Genome-Wide Thioredoxin System in Cardamine hupingshanensis: Role in Se Stress and Metabolism
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of ChTRX and ChTR Genes in C. hupingshanensis
2.2. Physicochemical Properties and Phylogenetic Analysis of ChTRXs and ChTRs
2.3. Chromosomal Localization and Domain Analysis of ChTRXs and ChTRs
2.4. Plant Materials and Sample Preparation
2.5. Gene Expression Analysis
2.6. Protein Modelling and Validation of ChTRX and ChTR
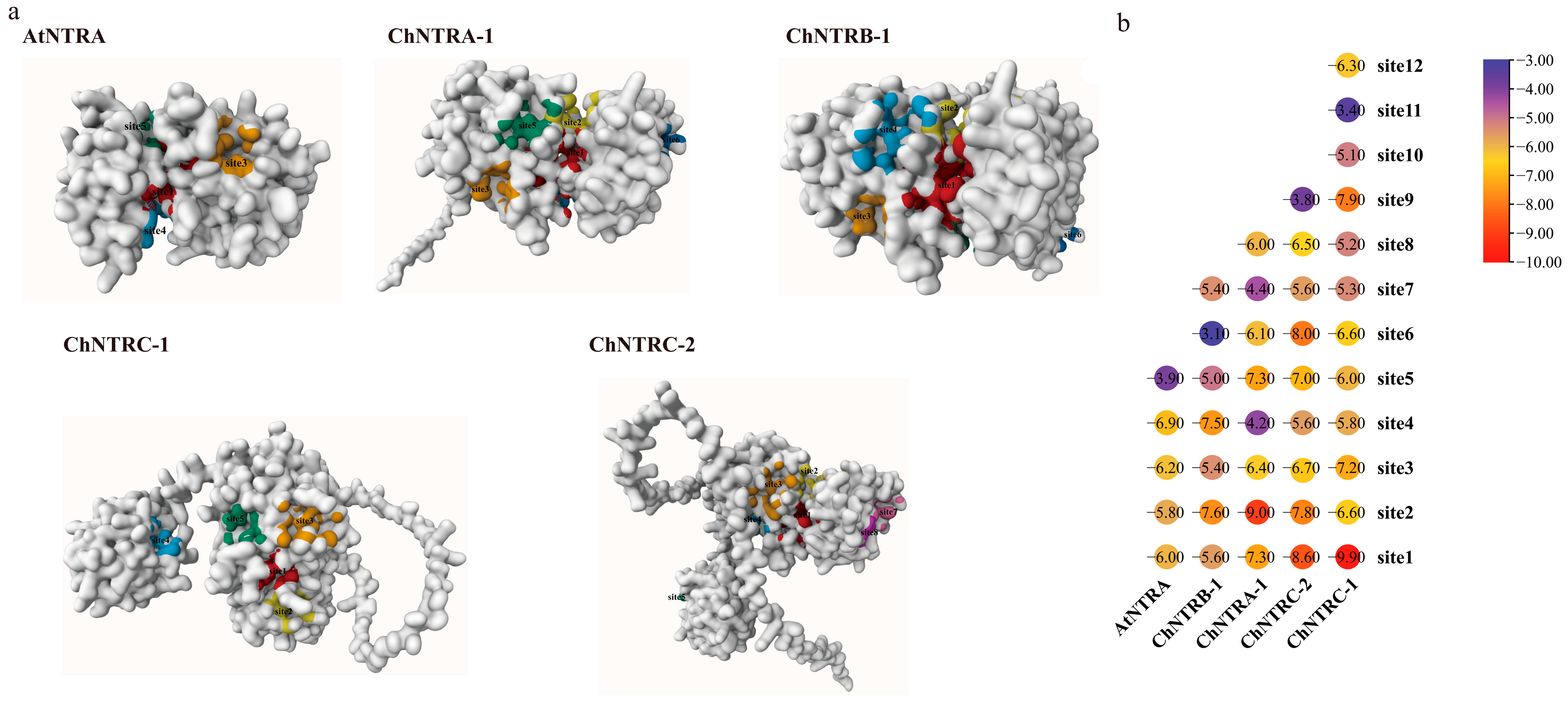
2.7. Ligand Preparation and Protein Docking
2.8. Statistical Analysis
3. Results
3.1. Identification and Analysis of ChTRX and ChTR Gene Families
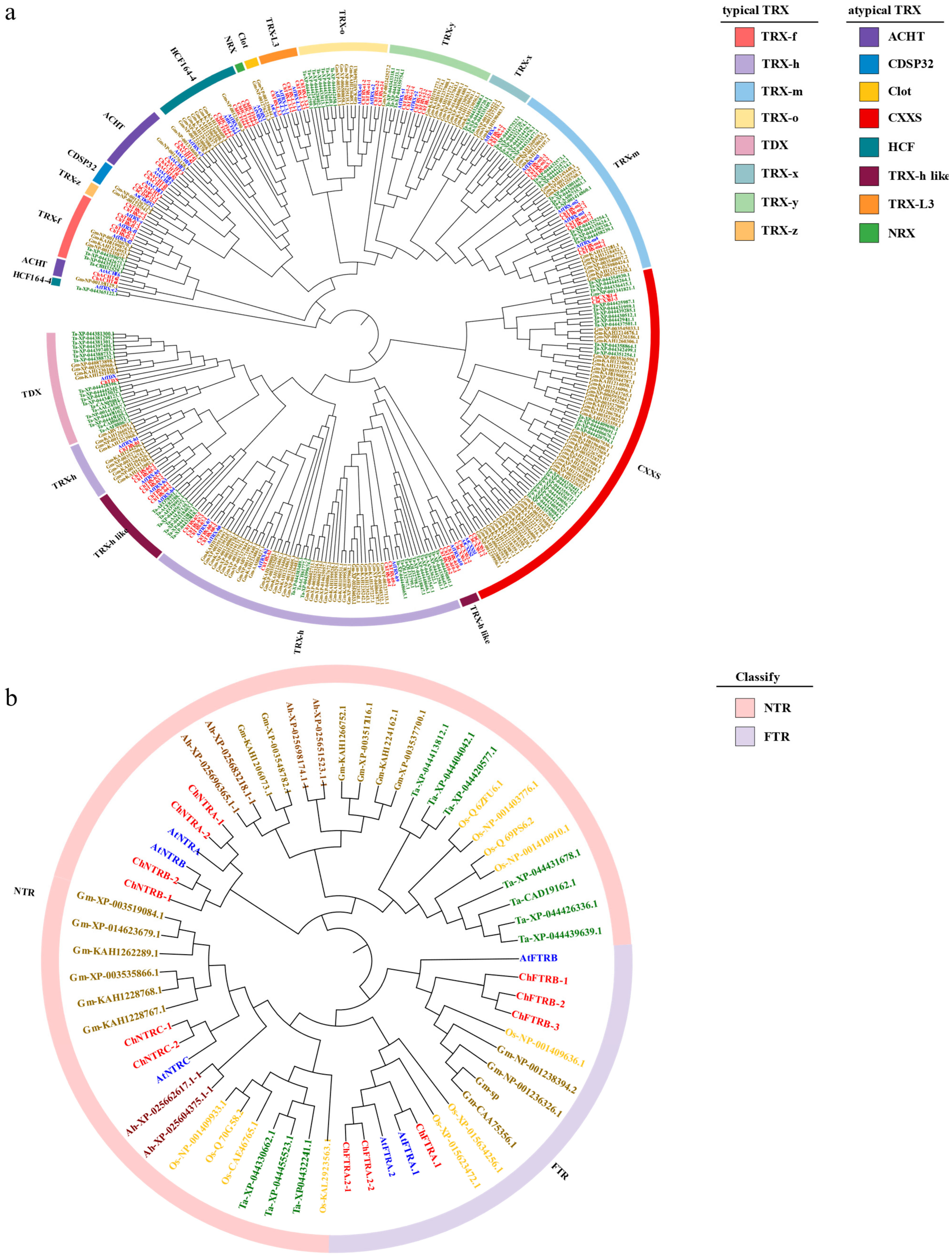
3.2. Phylogenetic Analysis of ChTRX and ChTR Gene Families
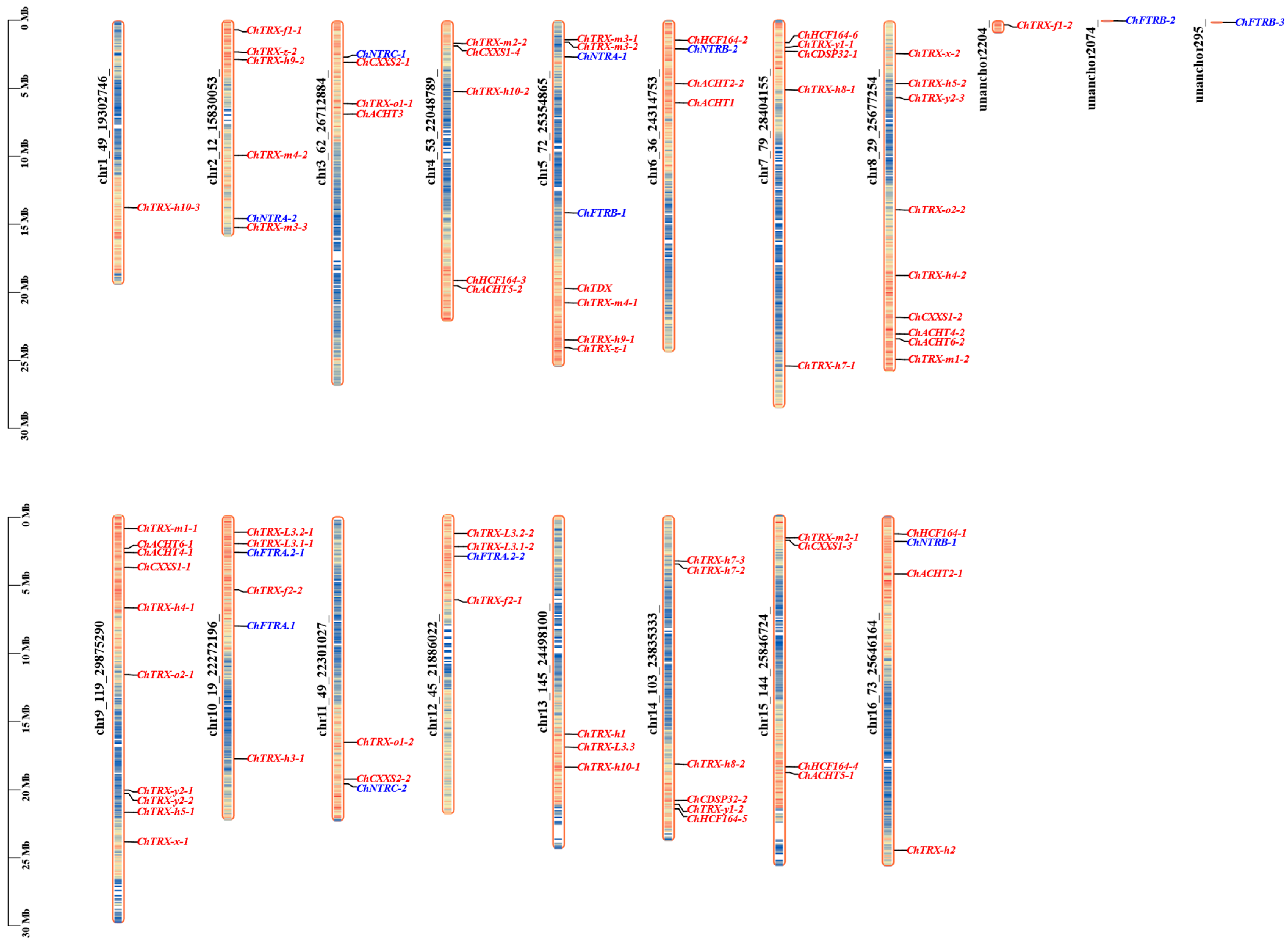
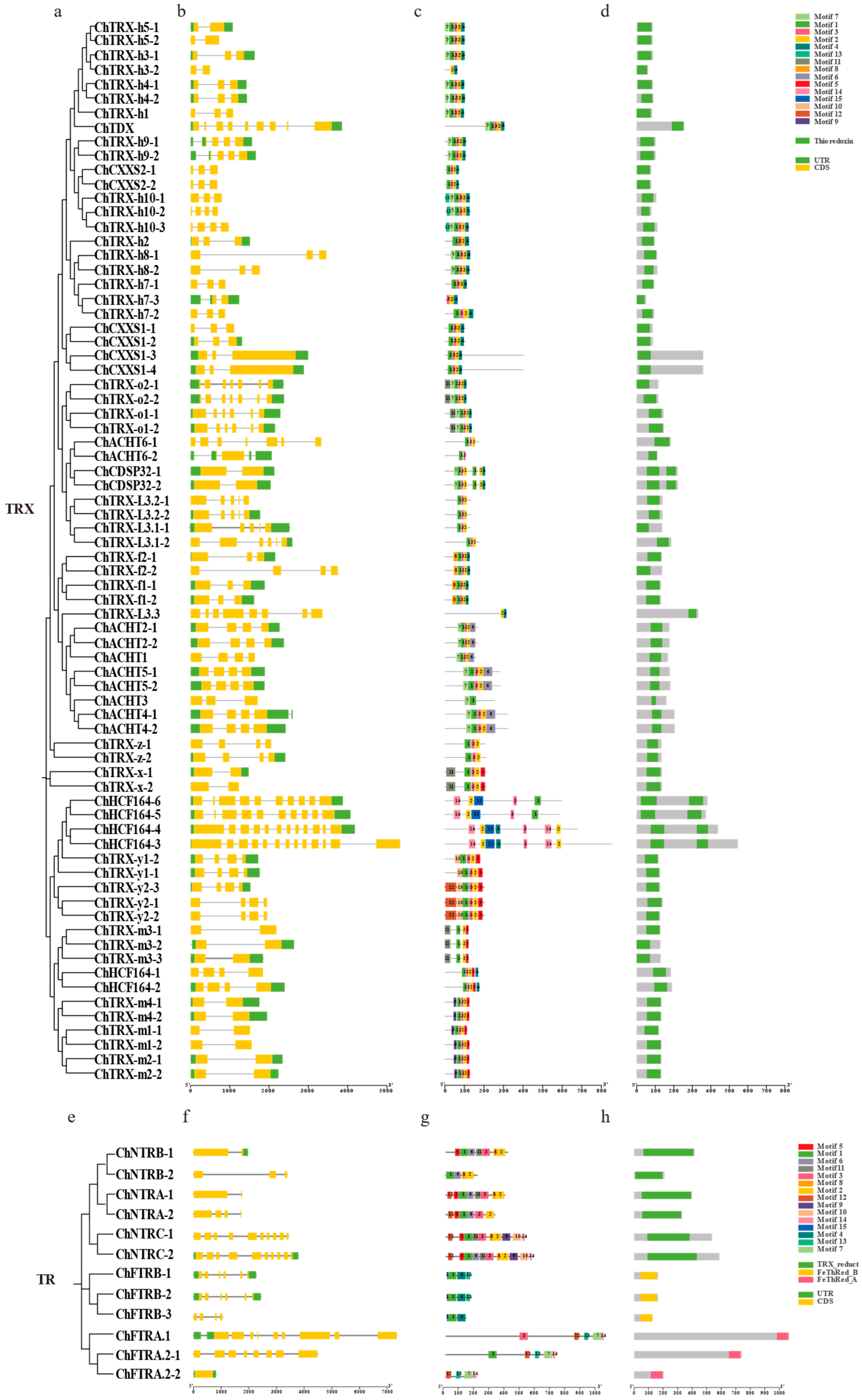
3.3. Chromosomal Localization and Domain Analysis of ChTRX and ChTR Gene Families
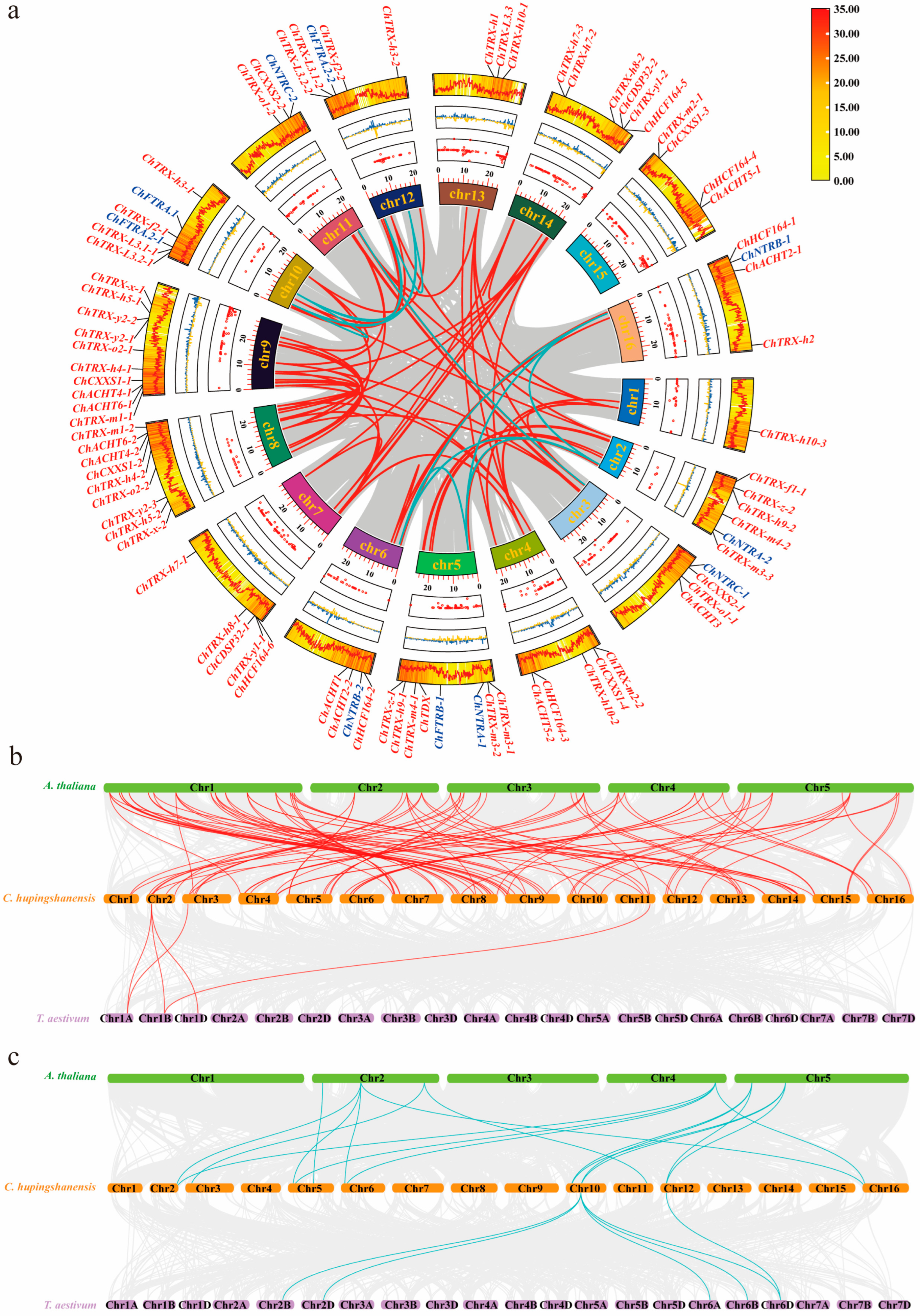
3.4. Synteny and Evolutionary Analysis of ChTRX and ChTR Gene Families
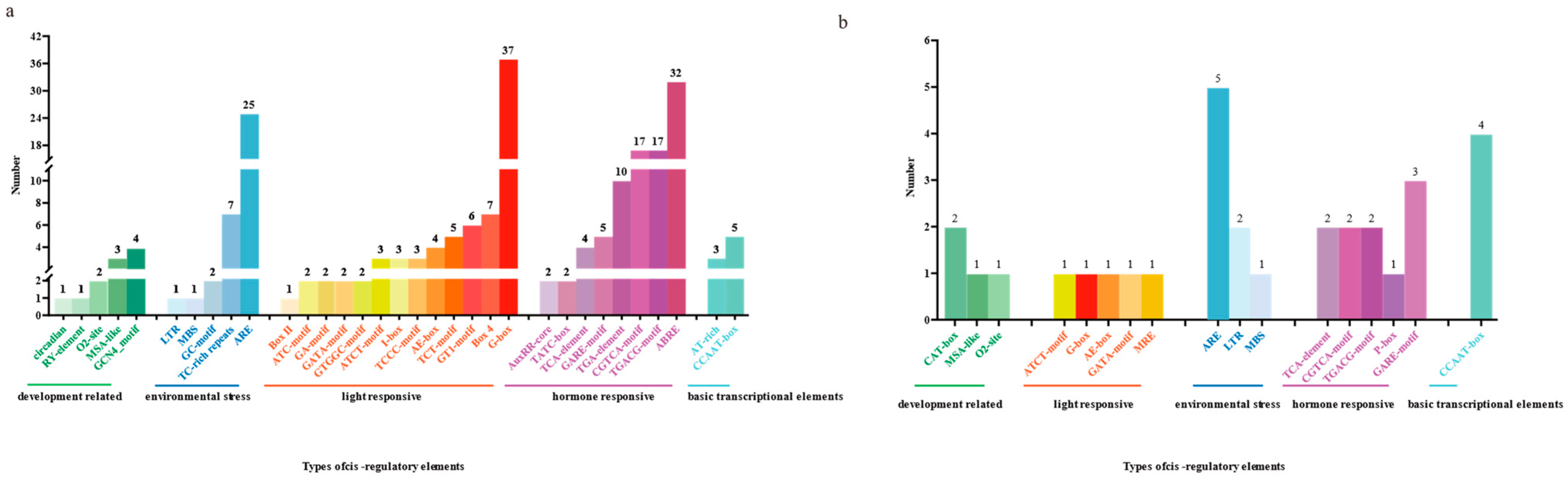
3.5. Analysis of Cis-Acting Elements in ChTRX and ChTR Gene Families
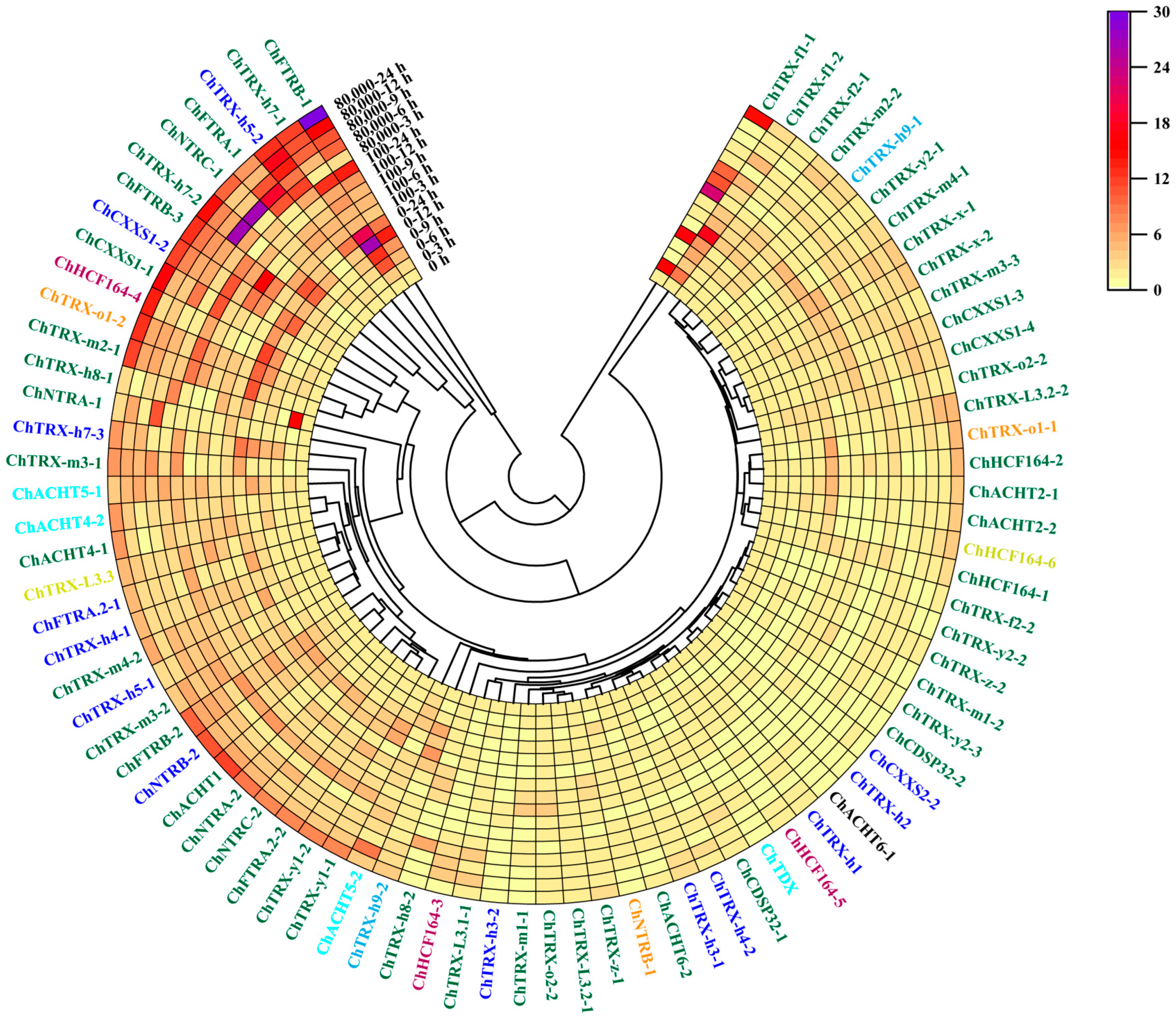
3.6. Expression Analysis of ChTRXs and ChTRs in Leaves Under Se Stress
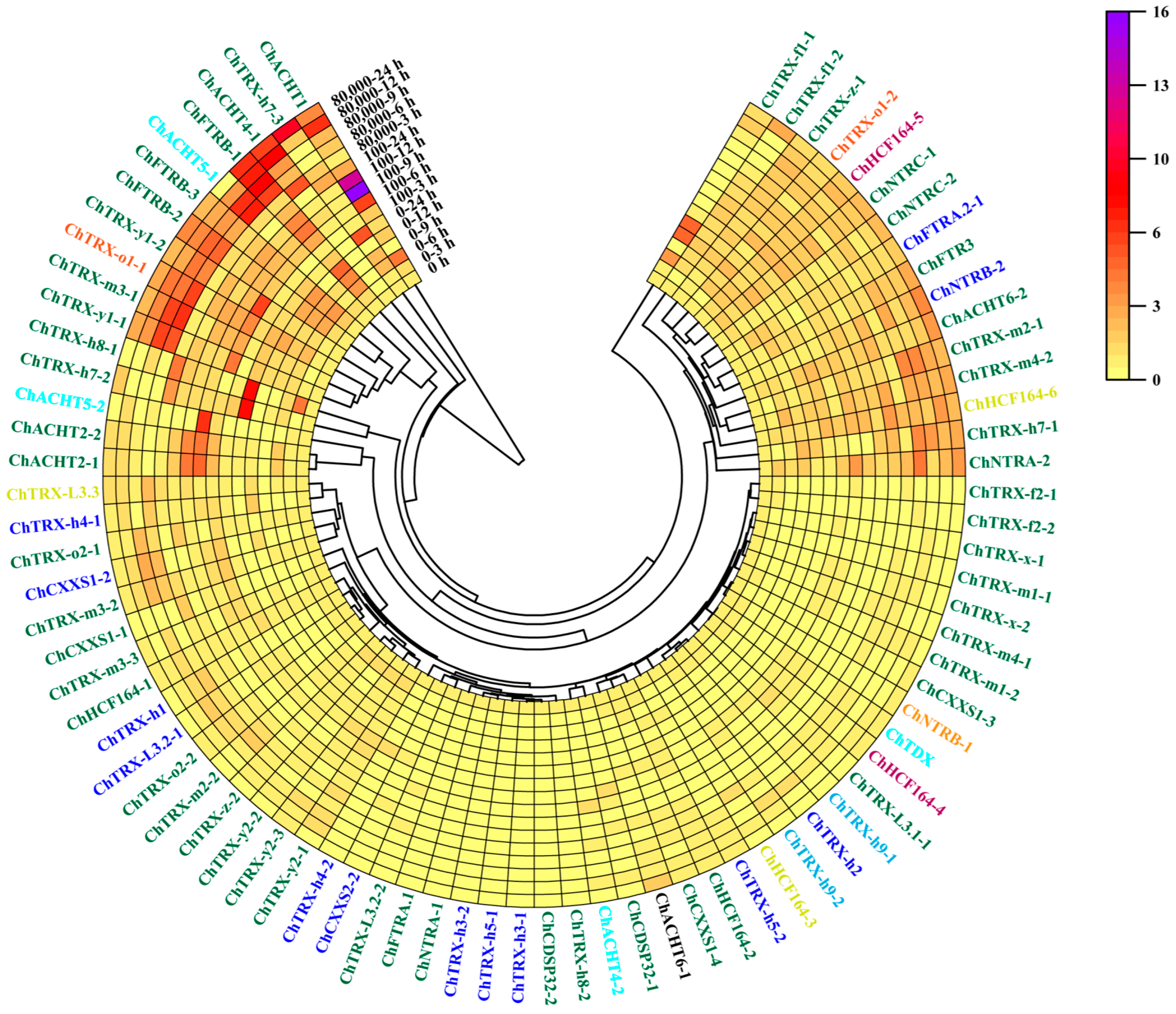
3.7. Expression Analysis of ChTRXs and ChTRs in Roots Under Se Stress
3.8. Secondary and Tertiary Structure Prediction of ChTRXs and ChTRs Proteins
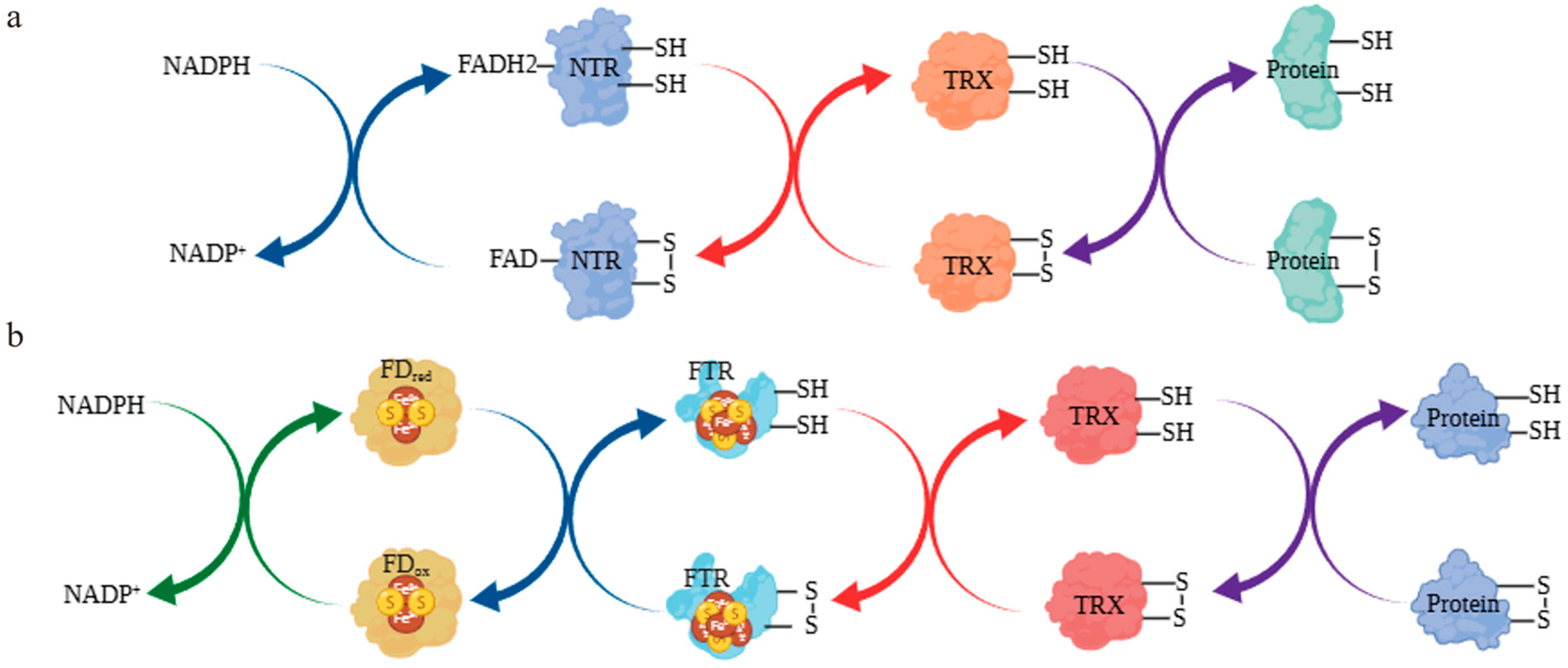
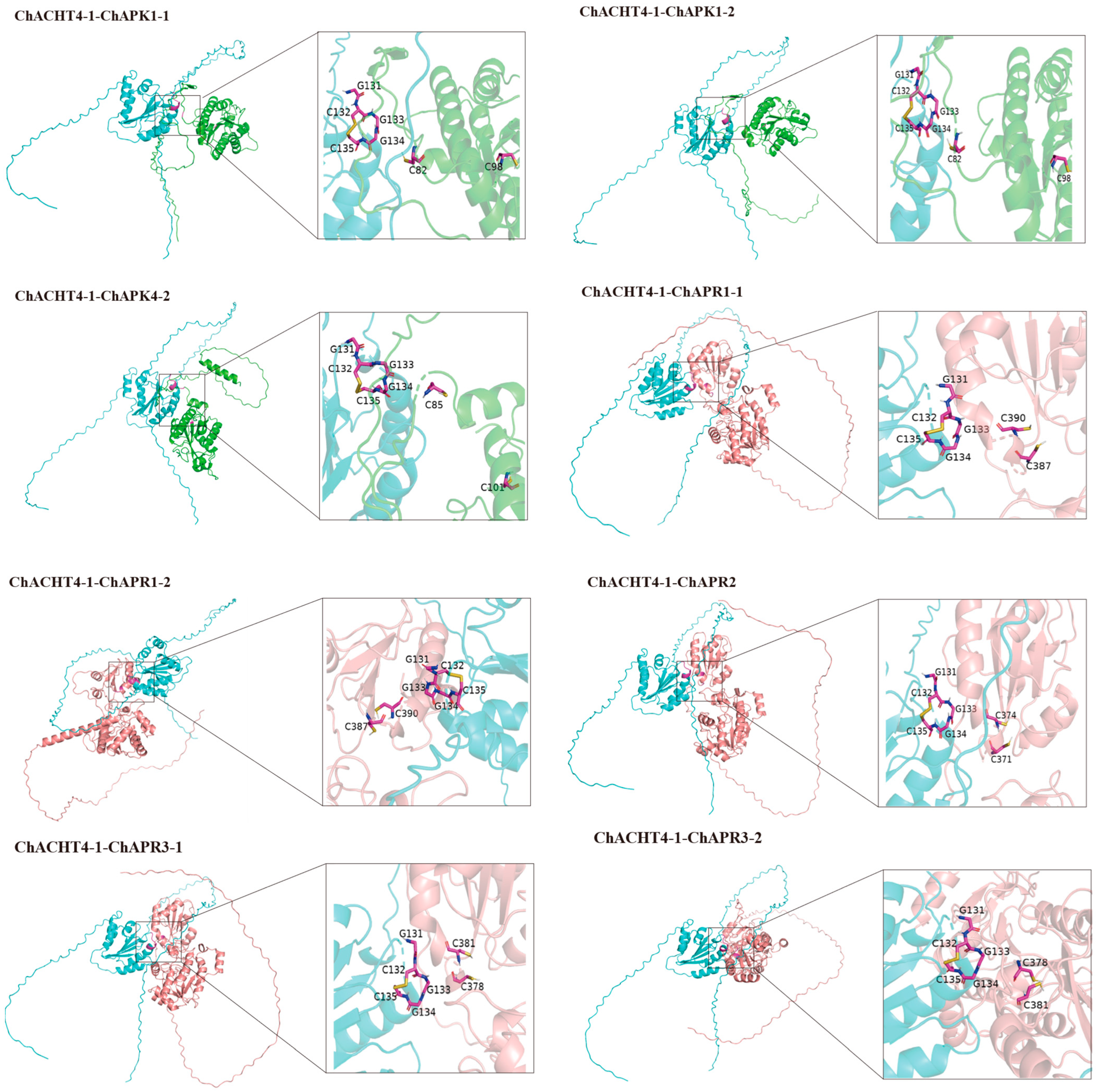
3.9. Simulation of the Thioredoxin System-Mediated Redox Regulation of the Se Metabolic Pathway ChAPK/ChAPR in C. hupingshanensis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gřešková, A.; Petřivalský, M. Thioredoxin System in Insects: Uncovering the Roles of Thioredoxins and Thioredoxin Reductase beyond the Antioxidant Defences. Insects 2024, 15, 797. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, A.; López-Martínez, R.; Martí, M.C.; Cano-Yelo, D.; Sevilla, F. The integration of TRX/GRX systems and phytohormonal signalling pathways in plant stress and development. Plant Physiol. Biochem. 2024, 207, 108298. [Google Scholar] [CrossRef]
- Chibani, K.; Saul, F.; Didierjean, C.; Rouhier, N.; Haouz, A. Structural snapshots along the reaction mechanism of the atypical poplar thioredoxin-like2.1. FEBS Lett. 2018, 592, 1030–1041. [Google Scholar] [CrossRef]
- Shaykholeslam Esfahani, E.; Shahpiri, A. Thioredoxin h isoforms from rice are differentially reduced by NADPH/thioredoxin or GSH/glutaredoxin systems. Int. J. Biol. Macromol. 2015, 74, 243–248. [Google Scholar] [CrossRef]
- Jacquot, J.P.; Lancelin, J.M.; Meyer, Y. Thioredoxins: Structure and function in plant cells. New Phytol. 1997, 136, 543–570. [Google Scholar] [CrossRef]
- Lee, S.R.; Bar-Noy, S.; Kwon, J.; Levine, R.L.; Stadtman, T.C.; Rhee, S.G. Mammalian thioredoxin reductase: Oxidation of the C-terminal cysteine/selenocysteine active site forms a thioselenide, and replacement of selenium with sulfur markedly reduces catalytic activity. Proc. Natl. Acad. Sci. USA 2000, 97, 2521–2526. [Google Scholar] [CrossRef]
- Gelhaye, E.; Rouhier, N.; Navrot, N.; Jacquot, J.P. The plant thioredoxin system. Cell. Mol. Life Sci. 2005, 62, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Madhu; Sharma, A.; Kaur, A.; Tyagi, S.; Upadhyay, S.K. Glutathione Peroxidases in Plants: Innumerable Role in Abiotic Stress Tolerance and Plant Development. J. Plant Growth Regul. 2023, 42, 598–613. [Google Scholar] [CrossRef]
- Guevara-Flores, A.; Nava-Balderas, G.; de Jesús Martínez-González, J.; Vásquez-Lima, C.; Rendón, J.L.; Del Arenal Mena, I.P. A Physiological Approach to Explore How Thioredoxin-Glutathione Reductase (TGR) and Peroxiredoxin (Prx) Eliminate H2O2 in Cysticerci of Taenia. Antioxidants 2024, 13, 444. [Google Scholar] [CrossRef] [PubMed]
- Peskin, A.V.; Meotti, F.C.; Kean, K.M.; Göbl, C.; Peixoto, A.S.; Pace, P.E.; Horne, C.R.; Heath, S.G.; Crowther, J.M.; Dobson, R.C.J.; et al. Modifying the resolving cysteine affects the structure and hydrogen peroxide reactivity of peroxiredoxin 2. J. Biol. Chem. 2021, 296, 100494. [Google Scholar] [CrossRef]
- Matsuzawa, A. Thioredoxin and redox signaling: Roles of the thioredoxin system in control of cell fate. Arch. Biochem. Biophys. 2017, 617, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Fiorentino, G.; Contursi, P.; Gallo, G.; Bartolucci, S.; Limauro, D. A peroxiredoxin of Thermus thermophilus HB27: Biochemical characterization of a new player in the antioxidant defence. Int. J. Biol. Macromol. 2020, 153, 608–615. [Google Scholar] [CrossRef]
- Souza, P.V.L. The redox power of thioredoxins: Evolution, mechanisms, and redox regulation of carbon–nitrogen metabolism in plants. Discov. Plants 2025, 2, 94. [Google Scholar] [CrossRef]
- Yokochi, Y.; Fukushi, Y.; Wakabayashi, K.I.; Yoshida, K.; Hisabori, T. Oxidative regulation of chloroplast enzymes by thioredoxin and thioredoxin-like proteins in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2021, 118, e2114952118. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, Y.; Kozaki, A.; Hatano, M. Link between light and fatty acid synthesis: Thioredoxin-linked reductive activation of plastidic acetyl-CoA carboxylase. Proc. Natl. Acad. Sci. USA 1997, 94, 11096–11101. [Google Scholar] [CrossRef]
- Hernández, M.L.; Jiménez-López, J.; Cejudo, F.J.; Pérez-Ruiz, J.M. 2-Cys peroxiredoxins contribute to thylakoid lipid unsaturation by affecting ω-3 fatty acid desaturase 8. Plant Physiol. 2024, 195, 1521–1535. [Google Scholar] [CrossRef] [PubMed]
- Stancill, J.S.; Corbett, J.A. The Role of Thioredoxin/Peroxiredoxin in the β-Cell Defense Against Oxidative Damage. Front. Endocrinol. 2021, 12, 718235. [Google Scholar] [CrossRef]
- Smiriglia, A.; Lorito, N.; Bacci, M.; Subbiani, A.; Bonechi, F.; Comito, G.; Kowalik, M.A.; Perra, A.; Morandi, A. Estrogen-dependent activation of TRX2 reverses oxidative stress and metabolic dysfunction associated with steatotic disease. Cell Death Dis. 2025, 16, 57. [Google Scholar] [CrossRef]
- Telman, W.; Dietz, K.J. Thiol redox-regulation for efficient adjustment of sulfur metabolism in acclimation to abiotic stress. J. Exp. Bot. 2019, 70, 4223–4236. [Google Scholar] [CrossRef]
- Maruyama-Nakashita, A.; Watanabe-Takahashi, A.; Inoue, E.; Yamaya, T.; Saito, K.; Takahashi, H. Sulfur-Responsive Elements in the 3′-Nontranscribed Intergenic Region Are Essential for the Induction of SULFATE TRANSPORTER 2;1 Gene Expression in Arabidopsis Roots under Sulfur Deficiency. Plant Cell 2015, 27, 1279–1296. [Google Scholar] [CrossRef]
- Maruyama-Nakashita, A.; Nakamura, Y.; Tohge, T.; Saito, K.; Takahashi, H. Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 2006, 18, 3235–3251. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, D.; Peffer, S.; Morano, K.A. Cytoplasmic redox imbalance in the thioredoxin system activates Hsf1 and results in hyperaccumulation of the sequestrase Hsp42 with misfolded proteins. bioRxiv 2023. [Google Scholar] [CrossRef]
- Wei, X.; Zhu, Y.; Xie, W.; Ren, W.; Zhang, Y.; Zhang, H.; Dai, S.; Huang, C.F. H2O2 negatively regulates aluminum resistance via oxidation and degradation of the transcription factor STOP1. Plant Cell 2024, 36, 688–708. [Google Scholar] [CrossRef]
- Tada, Y.; Spoel, S.H.; Pajerowska-Mukhtar, K.; Mou, Z.; Song, J.; Wang, C.; Zuo, J.; Dong, X. Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 2008, 321, 952–956. [Google Scholar] [CrossRef]
- Zhang, F.; Gao, Y.; Ma, M.; Li, L.; Wei, Y.; Fan, L.; Xie, Z.; Qi, K.; Wu, J.; Tao, S.; et al. PbNAC3 coordinates AsA generation and ABA biosynthesis to improve salt tolerance in pear. Plant J. 2025, 122, e70171. [Google Scholar] [CrossRef] [PubMed]
- Rayman, M.P. Selenium and human health. Lancet 2012, 379, 1256–1268. [Google Scholar] [CrossRef] [PubMed]
- Barchielli, G.; Capperucci, A.; Tanini, D. The Role of Selenium in Pathologies: An Updated Review. Antioxidants 2022, 11, 251. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.; Thounaojam, T.C.; Chowdhury, D.; Upadhyaya, H. The role of selenium and nano selenium on physiological responses in plant: A review. Plant Growth Regul. 2023, 100, 409–433. [Google Scholar] [CrossRef]
- Gui, J.Y.; Rao, S.; Huang, X.; Liu, X.; Cheng, S.; Xu, F. Interaction between selenium and essential micronutrient elements in plants: A systematic review. Sci. Total Environ. 2022, 853, 158673. [Google Scholar] [CrossRef]
- Cui, L.; Zhao, J.; Chen, J.; Zhang, W.; Gao, Y.; Li, B.; Li, Y.-F. Translocation and transformation of selenium in hyperaccumulator plant Cardamine enshiensis from Enshi, Hubei, China. Plant Soil 2018, 425, 577–588. [Google Scholar] [CrossRef]
- Valdez Barillas, J.R.; Quinn, C.F.; Freeman, J.L.; Lindblom, S.D.; Fakra, S.C.; Marcus, M.A.; Gilligan, T.M.; Alford, É.R.; Wangeline, A.L.; Pilon-Smits, E.A. Selenium distribution and speciation in the hyperaccumulator Astragalus bisulcatus and associated ecological partners. Plant Physiol. 2012, 159, 1834–1844. [Google Scholar] [CrossRef]
- Cappa, J.J.; Cappa, P.J.; El Mehdawi, A.F.; McAleer, J.M.; Simmons, M.P.; Pilon-Smits, E.A. Characterization of selenium and sulfur accumulation across the genus Stanleya (Brassicaceae): A field survey and common-garden experiment. Am. J. Bot. 2014, 101, 830–839. [Google Scholar] [CrossRef]
- Tangjaidee, P.; Swedlund, P.; Xiang, J.; Yin, H.; Quek, S.Y. Selenium-enriched plant foods: Selenium accumulation, speciation, and health functionality. Front. Nutr. 2022, 9, 962312. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Bhuyan, M.; Raza, A.; Hawrylak-Nowak, B.; Matraszek-Gawron, R.; Nahar, K.; Fujita, M. Selenium Toxicity in Plants and Environment: Biogeochemistry and Remediation Possibilities. Plants 2020, 9, 1711. [Google Scholar] [CrossRef]
- Chen, Y.; Li, Y.; Luo, G.; Luo, C.; Xiao, Z.; Lu, Y.; Xiang, Z.; Hou, Z.; Xiao, Q.; Zhou, Y.; et al. Gene identification, expression analysis, and molecular docking of SAT and OASTL in the metabolic pathway of selenium in Cardamine hupingshanensis. Plant Cell Rep. 2024, 43, 148. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Yuan, Q.; Chen, A.; Zeng, X.; Lu, Y.; Xiang, Z.; Hou, Z.; Tang, Q.; Zhou, Y. Sulfotransferases in Cardamine hupingshanensis: Identification, expression, and regulatory role in selenium metabolism under selenium stress. Plant Sci. 2025, 359, 112670. [Google Scholar] [CrossRef]
- Li, J.; Huang, C.; Lai, L.; Wang, L.; Li, M.; Tan, Y.; Zhang, T. Selenium hyperaccumulator plant Cardamine enshiensis: From discovery to application. Environ. Geochem. Health 2023, 45, 5515–5529. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Zhou, Y.; Tang, Q.; Wu, M.; Mou, D.; Liu, H.; Wang, S.; Zhang, C.; Ding, L.; Luo, J. Comparative transcriptomics provides novel insights into the mechanisms of selenium tolerance in the hyperaccumulator plant Cardamine hupingshanensis. Sci. Rep. 2018, 8, 2789. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhang, Y.; Skolnick, J. Scoring function for automated assessment of protein structure template quality. Proteins 2004, 57, 702–710. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, Y. How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics 2010, 26, 889–895. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Chibani, K.; Pucker, B.; Dietz, K.J.; Cavanagh, A. Genome-wide analysis and transcriptional regulation of the typical and atypical thioredoxins in Arabidopsis thaliana. FEBS Lett. 2021, 595, 2715–2730. [Google Scholar] [CrossRef] [PubMed]
- Magadum, S.; Banerjee, U.; Murugan, P.; Gangapur, D.; Ravikesavan, R. Gene duplication as a major force in evolution. J. Genet. 2013, 92, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Troukhan, M.; Tatarinova, T.; Bouck, J.; Flavell, R.B.; Alexandrov, N.N. Genome-wide discovery of cis-elements in promoter sequences using gene expression. Omics 2009, 13, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Gao, L.; Liu, J.; Guan, W.; Xie, J.; Zeng, X.; Chen, Y.; Lu, Y.; Hou, Z.; Xiang, Z.; et al. Genome-wide identification, molecular docking and expression analysis of enzymes involved in the primary and secondary metabolic branching points of the selenium metabolic pathway in Cardamine hupingshanensis. BMC Plant Biol. 2025, 25, 680. [Google Scholar] [CrossRef]
- Honorato, R.V.; Trellet, M.E.; Jiménez-García, B.; Schaarschmidt, J.J.; Giulini, M.; Reys, V.; Koukos, P.I.; Rodrigues, J.P.G.L.M.; Karaca, E.; van Zundert, G.C.P.; et al. The HADDOCK2.4 web server for integrative modeling of biomolecular complexes. Nat. Protoc. 2024, 19, 3219–3241. [Google Scholar] [CrossRef]
- Dai, S.; Saarinen, M.; Ramaswamy, S.; Meyer, Y.; Jacquot, J.P.; Eklund, H. Crystal structure of Arabidopsis thaliana NADPH dependent thioredoxin reductase at 2.5 A resolution. J. Mol. Biol. 1996, 264, 1044–1057. [Google Scholar] [CrossRef]
- Marchetti, G.M.; Füsser, F.; Singh, R.K.; Brummel, M.; Koch, O.; Kümmel, D.; Hippler, M. Structural analysis revealed a novel conformation of the NTRC reductase domain from Chlamydomonas reinhardtii. J. Struct. Biol. 2022, 214, 107829. [Google Scholar] [CrossRef] [PubMed]
- Geigenberger, P.; Thormählen, I.; Daloso, D.M.; Fernie, A.R. The Unprecedented Versatility of the Plant Thioredoxin System. Trends Plant Sci. 2017, 22, 249–262. [Google Scholar] [CrossRef]
- Souza, P.V.L.; Vieira-Neto, A.E. Redefining Redox Regulation in Plants: The Evolving Role of Thioredoxins and TRX-Like 2.2 in Plant Metabolism. J. Evol. Biochem. Physiol. 2025, 61, 298–312. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, T.; Yan, F.; Wang, L.; Tang, Y.; Wang, Y.; Zhang, C. Genome-Wide Identification and Expression Analysis of Thioredoxin (Trx) Genes in Seed Development of Vitis vinifera. J. Plant Growth Regul. 2022, 41, 3030–3045. [Google Scholar] [CrossRef]
- Huang, C.; Ying, H.; Yang, X.; Gao, Y.; Li, T.; Wu, B.; Ren, M.; Zhang, Z.; Ding, J.; Gao, J.; et al. The Cardamine enshiensis genome reveals whole genome duplication and insight into selenium hyperaccumulation and tolerance. Cell Discov. 2021, 7, 62. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Li, Y.; Qin, Z.; Guo, S.; Li, Y.; Miao, Y.; Song, C.; Chen, S.; Dai, S. Plant Chloroplast Stress Response: Insights from Thiol Redox Proteomics. Antioxid. Redox Signal. 2020, 33, 35–57. [Google Scholar] [CrossRef]
- Xiao, Z.; Lu, Y.; Zou, Y.; Zhang, C.; Ding, L.; Luo, K.; Tang, Q.; Zhou, Y. Gene Identification, expression analysis and molecular docking of ATP sulfurylase in the selenization pathway of Cardamine hupingshanensis. BMC Plant Biol. 2022, 22, 491. [Google Scholar] [CrossRef]
- Martí, M.C.; Jiménez, A.; Sevilla, F. Thioredoxin Network in Plant Mitochondria: Cysteine S-Posttranslational Modifications and Stress Conditions. Front. Plant Sci. 2020, 11, 571288. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | pTM | Alpha Helix | Beta Turn | Extended Strand | Random Coil |
|---|---|---|---|---|---|
| ChTRX-y1-1 | 0.56 | 27.91% | 4.65% | 11.63% | 55.81% |
| ChTRX-y1-2 | 0.60 | 29.81% | 4.97% | 12.42% | 52.80% |
| ChTRX-m3-1 | 0.60 | 27.59% | 4.02% | 16.67% | 51.72% |
| ChACHT1 | 0.57 | 30.13% | 3.49% | 9.17% | 57.21% |
| ChACHT4-1 | 0.54 | 26.26% | 3.24% | 7.91% | 62.59% |
| ChNTRA-1 | 0.83 | 29.30% | 7.32% | 21.41% | 41.97% |
| ChNTRB-1 | 0.83 | 30.38% | 6.45% | 20.97% | 42.20% |
| ChNTRC-1 | 0.46 | 38.54% | 5.62% | 17.50% | 38.33% |
| ChNTRC-2 | 0.66 | 33.33% | 6.48% | 18.86% | 41.33% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Xue, H.; Lu, Y.; Xiang, Z.; Hou, Z.; Zhou, Y.; Tang, Q. Genome-Wide Thioredoxin System in Cardamine hupingshanensis: Role in Se Stress and Metabolism. Biology 2025, 14, 1404. https://doi.org/10.3390/biology14101404
Li Y, Xue H, Lu Y, Xiang Z, Hou Z, Zhou Y, Tang Q. Genome-Wide Thioredoxin System in Cardamine hupingshanensis: Role in Se Stress and Metabolism. Biology. 2025; 14(10):1404. https://doi.org/10.3390/biology14101404
Chicago/Turabian StyleLi, Yao, Huanqiu Xue, Yanke Lu, Zhixin Xiang, Zhi Hou, Yifeng Zhou, and Qiaoyu Tang. 2025. "Genome-Wide Thioredoxin System in Cardamine hupingshanensis: Role in Se Stress and Metabolism" Biology 14, no. 10: 1404. https://doi.org/10.3390/biology14101404
APA StyleLi, Y., Xue, H., Lu, Y., Xiang, Z., Hou, Z., Zhou, Y., & Tang, Q. (2025). Genome-Wide Thioredoxin System in Cardamine hupingshanensis: Role in Se Stress and Metabolism. Biology, 14(10), 1404. https://doi.org/10.3390/biology14101404





