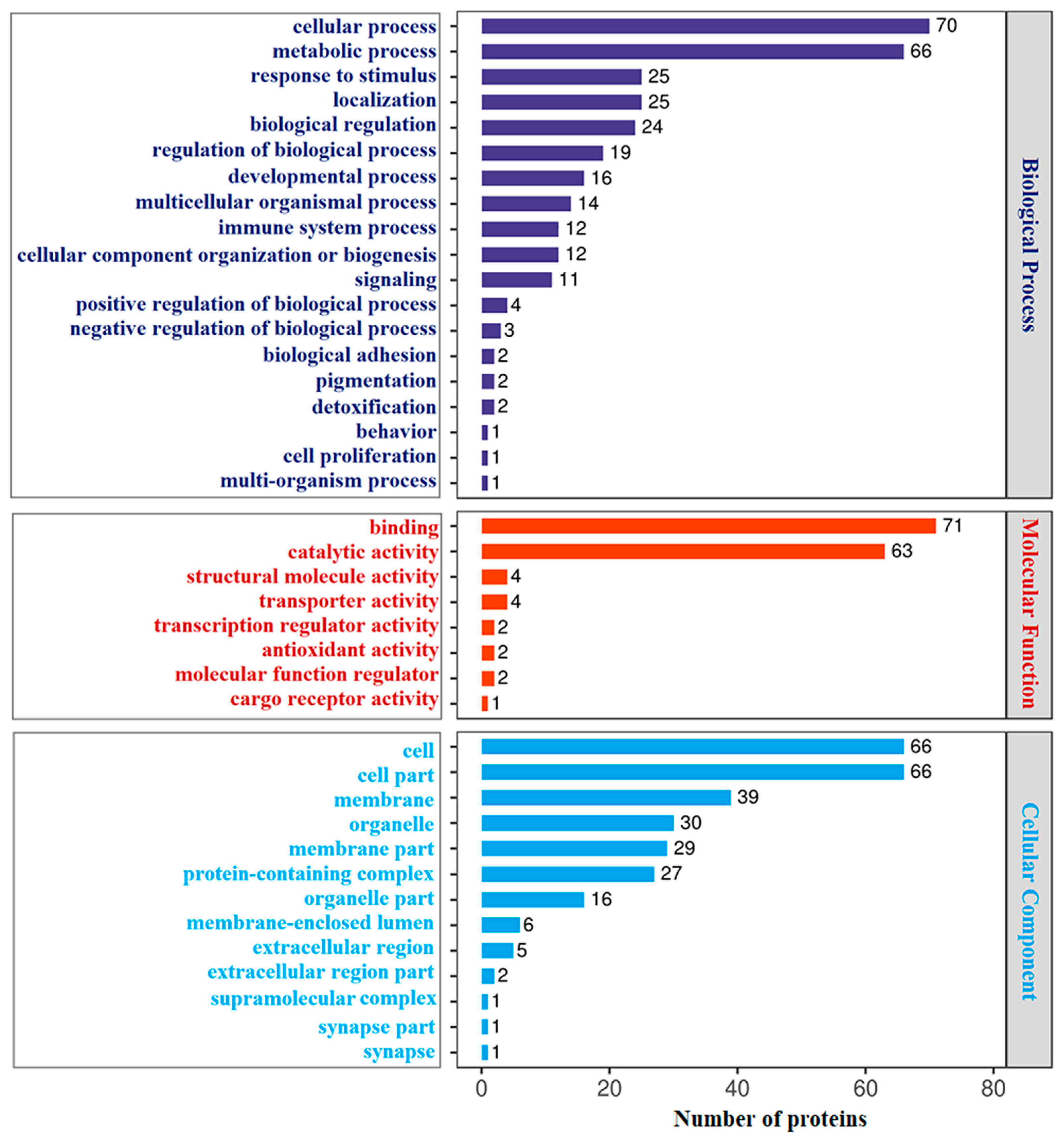
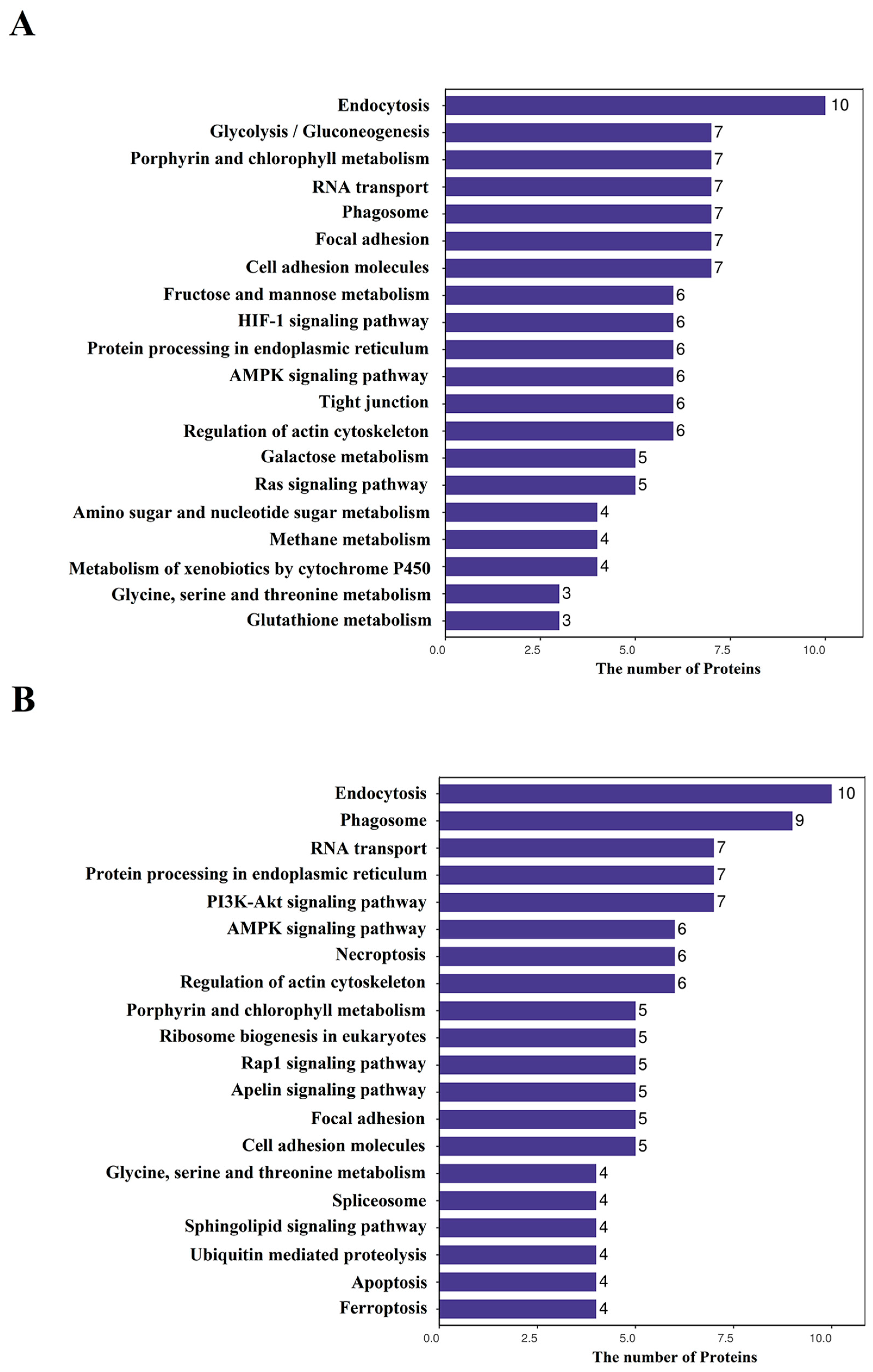
3.2. GO and KEGG Analyses
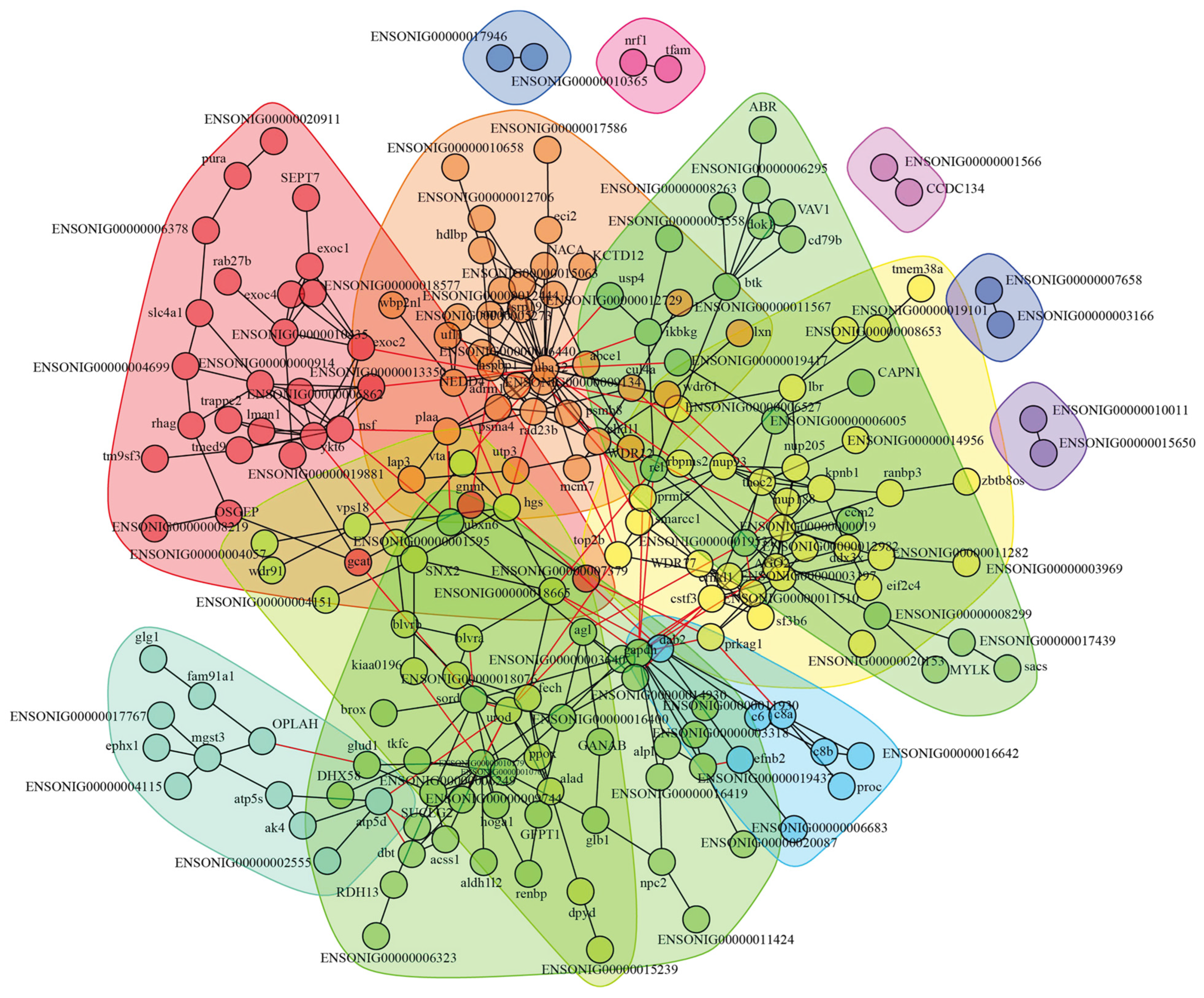
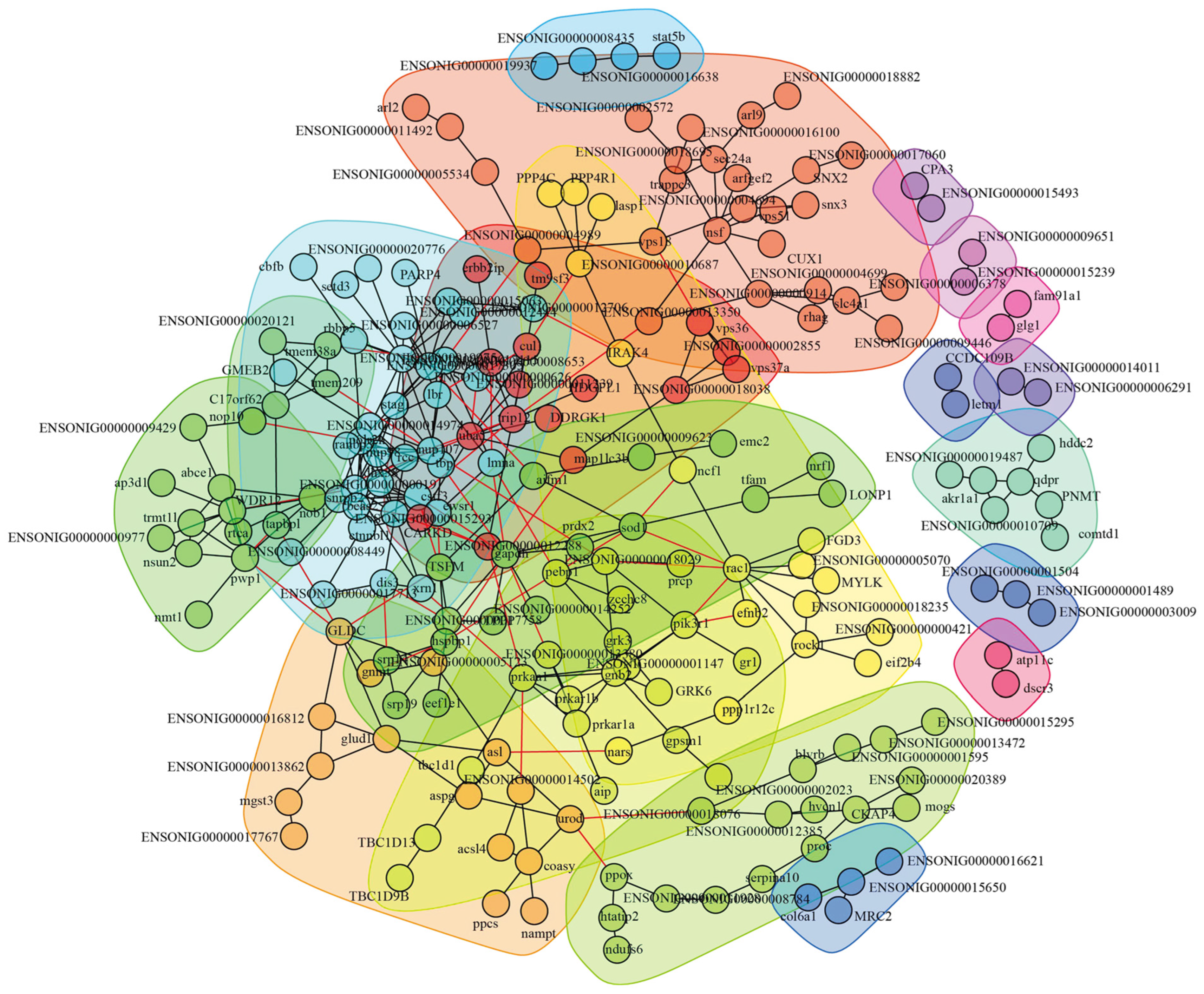
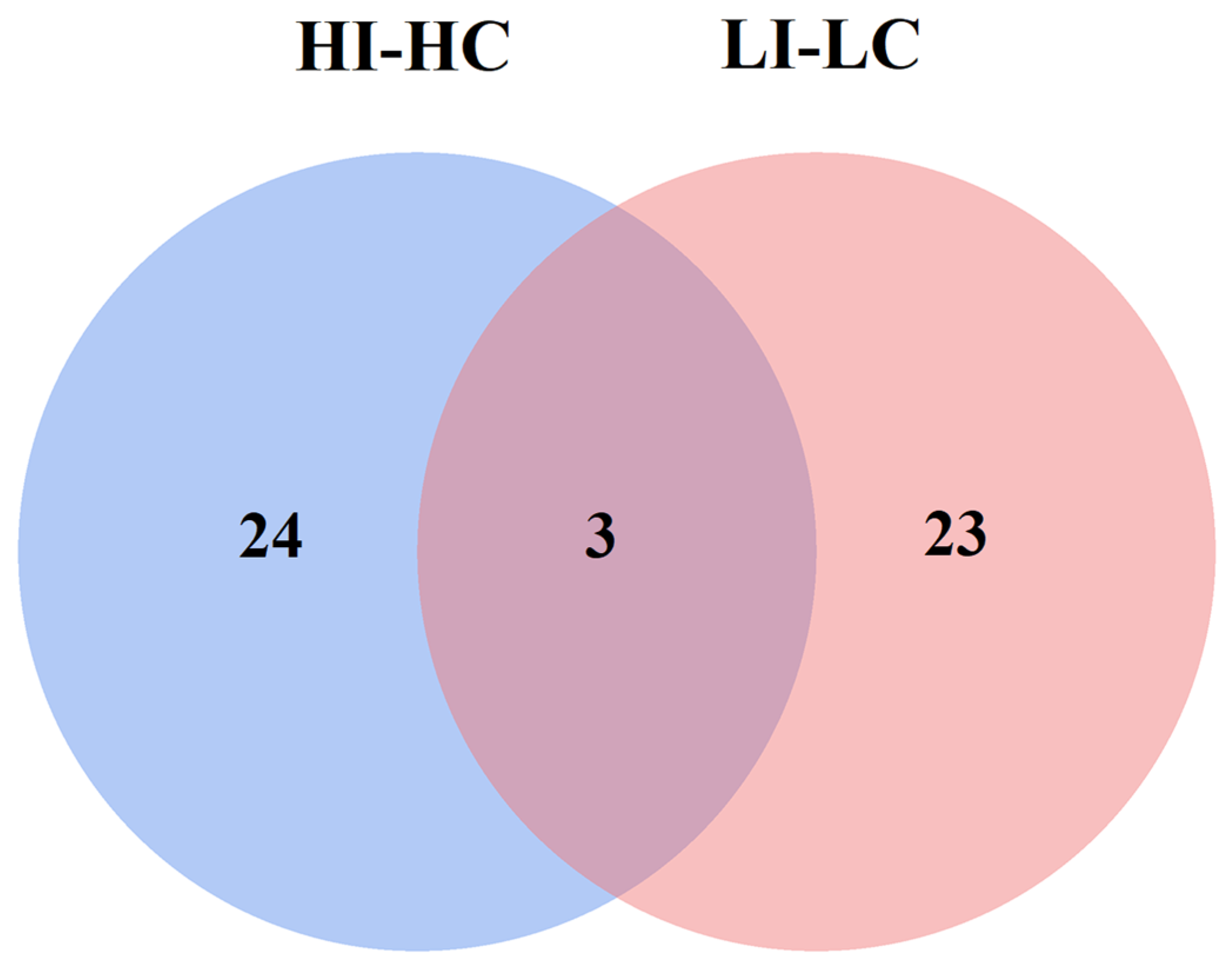
3.3. Construction of Interaction Network and Key Proteins Identification
3.4. Special Key Proteins Analysis in HI-HC Group
3.5. Special Key Proteins Analysis in LI-LC Group
3.6. Shared Key Proteins Analysis Between HI-HC and LI-LC Groups
3.7. Key Proteins Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Agarwal, D.; Shanmugam, S.A.; Kathirvelpandian, A.; Eswaran, S.; Rather, M.A.; Rakkannan, G. Unraveling the impact of climate change on fish physiology: A focus on temperature and salinity dynamics. J. Appl. Ichthyol. 2024, 2024, 5782274. [Google Scholar] [CrossRef]
- Jiang, G.; Xue, Y.; Huang, X. Temperature-induced sex differentiation in river prawn (Macrobrachium nipponense): Mechanisms and effects. Int. J. Mol. Sci. 2024, 25, 1207. [Google Scholar] [CrossRef]
- Ma, S.B.; Lv, Y.B.; Hou, L.; Jia, Z.M.; Lin, S.; Wang, S.D.; He, X.G.; Hou, J. Effect of acute temperature stress on energy metabolism, immune performance and gut microbiome of largemouth bass (Micropterus salmoides). AAF 2025, 10, 260–270. [Google Scholar] [CrossRef]
- Jung, M.H.; Park, C.S.; Kole, S.; Ryu, J.W.; Jung, S.J. Water temperature and immunization period required to establish immunity against the viral hemorrhagic septicemia virus vaccine in olive flounder (Paralichthys olivaceus). Aquaculture 2024, 591, 741118. [Google Scholar] [CrossRef]
- Alcorn, S.W.; Murra, A.L.; Pascho, R.J. Effects of rearing temperature on immune functions in sockeye salmon (Oncorhynchus nerka). Fish. Shellfish. Immunol. 2002, 12, 303–334. [Google Scholar] [CrossRef]
- Ndong, D.; Chen, Y.Y.; Lin, Y.H.; Vaseeharan, B.; Chen, J.C. The immune response of tilapia Oreochromis mossambicus and its susceptibility to Streptococcus iniae under stress in low and high temperatures. Fish. Shellfish. Immunol. 2007, 22, 686–694. [Google Scholar] [CrossRef] [PubMed]
- Chebaani, N.; Guardiola, F.A.; Sihem, M.; Nabil, A.; Oumouna, M.; Meseguer, J.; Esteban, M.A.; Cuesta, A. Innate humoral immune parameters in Tilapia zillii under acute stress by low temperature and crowding. Fish. Physiol. Biochem. 2014, 40, 797–804. [Google Scholar] [CrossRef] [PubMed]
- Nikoskelainen, S.; Bylund, G.; Lilius, E.M. Effect of environmental temperature on rainbow trout (Oncorhynchus mykiss) innate immunity. Dev. Comp. Immunol. 2004, 28, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Morvan, C.L.; Troutaud, D.; Deschaux, P. Differential effects of temperature on specific and nonspecific immune defences in fish. J. Exp. Biol. 1998, 201, 165–168. [Google Scholar] [CrossRef]
- Cushing, J.E., Jr. An effect of temperature upon antibody-production in fish. J. Immunol. 1942, 45, 123–126. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, S.K.; Hur, Y.B. Temperature-mediated changes in stress responses, acetylcholinesterase, and immune responses of juvenile olive flounder Paralichthys olivaceus in a bio-floc environment. Aquaculture 2019, 506, 453–458. [Google Scholar] [CrossRef]
- Dominguez, M.; Takemura, A.; Tsuchiya, M.; Nakamura, S. Impact of different environmental factors on the circulating immunoglobulin levels in the Nile tilapia, Oreochromis niloticus. Aquaculture 2004, 241, 491–500. [Google Scholar] [CrossRef]
- Hori, T.S.; Gamperl, A.K.; Booman, M.; Nash, G.W.; Rise, M.L. A moderate increase in ambient temperature modulates the Atlantic cod (Gadus morhua) spleen transcriptome response to intraperitoneal viral mimic injection. BMC Genom. 2012, 13, 431. [Google Scholar] [CrossRef] [PubMed]
- Polinski, M.; Bridle, A.; Nowak, B. Temperature-induced transcription of inflammatory mediators and the influence of Hsp70 following LPS stimulation of southern bluefin tuna peripheral blood leukocytes and kidney homogenates. Fish. Shellfish. Immunol. 2013, 34, 1147–1157. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.H.; Ma, A.J.; Wang, X.A. Interaction of temperature and salinity on the expression of immunity factors in different tissues of juvenile turbot Scophthalmus maximus based on response surface methodology. Chin. J. Oceanol. Limn. 2015, 33, 28–36. [Google Scholar] [CrossRef]
- Raida, M.K.; Buchmann, K. Temperature-dependent expression of immune-relevant genes in rainbow trout following Yersinia ruckeri vaccination. Dis. Aquat. Organ. 2007, 77, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Kaneshige, N.; Jirapongpairoj, W.; Hirono, I.; Kondo, H. Temperature-dependent regulation of gene expression in Japanese flounder Paralichthys olivaceus kidney after Edwardsiella tarda formalin-killed cells. Fish. Shellfish. Immunol. 2016, 59, 298–304. [Google Scholar] [CrossRef]
- Xu, T.; Zhang, X.H. Edwardsiella tarda: An intriguing problem in aquaculture. Aquaculture 2014, 431, 129–135. [Google Scholar] [CrossRef]
- Han, H.J.; Kim, D.H.; Lee, D.C.; Kim, S.M.; Park, S.I. Pathogenicity of Edwardsiella tarda to olive flounder, Paralichthys olivaceus (Temminck & Schlegel). J. Fish. Dis. 2006, 29, 601–609. [Google Scholar]
- Chakraborty, S.; Li, M.; Chatterjee, C.; Sivaraman, J.; Leung, K.Y.; Mok, Y.K. Temperature and Mg2+ sensing by a novel PhoP-PhoQ two-component system for regulation of virulence in Edwardsiella tarda. J. Biol. Chem. 2010, 285, 38876–38888. [Google Scholar] [CrossRef]
- Seikai, T. Flounder culture and its challenges in Asia. Rev. Fish. Sci. 2002, 10, 421–432. [Google Scholar] [CrossRef]
- Avunje, S.; Oh, M.J.; Jung, S.J. Impaired TLR2 and TLR7 response in olive flounder infected with viral haemorrhagic septicaemia virus at host susceptible 15 °C but high at non-susceptible 20 °C. Fish. Shellfish. Immunol. 2013, 34, 1236–1243. [Google Scholar] [CrossRef]
- Jacobsen, K.L.; Griffin, M.; Phinney, B.S.; Salemi, M.; Yazdi, Z.; Balami, S.; Older, C.E.; Soto, E. Temperature—dependent alterations in the proteome of the emergent fish pathogen Edwardsiella piscicida. J. Fish. Dis. 2024, 48, e14017. [Google Scholar] [CrossRef]
- Gobena, S.; Admassu, B.; Kinde, M.Z.; Gessese, A.T. Proteomics and its current application in biomedical area: Concise review. J. TSW 2024, 2024, 4454744. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Sun, Y.Y.; Li, X.P.; Jiang, S.; Sun, L. Particle and bacteria uptake by Japanese flounder (Paralichthys olivaceus) red blood cells: Size dependence and pathway specificity. Tissue Cell 2019, 61, 79–88. [Google Scholar] [CrossRef]
- Li, M.F.; Sun, L. Characterization of a teleost membrane-associated protein that is involved in the regulation of complement activation and bacterial infection. Dev. Comp. Immunol. 2018, 79, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Sun, K.; Sun, L. Regulation of autoinducer 2 production and luxS expression in a pathogenic Edwardsiella tarda strain. Microbiol. 2008, 154, 2060–2069. [Google Scholar] [CrossRef]
- Li, P.; Tian, M.; Hu, H.; Yin, Y.; Guan, X.; Ding, C.; Wang, S.H.; Yu, S.Q. Lable-free based comparative proteomic analysis of secretory proteins of rough Brucella mutants. J. Proteom. 2019, 195, 66–75. [Google Scholar] [CrossRef]
- Battisti, I.; Ebinezer, L.B.; Lomolino, G.; Masi, A.; Arrigoni, G. Protein profile of commercial soybean milks analyzed by label-free quantitative proteomics. Food Chem. 2021, 352, 129299. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yu, Y.; Maha, I.F.; Kong, J.; Xie, X.; Yin, F. Label-free quantitative proteomics analysis of skin of yellow drum (Nibea albiflora) reveals immune mechanism against Cryptocaryon irritans. Fish. Shellfish. Immunol. 2020, 101, 284–290. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Fang, H.; Shao, X.Q.; Liu, M.; Jiang, K.Y.; Wang, M.Q.; Wang, B.J.; Wang, L. Exploration of the influence of surface proteins on the probiotic activity of Lactobacillus pentosus HC-2 in the Litopenaeus vannamei midgut via label-free quantitative proteomic analysis. Fish. Shellfish. Immunol. 2019, 95, 368–382. [Google Scholar] [CrossRef]
- Sun, B.; Li, X.P.; Ning, X.H.; Sun, L. Transcriptome analysis of Paralichthys olivaceus erythrocytes reveals profound immune responses induced by Edwardsiella tarda infection. Int. J. Mol. Sci. 2020, 21, 3094. [Google Scholar] [CrossRef]
- Zheng, W.; Sun, L. Evaluation of housekeeping genes as references for quantitative real time RT-PCR analysis of gene expression in Japanese flounder (Paralichthys olivaceus). Fish. Shellfish. Immunol. 2011, 30, 638–645. [Google Scholar] [CrossRef]
- Wang, J.; Sun, L. Edwardsiella tarda-regulated proteins in Japanese flounder (Paralichthys olivaceus): Identification and evaluation of antibacterial potentials. J. Proteom. 2015, 124, 1–10. [Google Scholar] [CrossRef]
- Zhang, Z.G.; Jiang, J.F.; Liu, X.L.; Wu, H.M. Preliminary study on the causes of mass mortality in cultured flounder (Paralichthys olivaceus). Heilongjiang Anim. Sci. Vet. Med. 2017, 1, 194–197. (In Chinese) [Google Scholar]
- Zhang, Y.Y.; Wang, W.N.; Wang, J.X.; Wang, A.L. Research on Edwardsiellosis in Japanese Flounder. Fish. Sci. 2003, 1, 34–36. (In Chinese) [Google Scholar]
- Sun, B.; Sun, B.G.; Zhang, B.B.; Sun, L. Temperature induces metabolic reprogramming in fish during bacterial infection. Front. Immunol. 2022, 13, 1010948. [Google Scholar] [CrossRef] [PubMed]
- Settembre, C.; Ballabio, A. Lysosomal adaptation: How the lysosome responds to external cues. Cold Spring Harb. Perspect. Biol. 2014, 6, a016907. [Google Scholar] [CrossRef]
- Peltan, A.; Briggs, L.; Matthews, G.; Sweeney, S.T.; Smith, D.F. Identification of Drosophila gene products required for phagocytosis of Leishmania donovani. PLoS ONE 2012, 7, e51831. [Google Scholar] [CrossRef]
- Hasegawa, H.; Zinsser, S.; Rhee, Y.Y.; Vik-Mo, E.O.; Davanger, S.; Hay, J.C. Mammalian ykt6 is a neuronal SNARE targeted to a specialized compartment by its profilin-like amino terminal domain. Mol. Biol. Cell 2003, 14, 698–720. [Google Scholar] [CrossRef] [PubMed]
- Komada, M.; Soriano, P. Hrs, a FYVE finger protein localized to early endosomes, is implicated in vesicular traffic and required for ventral folding morphogenesis. Genes. Dev. 1999, 13, 1475–1485. [Google Scholar] [CrossRef]
- Komada, M.; Masaki, R.; Yamamoto, A.; Kitamura, N. Hrs, a tyrosine kinase substrate with a conserved double zinc finger domain, is localized to the cytoplasmic surface of early endosomes. J. Biol. Chem. 1997, 272, 20538–20544. [Google Scholar] [CrossRef]
- Kawabata, H. Transferrin and transferrin receptors update. Free Radic. Biol. Med. 2019, 133, 46–54. [Google Scholar] [CrossRef]
- Yin, X.; Yang, Y.; Han, K.L.; Wu, L.T.; Wu, H.R.; Bian, X.; Wei, X.F.; Guo, Z.; Mu, L.L.; Ye, J.M. Purification and functional characterization of serum transferrin from Nile tilapia (Oreochromis niloticus). Fish. Shellfish. Immunol. 2019, 88, 36–46. [Google Scholar] [CrossRef]
- Yin, X.; Mu, L.; Wu, H.; Han, K.L.; Guo, Z.; Ye, J.M. Expression and functional analysis of Nile tilapia transferrin receptors (TfRs) in host resistance to pathogenic bacteria and iron ion metabolism. Fish. Shellfish. Immunol. 2020, 100, 407–417. [Google Scholar] [CrossRef]
- Zulkefli, K.L.; Houghton, F.J.; Gosavi, P.; Gleeson, P.A. A role for Rab11 in the homeostasis of the endosome-lysosomal pathway. Exp. Cell Res. 2019, 380, 55–68. [Google Scholar] [CrossRef]
- Kim, J.H.; Park, H.J.; Kim, K.W.; Hwang, I.K.; Kim, D.H.; Oh, C.W.; Lee, S.J.; Kang, J.C. Growth performance, oxidative stress, and non-specific immune responses in juvenile sablefish, Anoplopoma fimbria, by changes of water temperature and salinity. Fish. Physiol. Biochem. 2017, 43, 1421–1431. [Google Scholar] [CrossRef] [PubMed]
- Collazos, M.E.; Barriga, C.; Ortega, E. Enhanced granulocyte phagocytosis at low winter temperature and high summer temperature in the tench (Tinca tinca L.). Comp. Biochem. Physiol. Physiol. 1994, 109, 643–648. [Google Scholar] [CrossRef]
- Morvan, C.; Clerton, P.; Deschaux, P.; Troutaud, D. Effects of environmental temeperature on macrophage activities in carp. Fish. Shellfish. Immunol. 1997, 7, 209–212. [Google Scholar] [CrossRef]
- Li, M.F.; Zhang, H.Q. An overview of complement systems in teleosts. Dev. Comp. Immunol. 2022, 137, 104520. [Google Scholar] [CrossRef]
- Bubeck, D. The making of a macromolecular machine: Assembly of the membrane attack complex. Biochemistry 2014, 53, 1908–1915. [Google Scholar] [CrossRef] [PubMed]
- Uemura, T.; Yano, T.; Shiraishi, H.; Nakao, M. Purification and characterization of the eighth and ninth components of carp complement. Mol. Immunol. 1996, 33, 925–932. [Google Scholar] [CrossRef]
- Hernández, A.; Tort, L. Annual variation of complement, lysozyme and haemagglutinin levels in serum of the gilthead sea bream Sparus aurata. Fish. Shellfish. Immunol. 2003, 15, 479–481. [Google Scholar] [CrossRef]
- Wen, L.L.; Zhao, M.; Chi, H.; Sun, L. Histones and chymotrypsin-like elastases play significant roles in the antimicrobial activity of tongue sole neutrophil extracellular traps. Fish. Shellfish. Immunol. 2018, 72, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Gu, H.J.; Huang, H.Q.; Wang, H.Y.; Xia, Z.H.; Hu, Y.H. Characterization, expression, and antimicrobial activity of histones from Japanese flounder Paralichthys olivaceus. Fish. Shellfish. Immunol. 2020, 96, 235–244. [Google Scholar] [CrossRef]
- Brinkmann, V.; Reichard, U.; Goosmann, C.; Fauler, B.; Uhlemann, Y.; Weiss, D.S.; Weinrauch, Y.; Zychlinsky, A. Neutrophil extracellular traps kill bacteria. Science 2004, 303, 1532–1535. [Google Scholar] [CrossRef]
- Lee, D.Y.; Huang, C.M.; Nakatsuji, T.; Thiboutot, D.; Kang, S.A.; Monestier, M.; Gallo, R.L. Histone H4 is a major component of the antimicrobial action of human sebocytes. J. Investig. Dermatol. 2009, 129, 2489–2496. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Y.; Xin, L.; Beverley, S.M.; Carlsen, E.D.; Popov, V.; Chang, K.P.; Wang, M.; Soong, L. Differential microbicidal effects of human histone proteins H2A and H2B on Leishmania promastigotes and amastigotes. Infect. Immun. 2011, 79, 1124–1133. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, T.U. The structure inventory of the nuclear pore complex. J. Mol. Biol. 2016, 428, 1986–2000. [Google Scholar] [CrossRef]
- Kim, Y.H.; Han, M.E.; Oh, S.O. The molecular mechanism for nuclear transport and its application. Anat. Cell Biol. 2017, 50, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Takeda, A.; Sarma, N.J.; Abdul-Nabi, A.M.; Yaseen, N.R. Inhibition of CRM1-mediated nuclear export of transcription factors by leukemogenic NUP98 fusion proteins. J. Biol. Chem. 2010, 285, 16248–16257. [Google Scholar] [CrossRef] [PubMed]
- Hutten, S.; Kehlenbach, R.H. Nup214 is required for CRM1-dependent nuclear protein export in vivo. Mol. Cell Biol. 2006, 26, 6772–6785. [Google Scholar] [CrossRef]
- Boehmer, T.; Enninga, J.; Dales, S.; Blobel, G.; Zhong, H. Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proc. Natl. Acad. Sci. USA 2003, 100, 981–985. [Google Scholar] [CrossRef]
- Guan, Y.; Gao, X.; Tang, Q.; Huang, L.; Gao, S.; Yu, S.; Huang, J.; Li, J.; Zhou, D.; Zhang, Y.; et al. Nucleoporin 107 facilitates the nuclear export of Scn5a mRNA to regulate cardiac bioelectricity. J. Cell Mol. Med. 2019, 23, 1448–1457. [Google Scholar] [CrossRef]
- Fan, J.; Zhou, X.; Wang, Y.; Kuang, C.; Sun, Y.; Liu, X.; Toomre, D.; Xu, Y. Differential requirement for N-ethylmaleimide-sensitive factor in endosomal trafficking of transferrin receptor from anterograde trafficking of vesicular stomatitis virus glycoprotein G. FEBS Lett. 2017, 591, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Wei, Q.; Jiang, X.; Chen, N.; Zuo, X.; Zhao, H.; Liu, Y.; Liu, X.; Xie, L.; Yang, Y.; et al. CSTF3 contributes to platinum resistance in ovarian cancer through alternative polyadenylation of lncRNA NEAT1 and generating the short isoform NEAT11. Cell Death Dis. 2024, 15, 432. [Google Scholar] [CrossRef]
- Rahman, M.M.; Estifanos, B.; Glenn, H.L.; Gutierrez-Jensen, A.D.; Kibler, K.; Li, Y.; Jacobs, B.; McFadden, G.; Hogue, B.G. Effect of Exportin 1/XPO1 Nuclear Export Pathway Inhibition on Coronavirus Replication. Viruses 2025, 17, 284. [Google Scholar] [CrossRef]
- Li, M.F.; Sun, L.; Li, J. Edwardsiella tarda evades serum killing by preventing complement activation via the alternative pathway. Fish. Shellfish. Immunol. 2015, 43, 325–329. [Google Scholar] [CrossRef]
- Zhou, Z.J.; Sun, L. Edwardsiella tarda-induced inhibition of apoptosis: A strategy for intracellular survival. Front. Cell Infect. Microbiol. 2016, 6, 76. [Google Scholar] [CrossRef] [PubMed]
- Strauss, E.J.; Ghori, N.; Falkow, S. An Edwardsiella tarda strain containing a mutation in a gene with homology to shlB and hpmB is defective for entry into epithelial cells in culture. Infect. Immun. 1997, 65, 3924–3932. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Gene ID | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|---|
| ELAVL1 | XM_020107316.1 | GATGTTTGGGCCGTTTGGTG | CCCAGTCGATACCCGTTCAG |
| YKT6 | XM_020099185.1 | ATCTCGTGGCAAAGTCGGAA | AGCAGAAAGGCAGAGACGAG |
| SORD | XM_020080198.1 | TTGGCAGATTGGTAGTGAGTCT | AACCCGATAGGCCCTGAAGA |
| THOC2 | XM_020085721.1 | AATGAGGAAACCCCCACGTC | CACAACCATGACCAGCTTGC |
| HGS | XM_020094525.1 | GGCCCGTTACCTGAACAGAA | ACTGCTGCTCCACTATGCTG |
| TFRC | XM_020105708.1 | GCAGCTCTGTCCAACCATCT | ACTGGCTGTCGGTGAATTGT |
| C8A | XM_020099517.2 | ACGTGCAAGTGGGACTCAAA | GCGGGAGTACATCCCAAAGT |
| NUP107 | XM_020112821.1 | ATCGACTGGCTGCTGTTTGA | TCATCGAGTCCTCCGGAACT |
| NUP214 | XM_020081961.1 | AGGGTCTTTTCACAACACTGG | TCACTGGTGAGATCTTGACTG |
| H3 | XM_020109331.1 | GTGGTTCAGTTGTGCGTCC | CAGGTTGGCGTCTGAGAAC |
| macroH2A1 | XM_020086110.1 | CCCGACCAACTCCTCCATCTA | TGTGGCAGTGAATGACGTACT |
| TRIP12 | XM_020095042.1 | GCCCCTCAGCTGATGTGAAA | TGGAATTAGGCCGGTTGGAC |
| PWP1 | XM_020092036.1 | TCTGCTGCCAGCTTATCCAC | AGGAGTCATGTTGCCCACAG |
| UBA5 | XM_020087372.1 | CTCCAACCCGTACAGTCGTC | AGCAGCTTACCAATGCCACA |
| RAC1 | XM_020104358.1 | TGGGGCTAAGTAAACGCTGT | GGTGCAGCCTCAACAATCAGT |
| PRKAA1 | XM_020082451.1 | CAAAAGTCCACGTCCCACCT | CAAAAGTGCTTCCTGCGTCC |
| α-Tubulin | XM_020111916.1 | TGACATCACAAACGCCTGCTTC | GCACCACATCTCCACGGTACAG |
| HI-HC Group | LI-LC Group | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein ID | Name | PPI Degree | Fold Change | p-Value | Regulation | Protein ID | Name | PPI Degree | Fold Change | p-Value | Regulation |
| XP_019954006.1 | UBA52 | 32 | +∞ | NA | Up | XP_019964890.1 | H3 | 12 | +∞ | NA | Up |
| XP_019953060.1 | ALDH18A1 | 11 | +∞ | NA | Up | XP_019966822.1 | XPO1 | 11 | +∞ | NA | Up |
| XP_019938172.1 | RAD23B | 11 | −2.12 | 1.46 × 10−3 | Down | XP_019941669.1 | macroH2A1 | 11 | +∞ | NA | Up |
| XP_019962875.1 | ELAVL1 | 10 | +∞ | NA | Up | XP_019968380.1 | NUP107 | 11 | −∞ | NA | Down |
| XP_019954477.1 | CSTF3 | 10 | +∞ | NA | Up | XP_019950601.1 | TRIP12 | 10 | +∞ | NA | Up |
| XP_019935757.1 | SORD | 10 | +∞ | NA | Up | XP_019937520.1 | NUP214 | 10 | −∞ | NA | Down |
| XP_019966822.1 | XPO1 | 9 | −∞ | NA | Down | XP_019959917.1 | RAC1 | 10 | +∞ | NA | Up |
| XP_019954744.1 | YKT6 | 9 | +∞ | NA | Up | XP_019954477.1 | CSTF3 | 9 | +∞ | NA | Up |
| XP_019933706.1 | NSF | 9 | +∞ | NA | Up | XP_019954372.1 | DHX38 | 9 | +∞ | NA | Up |
| XP_019950084.1 | HGS | 8 | +∞ | NA | Up | XP_019947595.1 | PWP1 | 9 | +∞ | NA | Up |
| XP_019935975.1 | CHD1L | 8 | −∞ | NA | Down | XP_019954034.1 | POLR2H | 9 | −2.06 | 2.10 × 10−2 | Down |
| XP_019941280.1 | THOC2 | 8 | +2.07 | 2.81 × 10−2 | Up | XP_019969351.1 | WDR12 | 9 | −∞ | NA | Down |
| XP_019964808.1 | AGL | 7 | +∞ | NA | Up | XP_019942931.1 | UBA5 | 8 | −∞ | NA | Down |
| XP_019958157.1 | EXOC2 | 7 | +∞ | NA | Up | XP_019938010.1 | PRKAA1 | 8 | −∞ | NA | Down |
| XP_019969379.1 | KPNB1 | 7 | −2.28 | 3.54 × 10−2 | Down | XP_019933706.1 | NSF | 8 | +∞ | NA | Up |
| XP_019940542.1 | ROCK2 | 7 | +∞ | NA | Up | XP_019966497.1 | NUP98 | 7 | +∞ | NA | Up |
| XP_019961267.1 | TFRC | 7 | +∞ | NA | Up | XP_019963564.1 | CTNNBL1 | 7 | +∞ | NA | Up |
| XP_019936097.1 | ATP5D | 6 | +∞ | NA | Up | XP_019954792.1 | GLDC | 7 | +2.84 | 1.20 × 10−2 | Up |
| XP_019935808.1 | CUL4A | 6 | −∞ | NA | Down | XP_019957398.1 | SUN1 | 7 | −∞ | NA | Down |
| XP_019963351.1 | ALDH3B1 | 6 | +∞ | NA | Up | XP_019953698.1 | STAG1 | 6 | +∞ | NA | Up |
| XP_019937551.1 | NUP188 | 6 | −∞ | NA | Down | XP_019968563.1 | NOB1 | 6 | −∞ | NA | Down |
| XP_019946175.1 | TOP2B | 6 | +∞ | NA | Up | XP_019937802.1 | PIK3R1 | 6 | +∞ | NA | Up |
| XP_019965190.1 | RAB11B | 6 | +∞ | NA | Up | XP_019941399.1 | SEC24A | 6 | +∞ | NA | Up |
| XP_019951363.1 | PLAA | 6 | −∞ | NA | Down | XP_019965029.1 | TSFM | 6 | −∞ | NA | Down |
| XP_019955076.1 | C8A | 6 | +∞ | NA | Up | XP_019961505.1 | HSPBP1 | 6 | +∞ | NA | Up |
| XP_019964134.1 | CRNKL1 | 6 | +∞ | NA | Up | XP_019966512.1 | CUL1 | 6 | −∞ | NA | Down |
| XP_019965019.1 | GCK | 6 | +2.03 | 2.37 × 10−2 | Up | ||||||
| HI-HC Group (Special Protein) | LI-LC Group (Special Protein) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein ID | Name | PPI Degree | Fold Change | p-Value | Regulation | Protein ID | Name | PPI Degree | Fold Change | p-Value | Regulation |
| Endocytosis | Histone | ||||||||||
| XP_019954744.1 | YKT6 | 9 | +∞ | NA | Up | XP_019964890.1 | H3 | 12 | +∞ | NA | Up |
| XP_019950084.1 | HGS | 8 | +∞ | NA | Up | XP_019941669.1 | macroH2A1 | 11 | +∞ | NA | Up |
| XP_019961267.1 | TFRC | 7 | +∞ | NA | Up | RNA transport | |||||
| XP_019965190.1 | RAB11B | 6 | +∞ | NA | Up | XP_019968380.1 | NUP107 | 11 | −∞ | NA | Down |
| Complement | XP_019937520.1 | NUP214 | 10 | −∞ | NA | Down | |||||
| XP_019955076.1 | C8A | 6 | +∞ | NA | Up | XP_019966497.1 | NUP98 | 7 | +∞ | NA | Up |
| Other proteins | Other proteins | ||||||||||
| XP_019962875.1 | ELAVL1 | 10 | +∞ | NA | Up | XP_019959917.1 | RAC1 | 10 | +∞ | NA | Up |
| XP_019940542.1 | ROCK2 | 7 | +∞ | NA | Up | XP_019954034.1 | POLR2H | 9 | −2.06 | 2.10 × 10−2 | Down |
| XP_019958157.1 | EXOC2 | 7 | +∞ | NA | Up | XP_019938010.1 | PRKAA1 | 8 | −∞ | NA | Down |
| XP_019937802.1 | PIK3R1 | 6 | +∞ | NA | Up | ||||||
| Shared protein | |||||||||||
| HI-HC group | LI-LC group | ||||||||||
| Protein ID | Name | PPI degree | Fold change | p-value | Regulation | Protein ID | Name | PPI degree | Fold change | p-value | Regulation |
| XP_019933706.1 | NSF | 9 | +∞ | NA | Up | XP_019933706.1 | NSF | 8 | +∞ | NA | Up |
| XP_019954477.1 | CSTF3 | 10 | +∞ | NA | Up | XP_019954477.1 | CSTF3 | 9 | +∞ | NA | Up |
| XP_019966822.1 | XPO1 | 9 | −∞ | NA | Down | XP_019966822.1 | XPO1 | 11 | +∞ | NA | Up |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Luo, L.; Zhang, B.; Zhou, X.; Zhang, K.; Zhang, P.; Sun, B. Comparative Proteomics Analysis Reveals Differential Immune Responses of Paralichthys olivaceus to Edwardsiella tarda Infection Under High and Low Temperature. Biology 2025, 14, 1417. https://doi.org/10.3390/biology14101417
Chen X, Luo L, Zhang B, Zhou X, Zhang K, Zhang P, Sun B. Comparative Proteomics Analysis Reveals Differential Immune Responses of Paralichthys olivaceus to Edwardsiella tarda Infection Under High and Low Temperature. Biology. 2025; 14(10):1417. https://doi.org/10.3390/biology14101417
Chicago/Turabian StyleChen, Xiaojuan, Lejia Luo, Beibei Zhang, Xiaowei Zhou, Kaipeng Zhang, Panpan Zhang, and Bin Sun. 2025. "Comparative Proteomics Analysis Reveals Differential Immune Responses of Paralichthys olivaceus to Edwardsiella tarda Infection Under High and Low Temperature" Biology 14, no. 10: 1417. https://doi.org/10.3390/biology14101417
APA StyleChen, X., Luo, L., Zhang, B., Zhou, X., Zhang, K., Zhang, P., & Sun, B. (2025). Comparative Proteomics Analysis Reveals Differential Immune Responses of Paralichthys olivaceus to Edwardsiella tarda Infection Under High and Low Temperature. Biology, 14(10), 1417. https://doi.org/10.3390/biology14101417








