The Role of Protein Arginine Methylation as a Post-Translational Modification in Cellular Homeostasis and Disease
Simple Summary
Abstract
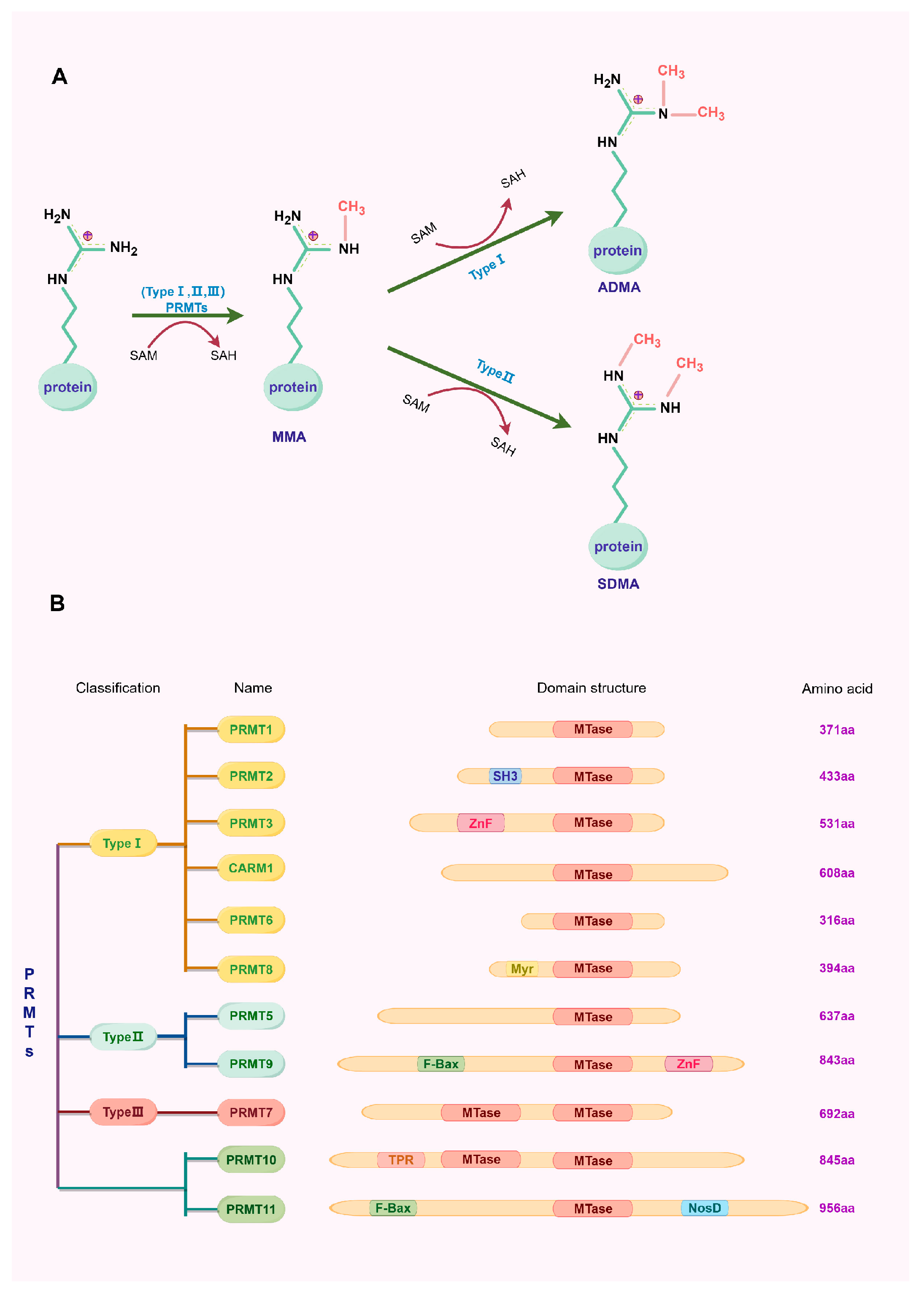
1. Introduction
2. Type I Protein Arginine Methyltransferase
2.1. PRMT1
| Cancer Type | Role(s) in Cancer | Mechanism of Action | Ref. |
|---|---|---|---|
| Acute myelogenous leukemia | Cell transformation | PRMT1 + KDM4C→epigenetic reprogramming↑→AML cell transformation↑ | [28] |
| Cell survival and proliferation | PRMT1→methylation of FLT3→survival and proliferation of FLT3-ITD AML cell↑ | [29] | |
| Breast cancer | Cell proliferation/viability and cell cycle progression | PRMT1→methylation of ERα→interacts with Src/FAK and p85→cell proliferation and survival | |
| PRMT1→EZH2 methylation→inhibits P16 and P21 transcriptional expression→promotes cell cycle progression | [30] | ||
| Colorectal cancer | Promotion of glycolysis, proliferation, and tumorigenesis | PRMT1→meR206-PGK1→pS203-PGK1↑→glycolytic activity and CRC tumorigenesis | [31] |
| Cell proliferative, colony-formative, and migratory abilities | PRMT1→H4R3me2a→recruits SMARCA4→EGFR signaling↑→proliferative, colony-formative, and migratory abilities↑ | [32] | |
| Gastric cancer | Cell migration and metastasis | PRMT1→ recruits MLXIP→β-catenin transcription and the β-catenin signaling pathway↑→GC cell migration and metastasis | [33] |
| Hepatocellular carcinoma | Cell proliferation and xenograft tumor growth | CDK5→phosphorylation of PRMT1→methylation of WDR24→mTORC1 pathway↑ | [34] |
| Human melanoma | Tumor growth and metastasis | PRMT1→methylation of ALCAM→tumor growth and metastasis | [35] |
| Lung cancer | DNA repair ability and chemotherapeutic drug resistance | PRMT1→methylation of FEN1→DNA repair ability and drug resistance | [36] |
| Cell invasion and drug resistance | PRMT1→methylation of Twist1 and p120-catenin expression↑→transcription of Kaiso↑→EMT in Osimertinib-resistant cells↑ | [37] | |
| Multiple myeloma | Cell proliferation and apoptosis | PRMT1→methylation of WTAP→NDUFS6 m6A modification →MM cell proliferation and OCR levels ↑ cell apoptosis and ROS levels↓ | [38] |
| Ovarian cancer | Cell migration and invasion | PRMT1→Methylation of BRD4→phosphorylation of BRD4→migration and invasion | [39] |
| Pancreatic cancer | Apoptosis | stress response →PRMT1→methylation of p14ARF→p53—independent apoptosis | [40] |
| Prostate cancer | PCa cell growth | PRMT1 + circ_0094606→methylation of ILF3→stability of IL-8 mRNA↑→M2 polarization of macrophages→PCa growth↑ | [41] |
2.2. PRMT2
2.3. PRMT3
2.4. PRMT4 (CARM1)
2.5. PRMT6
2.6. PRMT8
3. Type II Protein Arginine Methyltransferase
3.1. PRMT5
| Cancer Type | Role(s) in Cancer | Mechanism of Action | Ref. |
|---|---|---|---|
| Breast cancer | Inhibition of ferroptosis of breast cancer cells | PRMT5→KEAP1/R596me2→KEAP1 ubiquitination (by TRIM25)↓→NRF2/HMOX1 expression↓→ ferroptosis of TNBC cells↓ | [129] |
| Attenuation of autophagy | PRMT5→methylation of ULK1→kinase activity and basal autophagic function↓ | [131] | |
| Inhibition of cell proliferation | tamoxifen→PRMT5→methylation of Erα→transcription and cell proliferation↓ | [132] | |
| Cervical cancer | Promotion of cancer progression | PRMT5→H3R2me2s→transcription of STAT1↑→PD-L1 expression↑→development of cervical cancer↑ | [130] |
| Chronic lymphocytic leukemia | Promotion of cancer progression | PRMT5→activates oncogenic signaling pathways→CLL development↑→Richter’s transformation | [133] |
| Colorectal cancer | Promotion of cancer cell dissemination | PRMT5→ methylation of SMAD4→activation of TGF-β signaling→promotion of CRC dissemination | [134] |
| Promotion of cell proliferation, migration and invasion | PRMT5 interacts with MCM7→cell proliferation, migration, and invasion ↑ | [135] | |
| cell proliferation | PRMT5→H3R8Me2s→activates LDHA→glycolysis↑→cell proliferation | [136] | |
| Promotion of CRC development | PRMT5 and EZH2→H3K27me3, H4R3me2s and H3R8me2s→CDKN2B (p15INK4b) expression↓→promotes CRC development | [137] | |
| Glioma | Malignant phenotype | PRMT5→ERK1/2 pathway→malignant phenotype of glioma cells↑ | [138] |
| Promotion of self-renewal and proliferation | PRMT5→PTEN–AKT axis→GBM neurosphere self-renewal and proliferation↑ | [139] | |
| Hepatocellular carcinoma | Activation of de novo lipogenesis and tumorigenesis | PRMT5→methylation of SREBP1a→activates de novo lipogenesis and tumorigenesis | [140] |
| Liver cancer | Cell proliferation | PRMT5→ERK signaling→BTG2 expression↓→liver cancer cell proliferation | [141] |
| Lung cancer | Cell proliferation | PRMT5→methylation of KLF5→inhibit KLF5 degradation→promotes the maintenance and proliferation of lung cancer cells | [142] |
| Growth inhibition | PRMT5 inhibition + anti-PD-L1→inhibits the growth of lung cancer cells and activates CD8+ T cell immune surveillance | [143] | |
| Cell proliferation and metastasis | PRMT5→activation of the FGFR3/Akt signaling axis→lung cancer cell proliferation and metastasis | [144] | |
| Lymphoma | Cell survival | PRMT5→promotes WNT/β-CATENIN and AKT/GSK3β proliferative signaling→survival of lymphoma cells↑ | [145] |
| Ovarian cancer | Tumor growth | PRMT5→methylation of ENO1→promotes active ENO1 dimer formation→glycolysis flux↑→tumor growth↑ | [146] |
| Pancreatic cancer | Aerobic glycolysis and cell proliferation | PRMT5→expression of the tumor suppressor FBW7↓→stabilization of cMyc↑→aerobic glycolysis and pancreatic cancer cell proliferation↑ | [125] |
| Promotion of EMT | PRMT5→autophosphorylation of EGFR (Y1068 and Y1172)↑→activates Akt--β--catenin axis→promotes EMT | [147] | |
| Prostate cancer | Promotion of cancer progression | circSPON2/miR-331-3p axis→PRMT5→H4R3me2s and H3R8me2s→CAMK2N1↓→PCa progression↑ | [148] |
| Cell growth | PRMT5 + Sp1 + Brg1→H4R3me2s→activates transcription of AR→prostate cancer cell growth↑ | [149] |
3.2. PRMT9
4. Type III Protein Arginine Methyltransferase
PRMT7
| PRMT Isoform | Knockout Type | Model Description | Viability/Lethality | Key Tissue/Cellular Phenotypes | Ref. |
|---|---|---|---|---|---|
| PRMT1 | Conventional Homozygous KO | Gene trap | Embryotoxicity | [16] | |
| Conditional Homozygous KO | ERT2-Cre; endothelial cell-specific knockout | Viability | Worsening of pulmonary hemorrhage | [176] | |
| Conditional Homozygous KO | Mx1-Cre; adult hematopoietic cell-specific knockout | About one in eight mice died within five months | Prominent effect on adult hematopoiesis | [177] | |
| Conditional Homozygous KO | Ngn3-Cre; germ cell-specific knockout | Viability | Prmt1 KO male mice were completely infertile; female KO mice were fertile | [178] | |
| Conditional Homozygous KO | Vil-CreERT2; intestinal epithelium-specific knockout | Viability | Intestinal cell proliferation and crypt elongation increased in mice aged 8–12 weeks | [179] | |
| Conditional Homozygous KO | Myh6-cre; myocardium-specific knockout | Mouse lethality began at four weeks of age and nearly all died within two months | [180] | ||
| Conditional Heterozygous KO | Myh6-cre; myocardium-specific knockout | 25 percent of mice were dead within two months | [180] | ||
| Conditional Heterozygous KO | Myl1-Cre; skeletal muscle-specific knockout | Viability | Muscle loss | [181] | |
| Conventional Homozygous KO | Cre/loxP recombination system | Embryos did not survive to 7.5 days | [17] | ||
| Conditional Homozygous KO | Cre/loxP recombination system; epidermis-specific knockout | Small body size, thin clinical epidermis, and perinatal mortality | [182] | ||
| Conditional Homozygous KO | Flp-Cre; hepatocyte-specific knockout | Viability | [183] | ||
| PRMT2 | Conventional Homozygous KO | Standard gene targeting | Viability | There is no obvious difference in the display form | [45] |
| PRMT3 | Conventional Homozygous KO | Standard gene targeting | Viability; mouse embryos targeted to disrupt PRMT3 were smaller but survived after birth and reached normal mouse size as adults | [57] | |
| Conditional Homozygous KO | Alb-Cre; liver-specific knockout | Viability | Inhibited tumor progression and increased CD8+ T cell infiltration in mouse tumors | [184] | |
| CARM1 | Conventional Homozygous KO | Standard gene targeting | Mouse embryos survived to birth and responded to stimuli but developed respiratory distress and died within 20 min | [185] | |
| PRMT5 | Conditional Homozygous KO | MyoDCre; myogenic lineage-specific knockout | Adult mice died prematurely | Muscle atrophy and early death | [186] |
| Conditional Homozygous KO | Cre-loxP; medullary thymic epithelial cell-specific knockout | Viability | Smaller thymus in mice | [187] | |
| Conditional Homozygous KO | Myl1-CRE; skeletal muscle-specific knockout | Viability | Muscle mass, oxidative capacity, power generation, and exercise performance decreased in mice | [188] | |
| Conditional Homozygous KO | Pdx 1-CreER; islet-specific knockout | Viability | Decreased glucose tolerance in mice | [189] | |
| Conditional Homozygous KO | OC-Cre; osteoblast-specific knockout | Viability | Delayed socket healing in Prmt5 knockout mice | [190] | |
| Conventional Homozygous KO | Mx1-Cre | Two weeks after the first induction, the mice developed severe anemic pallor, and most of the mice with PRMT5 deficiency died within 1–2 days | Deletion of PRMT5 during adult hematopoiesis leads to severe cytopenias | [191] | |
| PRMT6 | Conventional Homozygous KO | CRISPR-Cas9 | Viability | Enhanced the body’s innate antiviral immunity | [87] |
| Conventional Homozygous KO | EIIa-Cre | Viability | [192] | ||
| PRMT7 | Conditional Homozygous KO | Cdh5-CreERT2; endothelial cell-specific knockout | Viability | Increased apoptosis and fibrosis impair heart recovery | [193] |
| Conventional Homozygous KO | Standard gene targeting | Viability | Muscles show a shift to glycolytic fiber types | [166] | |
| Conditional Homozygous KO | Myh6-Cre; cardiac-specific knockout | Viability; birth rate decreased by about half | Plays a protective role in Ang II-induced cardiomyopathy | [194] | |
| PRMT8 | Conventional Homozygous KO | Ayu1-Cre | Viability | Acetylcholine and choline decreased while PC levels increased | [95] |
| Conventional Homozygous KO | Standard gene targeting | Viability | In mice, the composition of phospholipids was changed, mitochondrial stress capacity was reduced, and neuroinflammatory markers were increased | [98] | |
| PRMT9 | Conventional Homozygous KO | CMV-Cre | Some died after birth | Sea horse neurons showed impaired excitatory synapse development | [195] |
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef]
- Larsen, S.C.; Sylvestersen, K.B.; Mund, A.; Lyon, D.; Mullari, M.; Madsen, M.V.; Daniel, J.A.; Jensen, L.J.; Nielsen, M.L. Proteome-wide analysis of arginine monomethylation reveals widespread occurrence in human cells. Sci. Signal 2016, 9, rs9. [Google Scholar] [CrossRef] [PubMed]
- Morales, Y.; Cáceres, T.; May, K.; Hevel, J.M. Biochemistry and regulation of the protein arginine methyltransferases (PRMTs). Arch. Biochem. Biophys. 2016, 590, 138–152. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Li, C. Evolutionarily conserved protein arginine methyltransferases in non-mammalian animal systems. FEBS J. 2012, 279, 932–945. [Google Scholar] [CrossRef] [PubMed]
- Low, J.K.; Wilkins, M.R. Protein arginine methylation in Saccharomyces cerevisiae. FEBS J. 2012, 279, 4423–4443. [Google Scholar] [CrossRef]
- Fulton, M.D.; Brown, T.; Zheng, Y.G. Mechanisms and Inhibitors of Histone Arginine Methylation. Chem. Rec. 2018, 18, 1792–1807. [Google Scholar] [CrossRef]
- Fuhrmann, J.; Clancy, K.W.; Thompson, P.R. Chemical biology of protein arginine modifications in epigenetic regulation. Chem. Rev. 2015, 115, 5413–5461. [Google Scholar] [CrossRef]
- Bedford, M.T.; Clarke, S.G. Protein arginine methylation in mammals: Who, what, and why. Mol. Cell 2009, 33, 1–13. [Google Scholar] [CrossRef]
- Di Lorenzo, A.; Bedford, M.T. Histone arginine methylation. FEBS Lett. 2011, 585, 2024–2031. [Google Scholar] [CrossRef]
- Li, Z.; Wang, D.; Wang, W.; Chen, X.; Tang, A.; Hou, P.; Li, M.; Zheng, J.; Bai, J. Macrophages-stimulated PRMT1-mediated EZH2 methylation promotes breast cancer metastasis. Biochem. Biophys. Res. Commun. 2020, 533, 679–684. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Bedford, M.T. PABP1 identified as an arginine methyltransferase substrate using high-density protein arrays. EMBO Rep. 2002, 3, 268–273. [Google Scholar] [CrossRef]
- McCabe, M.T.; Mohammad, H.P.; Barbash, O.; Kruger, R.G. Targeting Histone Methylation in Cancer. Cancer J. 2017, 23, 292–301. [Google Scholar] [CrossRef]
- Chen, H.; Lorton, B.; Gupta, V.; Shechter, D. A TGFβ-PRMT5-MEP50 axis regulates cancer cell invasion through histone H3 and H4 arginine methylation coupled transcriptional activation and repression. Oncogene 2016, 36, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Huang, Z.Q.; Xia, L.; Feng, Q.; Erdjument-Bromage, H.; Strahl, B.D.; Briggs, S.D.; Allis, C.D.; Wong, J.; Tempst, P.; et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 2001, 293, 853–857. [Google Scholar] [CrossRef] [PubMed]
- Boisvert, F.M.; Rhie, A.; Richard, S.; Doherty, A.J. The GAR motif of 53BP1 is arginine methylated by PRMT1 and is necessary for 53BP1 DNA binding activity. Cell Cycle 2005, 4, 1834–1841. [Google Scholar] [CrossRef]
- Pawlak, M.R.; Scherer, C.A.; Chen, J.; Roshon, M.J.; Ruley, H.E. Arginine N-methyltransferase 1 is required for early postimplantation mouse development, but cells deficient in the enzyme are viable. Mol. Cell Biol. 2000, 20, 4859–4869. [Google Scholar] [CrossRef]
- Yu, Z.; Chen, T.; Hébert, J.; Li, E.; Richard, S. A mouse PRMT1 null allele defines an essential role for arginine methylation in genome maintenance and cell proliferation. Mol. Cell Biol. 2009, 29, 2982–2996. [Google Scholar] [CrossRef]
- Mathioudaki, K.; Scorilas, A.; Ardavanis, A.; Lymberi, P.; Tsiambas, E.; Devetzi, M.; Apostolaki, A.; Talieri, M. Clinical evaluation of PRMT1 gene expression in breast cancer. Tumor Biol. 2011, 32, 575–582. [Google Scholar] [CrossRef]
- Papadokostopoulou, A.; Mathioudaki, K.; Scorilas, A.; Xynopoulos, D.; Ardavanis, A.; Kouroumalis, E.; Talieri, M. Colon cancer and protein arginine methyltransferase 1 gene expression. Anticancer. Res. 2009, 29, 1361–1366. [Google Scholar]
- Le Romancer, M.; Treilleux, I.; Leconte, N.; Robin-Lespinasse, Y.; Sentis, S.; Bouchekioua-Bouzaghou, K.; Goddard, S.; Gobert-Gosse, S.; Corbo, L. Regulation of estrogen rapid signaling through arginine methylation by PRMT1. Mol. Cell 2008, 31, 212–221. [Google Scholar] [CrossRef]
- Yamagata, K.; Daitoku, H.; Takahashi, Y.; Namiki, K.; Hisatake, K.; Kako, K.; Mukai, H.; Kasuya, Y.; Fukamizu, A. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol. Cell 2008, 32, 221–231. [Google Scholar] [CrossRef]
- Sakamaki, J.; Daitoku, H.; Ueno, K.; Hagiwara, A.; Yamagata, K.; Fukamizu, A. Arginine methylation of BCL-2 antagonist of cell death (BAD) counteracts its phosphorylation and inactivation by Akt. Proc. Natl. Acad. Sci. USA 2011, 108, 6085–6090. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhao, Y.; Zhang, J.; Lu, Y.; Liu, X.; Geng, P.; Huang, B.; Zhang, Y.; Lu, J. The dual function of PRMT1 in modulating epithelial-mesenchymal transition and cellular senescence in breast cancer cells through regulation of ZEB1. Sci. Rep. 2016, 6, 19874. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Bu, X.; Chu, C.; Dai, X.; Asara, J.M.; Sicinski, P.; Freeman, G.J.; Wei, W. PRMT1 mediated methylation of cGAS suppresses anti-tumor immunity. Nat. Commun. 2023, 14, 2806. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Nie, H.; Zhao, X.; Chen, K.; Peng, C.; Zhang, X.; Guo, H.; Chen, Y.; Yang, X.; Wang, D.; et al. Tumor-derived PRMT1 suppresses macrophage antitumor activity by inhibiting cGAS/STING signaling in gastric cancer cells. Cell Death Dis. 2025, 16, 649. [Google Scholar] [CrossRef]
- Tao, H.; Jin, C.; Zhou, L.; Deng, Z.; Li, X.; Dang, W.; Fan, S.; Li, B.; Ye, F.; Lu, J.; et al. PRMT1 Inhibition Activates the Interferon Pathway to Potentiate Antitumor Immunity and Enhance Checkpoint Blockade Efficacy in Melanoma. Cancer Res. 2024, 84, 419–433. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, Z.; Song, C.; Wu, S.; Xie, J.; Zou, Y.; Xie, X.; Wu, T.; Yang, H.; Tang, H. PRMT1-Mediated PARP1 Methylation Drives Lung Metastasis and Chemoresistance via P65 Activation in Triple-Negative Breast Cancer. Research 2025, 8, 0854. [Google Scholar] [CrossRef]
- Cheung, N.; Fung, T.K.; Zeisig, B.B.; Holmes, K.; Rane, J.K.; Mowen, K.A.; Finn, M.G.; Lenhard, B.; Chan, L.C.; So, C.W. Targeting Aberrant Epigenetic Networks Mediated by PRMT1 and KDM4C in Acute Myeloid Leukemia. Cancer Cell 2016, 29, 32–48. [Google Scholar] [CrossRef]
- He, X.; Zhu, Y.; Lin, Y.C.; Li, M.; Du, J.; Dong, H.; Sun, J.; Zhu, L.; Wang, H.; Ding, Z.; et al. PRMT1-mediated FLT3 arginine methylation promotes maintenance of FLT3-ITD(+) acute myeloid leukemia. Blood 2019, 134, 548–560. [Google Scholar] [CrossRef]
- Li, Z.; Wang, D.; Chen, X.; Wang, W.; Wang, P.; Hou, P.; Li, M.; Chu, S.; Qiao, S.; Zheng, J.; et al. PRMT1-mediated EZH2 methylation promotes breast cancer cell proliferation and tumorigenesis. Cell Death Dis. 2021, 12, 1080. [Google Scholar] [CrossRef]
- Liu, H.; Chen, X.; Wang, P.; Chen, M.; Deng, C.; Qian, X.; Bai, J.; Li, Z.; Yu, X. PRMT1-mediated PGK1 arginine methylation promotes colorectal cancer glycolysis and tumorigenesis. Cell Death Dis. 2024, 15, 170. [Google Scholar] [CrossRef] [PubMed]
- Yao, B.; Gui, T.; Zeng, X.; Deng, Y.; Wang, Z.; Wang, Y.; Yang, D.; Li, Q.; Xu, P.; Hu, R.; et al. PRMT1-mediated H4R3me2a recruits SMARCA4 to promote colorectal cancer progression by enhancing EGFR signaling. Genome Med. 2021, 13, 58. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Chen, S.; Peng, S.; Zhou, X.; Tang, H.; Liang, H.; Zhong, X.; Yang, H.; Ke, X.; Lü, M.; et al. PRMT1 promotes the proliferation and metastasis of gastric cancer cells by recruiting MLXIP for the transcriptional activation of the β-catenin pathway. Genes. Dis. 2023, 10, 2622–2638. [Google Scholar] [CrossRef] [PubMed]
- Yin, S.; Liu, L.; Ball, L.E.; Wang, Y.; Bedford, M.T.; Duncan, S.A.; Wang, H.; Gan, W. CDK5-PRMT1-WDR24 signaling cascade promotes mTORC1 signaling and tumor growth. Cell Rep. 2023, 42, 112316. [Google Scholar] [CrossRef]
- Li, L.; Zhang, Z.; Ma, T.; Huo, R. PRMT1 regulates tumor growth and metastasis of human melanoma via targeting ALCAM. Mol. Med. Rep. 2016, 14, 521–528. [Google Scholar] [CrossRef]
- He, L.; Hu, Z.; Sun, Y.; Zhang, M.; Zhu, H.; Jiang, L.; Zhang, Q.; Mu, D.; Zhang, J.; Gu, L.; et al. PRMT1 is critical to FEN1 expression and drug resistance in lung cancer cells. DNA Repair 2020, 95, 102953. [Google Scholar] [CrossRef]
- Racherla, K.S.; Dovalovsky, K.; Patel, M.; Harper, E.; Barnard, J.; Nasifuzzaman, S.M.; Smith, M.; Sikand, R.; Drinka, E.; Puri, N. PRMT-1 and p120-Catenin as EMT Mediators in Osimertinib Resistance in NSCLC. Cancers 2023, 15, 3461. [Google Scholar] [CrossRef]
- Jia, Y.; Yu, X.; Liu, R.; Shi, L.; Jin, H.; Yang, D.; Zhang, X.; Shen, Y.; Feng, Y.; Zhang, P.; et al. PRMT1 methylation of WTAP promotes multiple myeloma tumorigenesis by activating oxidative phosphorylation via m6A modification of NDUFS6. Cell Death Dis. 2023, 14, 512. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, H.; Ye, M.; Jiang, M.; Chen, X.; Song, G.; Ji, H.; Wang, Z.W.; Zhu, X. Methylation of BRD4 by PRMT1 regulates BRD4 phosphorylation and promotes ovarian cancer invasion. Cell Death Dis. 2023, 14, 624. [Google Scholar] [CrossRef]
- Repenning, A.; Happel, D.; Bouchard, C.; Meixner, M.; Verel-Yilmaz, Y.; Raifer, H.; Holembowski, L.; Krause, E.; Kremmer, E.; Feederle, R.; et al. PRMT1 promotes the tumor suppressor function of p14(ARF) and is indicative for pancreatic cancer prognosis. Embo J. 2021, 40, e106777. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, K.; Yang, D.; Liu, F.; Xu, X.; Feng, Y.; Wang, Y.; Zhu, S.; Gu, C.; Sheng, J.; et al. Hsa_circ_0094606 promotes malignant progression of prostate cancer by inducing M2 polarization of macrophages through PRMT1-mediated arginine methylation of ILF3. Carcinogenesis 2023, 44, 15–28. [Google Scholar] [CrossRef]
- Lakowski, T.M.; Frankel, A. Kinetic analysis of human protein arginine N-methyltransferase 2: Formation of monomethyl- and asymmetric dimethyl-arginine residues on histone H4. Biochem. J. 2009, 421, 253–261. [Google Scholar] [CrossRef]
- Meyer, R.; Wolf, S.S.; Obendorf, M. PRMT2, a member of the protein arginine methyltransferase family, is a coactivator of the androgen receptor. J. Steroid Biochem. Mol. Biol. 2007, 107, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Qi, C.; Chang, J.; Zhu, Y.; Yeldandi, A.V.; Rao, S.M.; Zhu, Y.J. Identification of protein arginine methyltransferase 2 as a coactivator for estrogen receptor alpha. J. Biol. Chem. 2002, 277, 28624–28630. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, T.; Boehm, M.; Olive, M.; Crook, M.F.; San, H.; Langenickel, T.; Nabel, E.G. The arginine methyltransferase PRMT2 binds RB and regulates E2F function. Exp. Cell Res. 2006, 312, 2040–2053. [Google Scholar] [CrossRef] [PubMed]
- Hussein, M.A.; Shrestha, E.; Ouimet, M.; Barrett, T.J.; Leone, S.; Moore, K.J.; Hérault, Y.; Fisher, E.A.; Garabedian, M.J. LXR-Mediated ABCA1 Expression and Function Are Modulated by High Glucose and PRMT2. PLoS ONE 2015, 10, e0135218. [Google Scholar] [CrossRef]
- Li, Y.Y.; Zhou, S.H.; Chen, S.S.; Zhong, J.; Wen, G.B. PRMT2 inhibits the formation of foam cell induced by ox-LDL in RAW 264.7 macrophage involving ABCA1 mediated cholesterol efflux. Biochem. Biophys. Res. Commun. 2020, 524, 77–82. [Google Scholar] [CrossRef]
- Vurusaner, B.; Thevkar-Nages, P.; Kaur, R.; Giannarelli, C.; Garabedian, M.J.; Fisher, E.A. Loss of PRMT2 in myeloid cells in normoglycemic mice phenocopies impaired regression of atherosclerosis in diabetic mice. Sci. Rep. 2022, 12, 12031. [Google Scholar] [CrossRef]
- Zhu, J.; Li, X.; Sun, X.; Zhou, Z.; Cai, X.; Liu, X.; Wang, J.; Xiao, W. Zebrafish prmt2 Attenuates Antiviral Innate Immunity by Targeting traf6. J. Immunol. 2021, 207, 2570–2580. [Google Scholar] [CrossRef]
- Hu, G.; Yan, C.; Xie, P.; Cao, Y.; Shao, J.; Ge, J. PRMT2 accelerates tumorigenesis of hepatocellular carcinoma by activating Bcl2 via histone H3R8 methylation. Exp. Cell Res. 2020, 394, 112152. [Google Scholar] [CrossRef]
- Bednarz-Misa, I.; Fleszar, M.G.; Fortuna, P.; Lewandowski, Ł.; Mierzchała-Pasierb, M.; Diakowska, D.; Krzystek-Korpacka, M. Altered L-Arginine Metabolic Pathways in Gastric Cancer: Potential Therapeutic Targets and Biomarkers. Biomolecules 2021, 11, 1086. [Google Scholar] [CrossRef]
- Zou, H.; Liu, Y.; Yang, X.; Zhang, Q.; Pan, Q.; Huang, J.; Guo, Y.; Zhou, Y.; Fang, S.; Chen, Z.S.; et al. PRMT2 promotes tumorigenic phenotypes through the Wnt signaling pathway and drives immune suppression in Colorectal cancer. Cancer Lett. 2025, 632, 217967. [Google Scholar] [CrossRef] [PubMed]
- Frankel, A.; Clarke, S. PRMT3 is a distinct member of the protein arginine N-methyltransferase family. Conferral of substrate specificity by a zinc-finger domain. J. Biol. Chem. 2000, 275, 32974–32982. [Google Scholar] [CrossRef] [PubMed]
- Hsu, M.C.; Tsai, Y.L.; Lin, C.H.; Pan, M.R.; Shan, Y.S.; Cheng, T.Y.; Cheng, S.H.; Chen, L.T.; Hung, W.C. Protein arginine methyltransferase 3-induced metabolic reprogramming is a vulnerable target of pancreatic cancer. J. Hematol. Oncol. 2019, 12, 79. [Google Scholar] [CrossRef] [PubMed]
- Bachand, F.; Silver, P.A. PRMT3 is a ribosomal protein methyltransferase that affects the cellular levels of ribosomal subunits. Embo J. 2004, 23, 2641–2650. [Google Scholar] [CrossRef]
- Swiercz, R.; Person, M.D.; Bedford, M.T. Ribosomal protein S2 is a substrate for mammalian PRMT3 (protein arginine methyltransferase 3). Biochem. J. 2005, 386, 85–91. [Google Scholar] [CrossRef]
- Swiercz, R.; Cheng, D.; Kim, D.; Bedford, M.T. Ribosomal protein rpS2 is hypomethylated in PRMT3-deficient mice. J. Biol. Chem. 2007, 282, 16917–16923. [Google Scholar] [CrossRef]
- Miyata, S.; Mori, Y.; Tohyama, M. PRMT3 is essential for dendritic spine maturation in rat hippocampal neurons. Brain Res. 2010, 1352, 11–20. [Google Scholar] [CrossRef]
- Singh, V.; Miranda, T.B.; Jiang, W.; Frankel, A.; Roemer, M.E.; Robb, V.A.; Gutmann, D.H.; Herschman, H.R.; Clarke, S.; Newsham, I.F. DAL-1/4.1B tumor suppressor interacts with protein arginine N-methyltransferase 3 (PRMT3) and inhibits its ability to methylate substrates in vitro and in vivo. Oncogene 2004, 23, 7761–7771. [Google Scholar] [CrossRef]
- Heller, G.; Geradts, J.; Ziegler, B.; Newsham, I.; Filipits, M.; Markis-Ritzinger, E.M.; Kandioler, D.; Berger, W.; Stiglbauer, W.; Depisch, D.; et al. Downregulation of TSLC1 and DAL-1 expression occurs frequently in breast cancer. Breast Cancer Res. Treat. 2007, 103, 283–291. [Google Scholar] [CrossRef]
- Hu, Y.; Su, Y.; He, Y.; Liu, W.; Xiao, B. Arginine methyltransferase PRMT3 promote tumorigenesis through regulating c-MYC stabilization in colorectal cancer. Gene 2021, 791, 145718. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Han, P.; Chen, Y.; Wang, H.; Wang, S.; Wang, M.; Liu, J.; Yan, W.; Tian, D.; Liu, M. Protein arginine methyltransferase 3 promotes glycolysis and hepatocellular carcinoma growth by enhancing arginine methylation of lactate dehydrogenase A. Clin. Transl. Med. 2022, 12, e686. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Wu, Z.; Liu, S.; Zuo, D.; Niu, Y.; Qiu, Y.; Qiao, L.; He, W.; Qiu, J.; Yuan, Y.; et al. Targeting PRMT3 impairs methylation and oligomerization of HSP60 to boost anti-tumor immunity by activating cGAS/STING signaling. Nat. Commun. 2024, 15, 7930. [Google Scholar] [CrossRef] [PubMed]
- Hupalowska, A.; Jedrusik, A.; Zhu, M.; Bedford, M.T.; Glover, D.M.; Zernicka-Goetz, M. CARM1 and Paraspeckles Regulate Pre-implantation Mouse Embryo Development. Cell 2018, 175, 1902–1916.e1913. [Google Scholar] [CrossRef]
- Sun, H.; Su, J.; Wu, T.; Wang, F.; Kang, J.; Zhang, J.; Xing, X.; Cheng, Y.; Zhang, Y. CARM1 is heterogeneous in mouse four-cell embryo and important to blastocyst development. Reproduction 2020, 159, 91–104. [Google Scholar] [CrossRef]
- Bao, J.; Rousseaux, S.; Shen, J.; Lin, K.; Lu, Y.; Bedford, M.T. The arginine methyltransferase CARM1 represses p300•ACT•CREMτ activity and is required for spermiogenesis. Nucleic Acids Res. 2018, 46, 4327–4343. [Google Scholar] [CrossRef]
- Wu, D.; He, J.; Zhang, W.; Wang, K.; Jin, S.; Li, J.; Gao, W. CARM1 promotes non-small cell lung cancer progression through upregulating CCNE2 expression. Aging 2020, 12, 10578–10593. [Google Scholar] [CrossRef]
- Veazey, K.J.; Cheng, D.; Lin, K.; Villarreal, O.D.; Gao, G.; Perez-Oquendo, M.; Van, H.T.; Stratton, S.A.; Green, M.; Xu, H.; et al. CARM1 inhibition reduces histone acetyltransferase activity causing synthetic lethality in CREBBP/EP300-mutated lymphomas. Leukemia 2020, 34, 3269–3285. [Google Scholar] [CrossRef]
- Peng, B.L.; Li, W.J.; Ding, J.C.; He, Y.H.; Ran, T.; Xie, B.L.; Wang, Z.R.; Shen, H.F.; Xiao, R.Q.; Gao, W.W.; et al. A hypermethylation strategy utilized by enhancer-bound CARM1 to promote estrogen receptor α-dependent transcriptional activation and breast carcinogenesis. Theranostics 2020, 10, 3451–3473. [Google Scholar] [CrossRef]
- Zhong, X.Y.; Yuan, X.M.; Xu, Y.Y.; Yin, M.; Yan, W.W.; Zou, S.W.; Wei, L.M.; Lu, H.J.; Wang, Y.P.; Lei, Q.Y. CARM1 Methylates GAPDH to Regulate Glucose Metabolism and Is Suppressed in Liver Cancer. Cell Rep. 2018, 24, 3207–3223. [Google Scholar] [CrossRef]
- Du, P.; Luo, K.; Li, G.; Zhu, J.; Xiao, Q.; Li, Y.; Zhang, X. PRMT4 promotes hepatocellular carcinoma progression by activating AKT/mTOR signaling and indicates poor prognosis. Int. J. Med. Sci. 2021, 18, 3588–3598. [Google Scholar] [CrossRef]
- Fedoriw, A.; Shi, L.; O’Brien, S.; Smitheman, K.N.; Wang, Y.; Hou, J.; Sherk, C.; Rajapurkar, S.; Laraio, J.; Williams, L.J.; et al. Inhibiting Type I Arginine Methyltransferase Activity Promotes T Cell-Mediated Antitumor Immune Responses. Cancer Immunol. Res. 2022, 10, 420–436. [Google Scholar] [CrossRef]
- Frankel, A.; Yadav, N.; Lee, J.; Branscombe, T.L.; Clarke, S.; Bedford, M.T. The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J. Biol. Chem. 2002, 277, 3537–3543. [Google Scholar] [CrossRef] [PubMed]
- Hyllus, D.; Stein, C.; Schnabel, K.; Schiltz, E.; Imhof, A.; Dou, Y.; Hsieh, J.; Bauer, U.M. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 2007, 21, 3369–3380. [Google Scholar] [CrossRef] [PubMed]
- Schneider, L.; Herkt, S.; Wang, L.; Feld, C.; Wesely, J.; Kuvardina, O.N.; Meyer, A.; Oellerich, T.; Häupl, B.; Seifried, E.; et al. PRMT6 activates cyclin D1 expression in conjunction with the transcription factor LEF1. Oncogenesis 2021, 10, 42. [Google Scholar] [CrossRef] [PubMed]
- Harrison, M.J.; Tang, Y.H.; Dowhan, D.H. Protein arginine methyltransferase 6 regulates multiple aspects of gene expression. Nucleic Acids Res. 2010, 38, 2201–2216. [Google Scholar] [CrossRef]
- Kim, S.; Kim, N.H.; Park, J.E.; Hwang, J.W.; Myung, N.; Hwang, K.T.; Kim, Y.A.; Jang, C.Y.; Kim, Y.K. PRMT6-mediated H3R2me2a guides Aurora B to chromosome arms for proper chromosome segregation. Nat. Commun. 2020, 11, 612. [Google Scholar] [CrossRef]
- Feng, J.; Dang, Y.; Zhang, W.; Zhao, X.; Zhang, C.; Hou, Z.; Jin, Y.; McNutt, M.A.; Marks, A.R.; Yin, Y. PTEN arginine methylation by PRMT6 suppresses PI3K-AKT signaling and modulates pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 2019, 116, 6868–6877. [Google Scholar] [CrossRef]
- Avasarala, S.; Wu, P.Y.; Khan, S.Q.; Yanlin, S.; Van Scoyk, M.; Bao, J.; Di Lorenzo, A.; David, O.; Bedford, M.T.; Gupta, V.; et al. PRMT6 Promotes Lung Tumor Progression via the Alternate Activation of Tumor-Associated Macrophages. Mol. Cancer Res. 2020, 18, 166–178. [Google Scholar] [CrossRef]
- Chan, L.H.; Zhou, L.; Ng, K.Y.; Wong, T.L.; Lee, T.K.; Sharma, R.; Loong, J.H.; Ching, Y.P.; Yuan, Y.F.; Xie, D.; et al. PRMT6 Regulates RAS/RAF Binding and MEK/ERK-Mediated Cancer Stemness Activities in Hepatocellular Carcinoma through CRAF Methylation. Cell Rep. 2018, 25, 690–701.e698. [Google Scholar] [CrossRef]
- Pan, R.; Yu, H.; Dai, J.; Zhou, C.; Ying, X.; Zhong, J.; Zhao, J.; Zhang, Y.; Wu, B.; Mao, Y.; et al. Significant association of PRMT6 hypomethylation with colorectal cancer. J. Clin. Lab. Anal. 2018, 32, e22590. [Google Scholar] [CrossRef]
- Okuno, K.; Akiyama, Y.; Shimada, S.; Nakagawa, M.; Tanioka, T.; Inokuchi, M.; Yamaoka, S.; Kojima, K.; Tanaka, S. Asymmetric dimethylation at histone H3 arginine 2 by PRMT6 in gastric cancer progression. Carcinogenesis 2019, 40, 15–26. [Google Scholar] [CrossRef]
- Jiang, N.; Li, Q.L.; Pan, W.; Li, J.; Zhang, M.F.; Cao, T.; Su, S.G.; Shen, H. PRMT6 promotes endometrial cancer via AKT/mTOR signaling and indicates poor prognosis. Int. J. Biochem. Cell Biol. 2020, 120, 105681. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.L.; Ng, K.Y.; Tan, K.V.; Chan, L.H.; Zhou, L.; Che, N.; Hoo, R.L.C.; Lee, T.K.; Richard, S.; Lo, C.M.; et al. CRAF Methylation by PRMT6 Regulates Aerobic Glycolysis-Driven Hepatocarcinogenesis via ERK-Dependent PKM2 Nuclear Relocalization and Activation. Hepatology 2020, 71, 1279–1296. [Google Scholar] [CrossRef] [PubMed]
- Che, N.; Ng, K.Y.; Wong, T.L.; Tong, M.; Kau, P.W.; Chan, L.H.; Lee, T.K.; Huen, M.S.; Yun, J.P.; Ma, S. PRMT6 deficiency induces autophagy in hostile microenvironments of hepatocellular carcinoma tumors by regulating BAG5-associated HSC70 stability. Cancer Lett. 2021, 501, 247–262. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liang, W.; Du, J.; Ma, J.; Liang, R.; Tao, M. PRMT6 functionally associates with PRMT5 to promote colorectal cancer progression through epigenetically repressing the expression of CDKN2B and CCNG1. Exp. Cell Res. 2023, 422, 113413. [Google Scholar] [CrossRef]
- Zhang, H.; Han, C.; Li, T.; Li, N.; Cao, X. The methyltransferase PRMT6 attenuates antiviral innate immunity by blocking TBK1-IRF3 signaling. Cell Mol. Immunol. 2019, 16, 800–809. [Google Scholar] [CrossRef]
- Jiang, Z.; Cheng, X.; Sun, Z.; Hu, J.; Xu, X.; Li, M.; Feng, Z.; Hu, C. Grass carp PRMT6 negatively regulates innate immunity by inhibiting the TBK1/IRF3 binding and cutting down IRF3 phosphorylation level. Dev. Comp. Immunol. 2022, 129, 104351. [Google Scholar] [CrossRef]
- Yu, P.; Xu, T.; Ma, W.; Fang, X.; Bao, Y.; Xu, C.; Huang, J.; Sun, Y.; Li, G. PRMT6-mediated transcriptional activation of ythdf2 promotes glioblastoma migration, invasion, and emt via the wnt-β-catenin pathway. J. Exp. Clin. Cancer Res. 2024, 43, 116. [Google Scholar] [CrossRef]
- Lee, J.; Sayegh, J.; Daniel, J.; Clarke, S.; Bedford, M.T. PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. J. Biol. Chem. 2005, 280, 32890–32896. [Google Scholar] [CrossRef]
- Sayegh, J.; Webb, K.; Cheng, D.; Bedford, M.T.; Clarke, S.G. Regulation of protein arginine methyltransferase 8 (PRMT8) activity by its N-terminal domain. J. Biol. Chem. 2007, 282, 36444–36453. [Google Scholar] [CrossRef] [PubMed]
- Dillon, M.B.; Rust, H.L.; Thompson, P.R.; Mowen, K.A. Automethylation of protein arginine methyltransferase 8 (PRMT8) regulates activity by impeding S-adenosylmethionine sensitivity. J. Biol. Chem. 2013, 288, 27872–27880. [Google Scholar] [CrossRef] [PubMed]
- Park, S.W.; Jun, Y.W.; Choi, H.E.; Lee, J.A.; Jang, D.J. Deciphering the molecular mechanisms underlying the plasma membrane targeting of PRMT8. BMB Rep. 2019, 52, 601–606. [Google Scholar] [CrossRef] [PubMed]
- Taneda, T.; Miyata, S.; Kousaka, A.; Inoue, K.; Koyama, Y.; Mori, Y.; Tohyama, M. Specific regional distribution of protein arginine methyltransferase 8 (PRMT8) in the mouse brain. Brain Res. 2007, 1155, 1–9. [Google Scholar] [CrossRef]
- Kim, J.D.; Park, K.E.; Ishida, J.; Kako, K.; Hamada, J.; Kani, S.; Takeuchi, M.A.-O.; Namiki, K.; Fukui, H.; Fukuhara, S.; et al. PRMT8 as a phospholipase regulates Purkinje cell dendritic arborization and motor coordination. Sci. Adv. 2015, 1, e1500615. [Google Scholar] [CrossRef]
- Lee, P.K.; Goh, W.W.; Sng, J.A.-O. Network-based characterization of the synaptic proteome reveals that removal of epigenetic regulator Prmt8 restricts proteins associated with synaptic maturation. J. Neurochem. 2017, 140, 613–628. [Google Scholar]
- Penney, J.; Seo, J.; Kritskiy, O.; Elmsaouri, S.; Gao, F.; Pao, P.C.; Su, S.C.; Tsai, L.H. Loss of Protein Arginine Methyltransferase 8 Alters Synapse Composition and Function, Resulting in Behavioral Defects. J. Neurosci. 2017, 37, 8655–8666. [Google Scholar] [CrossRef]
- Couto, E.S.A.; Wu, C.Y.; Clemons, G.A.; Acosta, C.H.; Chen, C.T.; Possoit, H.E.; Citadin, C.T.; Lee, R.H.; Brown, J.I.; Frankel, A.; et al. Protein arginine methyltransferase 8 modulates mitochondrial bioenergetics and neuroinflammation after hypoxic stress. J. Neurochem. 2021, 159, 742–761. [Google Scholar] [CrossRef]
- Simandi, Z.A.-O.; Pajer, K.; Karolyi, K.; Sieler, T.A.-O.X.; Jiang, L.A.-O.; Kolostyak, Z.A.-O.; Sari, Z.A.-O.; Fekecs, Z.; Pap, A.; Patsalos, A.A.-O.; et al. Arginine Methyltransferase PRMT8 Provides Cellular Stress Tolerance in Aging Motoneurons. J. Neurosci. 2018, 38, 7683–7700. [Google Scholar] [CrossRef]
- Lo, L.H.; Dong, R.; Lyu, Q.; Lai, K.O. The Protein Arginine Methyltransferase PRMT8 and Substrate G3BP1 Control Rac1-PAK1 Signaling and Actin Cytoskeleton for Dendritic Spine Maturation. Cell Rep. 2020, 31, 107744. [Google Scholar] [CrossRef]
- Jeong, H.C.; Park, S.J.; Choi, J.J.; Go, Y.H.; Hong, S.K.; Kwon, O.S.; Shin, J.G.; Kim, R.K.; Lee, M.O.; Lee, S.J.; et al. PRMT8 Controls the Pluripotency and Mesodermal Fate of Human Embryonic Stem Cells By Enhancing the PI3K/AKT/SOX2 Axis. Stem Cells 2017, 35, 2037–2049. [Google Scholar] [CrossRef]
- Lin, H.; Wang, B.; Yu, J.; Wang, J.; Li, Q.; Cao, B. Protein arginine methyltransferase 8 gene enhances the colon cancer stem cell (CSC) function by upregulating the pluripotency transcription factor. J. Cancer 2018, 9, 1394–1402. [Google Scholar] [CrossRef]
- Hernandez, S.J.; Dolivo, D.M.; Dominko, T. PRMT8 demonstrates variant-specific expression in cancer cells and correlates with patient survival in breast, ovarian and gastric cancer. Oncol. Lett. 2017, 13, 1983–1989. [Google Scholar] [CrossRef]
- Simandi, Z.; Czipa, E.; Horvath, A.; Koszeghy, A.; Bordas, C.; Póliska, S.; Juhász, I.; Imre, L.; Szabó, G.; Dezso, B.; et al. PRMT1 and PRMT8 regulate retinoic acid-dependent neuronal differentiation with implications to neuropathology. Stem Cells 2015, 33, 726–741. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Liu, Z.; Li, M.; Chen, C.; Wang, X. Increased glycolysis correlates with elevated immune activity in tumor immune microenvironment. EBioMedicine 2019, 42, 431–442. [Google Scholar] [CrossRef]
- Lin, Y.L.; Tsai, Y.J.; Liu, Y.F.; Cheng, Y.C.; Hung, C.M.; Lee, Y.J.; Pan, H.; Li, C. The critical role of protein arginine methyltransferase prmt8 in zebrafish embryonic and neural development is non-redundant with its paralogue prmt1. PLoS ONE 2013, 8, e55221. [Google Scholar] [CrossRef] [PubMed]
- Antonysamy, S.; Bonday, Z.; Campbell, R.M.; Doyle, B.; Druzina, Z.; Gheyi, T.; Han, B.; Jungheim, L.N.; Qian, Y.; Rauch, C.; et al. Crystal structure of the human PRMT5:MEP50 complex. Proc. Natl. Acad. Sci. USA 2012, 109, 17960–17965. [Google Scholar] [CrossRef] [PubMed]
- Ho, M.C.; Wilczek, C.; Bonanno, J.B.; Xing, L.; Seznec, J.; Matsui, T.; Carter, L.G.; Onikubo, T.; Kumar, P.R.; Chan, M.K.; et al. Structure of the arginine methyltransferase PRMT5-MEP50 reveals a mechanism for substrate specificity. PLoS ONE 2013, 8, e57008. [Google Scholar] [CrossRef]
- Tamburri, S.; Lavarone, E.; Fernández-Pérez, D.; Conway, E.; Zanotti, M.; Manganaro, D.; Pasini, D. Histone H2AK119 Mono-Ubiquitination Is Essential for Polycomb-Mediated Transcriptional Repression. Mol. Cell 2020, 77, 840–856.e845. [Google Scholar] [CrossRef]
- Lorton, B.M.; Harijan, R.K.; Burgos, E.S.; Bonanno, J.B.; Almo, S.C.; Shechter, D. A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5. Biochemistry 2020, 59, 3696–3708. [Google Scholar] [CrossRef]
- Tee, W.W.; Pardo, M.; Theunissen, T.W.; Yu, L.; Choudhary, J.S.; Hajkova, P.; Surani, M.A. Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes. Dev. 2010, 24, 2772–2777. [Google Scholar] [CrossRef]
- Blanc, R.S.; Richard, S. Arginine Methylation: The Coming of Age. Mol. Cell 2017, 65, 8–24. [Google Scholar] [CrossRef] [PubMed]
- Szewczyk, M.M.; Luciani, G.M.; Vu, V.; Murison, A.; Dilworth, D.; Barghout, S.H.; Lupien, M.; Arrowsmith, C.H.; Minden, M.D.; Barsyte-Lovejoy, D. PRMT5 regulates ATF4 transcript splicing and oxidative stress response. Redox Biol. 2022, 51, 102282. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.C.; Zheng, S.; Munro, S.; Liu, G.; Carr, S.M.; Moehlenbrink, J.; Lu, Y.C.; Stimson, L.; Khan, O.; Konietzny, R.; et al. Arginine methylation controls growth regulation by E2F-1. EMBO J. 2012, 31, 1785–1797. [Google Scholar] [CrossRef]
- Zheng, S.; Moehlenbrink, J.; Lu, Y.C.; Zalmas, L.P.; Sagum, C.A.; Carr, S.; McGouran, J.F.; Alexander, L.; Fedorov, O.; Munro, S.; et al. Arginine methylation-dependent reader-writer interplay governs growth control by E2F-1. Mol. Cell 2013, 52, 37–51. [Google Scholar] [CrossRef]
- Litzler, L.C.; Zahn, A.; Meli, A.P.; Hébert, S.; Patenaude, A.M.; Methot, S.P.; Sprumont, A.; Bois, T.; Kitamura, D.; Costantino, S.; et al. PRMT5 is essential for B cell development and germinal center dynamics. Nat. Commun. 2019, 10, 22. [Google Scholar] [CrossRef]
- Strobl, C.D.; Schaffer, S.; Haug, T.; Völkl, S.; Peter, K.; Singer, K.; Böttcher, M.; Mougiakakos, D.; Mackensen, A.; Aigner, M. Selective PRMT5 Inhibitors Suppress Human CD8(+) T Cells by Upregulation of p53 and Impairment of the AKT Pathway Similar to the Tumor Metabolite MTA. Mol. Cancer Ther. 2020, 19, 409–419. [Google Scholar] [CrossRef]
- Yan, F.; Alinari, L.; Lustberg, M.E.; Katherine Martin, L.; Cordero-Nieves, H.M.; Banasavadi-Siddegowda, Y.; Virk, S.; Barnholtz-Sloan, J.; Bell, E.H.; Wojton, J.; et al. Genetic validation of the protein arginine methyltransferase PRMT5 as a candidate therapeutic target in glioblastoma. Cancer Res. 2014, 74, 1752–1765. [Google Scholar] [CrossRef]
- Nicholas, C.; Yang, J.; Peters, S.B.; Bill, M.A.; Baiocchi, R.A.; Yan, F.; Sïf, S.; Tae, S.; Gaudio, E.; Wu, X.; et al. PRMT5 is upregulated in malignant and metastatic melanoma and regulates expression of MITF and p27(Kip1.). PLoS ONE 2013, 8, e74710. [Google Scholar] [CrossRef]
- Bao, X.; Zhao, S.; Liu, T.; Liu, Y.; Liu, Y.; Yang, X. Overexpression of PRMT5 promotes tumor cell growth and is associated with poor disease prognosis in epithelial ovarian cancer. J. Histochem. Cytochem. 2013, 61, 206–217. [Google Scholar] [CrossRef]
- Gu, Z.; Gao, S.; Zhang, F.; Wang, Z.; Ma, W.; Davis, R.E.; Wang, Z. Protein arginine methyltransferase 5 is essential for growth of lung cancer cells. Biochem. J. 2012, 446, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Chiang, K.; Zielinska, A.E.; Shaaban, A.M.; Sanchez-Bailon, M.P.; Jarrold, J.; Clarke, T.L.; Zhang, J.; Francis, A.; Jones, L.J.; Smith, S.; et al. PRMT5 Is a Critical Regulator of Breast Cancer Stem Cell Function via Histone Methylation and FOXP1 Expression. Cell Rep. 2017, 21, 3498–3513. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; Sohn, H.Y.; Yoon, S.Y.; Oh, J.H.; Yang, J.O.; Kim, J.H.; Song, K.S.; Rho, S.M.; Yoo, H.S.; Kim, Y.S.; et al. Identification of gastric cancer-related genes using a cDNA microarray containing novel expressed sequence tags expressed in gastric cancer cells. Clin. Cancer Res. 2005, 11, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Eckert, D.; Biermann, K.; Nettersheim, D.; Gillis, A.J.; Steger, K.; Jäck, H.M.; Müller, A.M.; Looijenga, L.H.; Schorle, H. Expression of BLIMP1/PRMT5 and concurrent histone H2A/H4 arginine 3 dimethylation in fetal germ cells, CIS/IGCNU and germ cell tumors. BMC Dev. Biol. 2008, 8, 106. [Google Scholar] [CrossRef]
- Qin, Y.; Hu, Q.; Xu, J.; Ji, S.; Dai, W.; Liu, W.; Xu, W.; Sun, Q.; Zhang, Z.; Ni, Q.; et al. PRMT5 enhances tumorigenicity and glycolysis in pancreatic cancer via the FBW7/cMyc axis. Cell Commun. Signal 2019, 17, 30. [Google Scholar] [CrossRef]
- Liu, M.; Yao, B.; Gui, T.; Guo, C.; Wu, X.; Li, J.; Ma, L.; Deng, Y.; Xu, P.; Wang, Y.; et al. PRMT5-dependent transcriptional repression of c-Myc target genes promotes gastric cancer progression. Theranostics 2020, 10, 4437–4452. [Google Scholar] [CrossRef]
- Karkhanis, V.; Alinari, L.; Ozer, H.G.; Chung, J.; Zhang, X.; Sif, S.; Baiocchi, R.A. Protein arginine methyltransferase 5 represses tumor suppressor miRNAs that down-regulate CYCLIN D1 and c-MYC expression in aggressive B-cell lymphoma. J. Biol. Chem. 2020, 295, 1165–1180. [Google Scholar] [CrossRef]
- Kim, H.; Kim, H.; Feng, Y.; Li, Y.; Tamiya, H.; Tocci, S.; Ronai, Z.A. PRMT5 control of cGAS/STING and NLRC5 pathways defines melanoma response to antitumor immunity. Sci. Transl. Med. 2020, 12, eaaz5683. [Google Scholar] [CrossRef]
- Wang, Z.; Li, R.; Hou, N.; Zhang, J.; Wang, T.; Fan, P.; Ji, C.; Zhang, B.; Liu, L.; Wang, Y.; et al. PRMT5 reduces immunotherapy efficacy in triple-negative breast cancer by methylating KEAP1 and inhibiting ferroptosis. J. Immunother. Cancer 2023, 11, e006890. [Google Scholar] [CrossRef]
- Jiang, Y.; Yuan, Y.; Chen, M.; Li, S.; Bai, J.; Zhang, Y.; Sun, Y.; Wang, G.; Xu, H.; Wang, Z.; et al. PRMT5 disruption drives antitumor immunity in cervical cancer by reprogramming T cell-mediated response and regulating PD-L1 expression. Theranostics 2021, 11, 9162–9176. [Google Scholar] [CrossRef]
- Brobbey, C.; Yin, S.; Liu, L.; Ball, L.E.; Howe, P.H.; Delaney, J.R.; Gan, W. Autophagy dictates sensitivity to PRMT5 inhibitor in breast cancer. Sci. Rep. 2023, 13, 10752. [Google Scholar] [CrossRef] [PubMed]
- Poulard, C.; Ha Pham, T.; Drouet, Y.; Jacquemetton, J.; Surmielova, A.; Kassem, L.; Mery, B.; Lasset, C.; Reboulet, J.; Treilleux, I.; et al. Nuclear PRMT5 is a biomarker of sensitivity to tamoxifen in ERα(+) breast cancer. EMBO Mol. Med. 2023, 15, e17248. [Google Scholar] [CrossRef] [PubMed]
- Hing, Z.A.; Walker, J.S.; Whipp, E.C.; Brinton, L.; Cannon, M.; Zhang, P.; Sher, S.; Cempre, C.B.; Brown, F.; Smith, P.L.; et al. Dysregulation of PRMT5 in chronic lymphocytic leukemia promotes progression with high risk of Richter’s transformation. Nat. Commun. 2023, 14, 97. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Yu, C.; Qiu, C.; Wu, Q.; Huang, C.; Li, X.; She, X.; Wan, K.; Liu, L.; Li, M.; et al. PRMT5 methylating SMAD4 activates TGF-β signaling and promotes colorectal cancer metastasis. Oncogene 2023, 42, 1572–1584. [Google Scholar] [CrossRef]
- Li, X.; Wang, X.; Zhao, J.; Wang, J.; Wu, J. PRMT5 promotes colorectal cancer growth by interaction with MCM7. J. Cell Mol. Med. 2021, 25, 3537–3547. [Google Scholar] [CrossRef]
- Shen, Y.; Zhao, P.; Dong, K.; Wang, J.; Li, H.; Li, M.; Li, R.; Chen, S.; Shen, Y.; Liu, Z.; et al. Tadalafil increases the antitumor activity of 5-FU through inhibiting PRMT5-mediated glycolysis and cell proliferation in colorectal cancer. Cancer Metab. 2022, 10, 22. [Google Scholar] [CrossRef]
- Yang, L.; Ma, D.W.; Cao, Y.P.; Li, D.Z.; Zhou, X.; Feng, J.F.; Bao, J. PRMT5 functionally associates with EZH2 to promote colorectal cancer progression through epigenetically repressing CDKN2B expression. Theranostics 2021, 11, 3742–3759. [Google Scholar] [CrossRef]
- Han, X.; Li, R.; Zhang, W.; Yang, X.; Wheeler, C.G.; Friedman, G.K.; Province, P.; Ding, Q.; You, Z.; Fathallah-Shaykh, H.M.; et al. Expression of PRMT5 correlates with malignant grade in gliomas and plays a pivotal role in tumor growth in vitro. J. Neurooncol. 2014, 118, 61–72. [Google Scholar] [CrossRef]
- Banasavadi-Siddegowda, Y.K.; Russell, L.; Frair, E.; Karkhanis, V.A.; Relation, T.; Yoo, J.Y.; Zhang, J.; Sif, S.; Imitola, J.; Baiocchi, R.; et al. PRMT5-PTEN molecular pathway regulates senescence and self-renewal of primary glioblastoma neurosphere cells. Oncogene 2017, 36, 263–274. [Google Scholar] [CrossRef]
- Liu, L.; Zhao, X.; Zhao, L.; Li, J.; Yang, H.; Zhu, Z.; Liu, J.; Huang, G. Arginine Methylation of SREBP1a via PRMT5 Promotes De Novo Lipogenesis and Tumor Growth. Cancer Res. 2016, 76, 1260–1272. [Google Scholar] [CrossRef]
- Jiang, H.; Zhu, Y.; Zhou, Z.; Xu, J.; Jin, S.; Xu, K.; Zhang, H.; Sun, Q.; Wang, J.; Xu, J. PRMT5 promotes cell proliferation by inhibiting BTG2 expression via the ERK signaling pathway in hepatocellular carcinoma. Cancer Med. 2018, 7, 869–882. [Google Scholar] [CrossRef]
- Zhou, H.; Chang, J.; Zhang, J.; Zheng, H.; Miao, X.; Mo, H.; Sun, J.; Jia, Q.; Qi, G. PRMT5 activates KLF5 by methylation to facilitate lung cancer. J. Cell Mol. Med. 2024, 28, e17856. [Google Scholar] [CrossRef]
- Hu, R.; Zhou, B.; Chen, Z.; Chen, S.; Chen, N.; Shen, L.; Xiao, H.; Zheng, Y. PRMT5 Inhibition Promotes PD-L1 Expression and Immuno-Resistance in Lung Cancer. Front. Immunol. 2021, 12, 722188. [Google Scholar] [CrossRef]
- Zheng, Y.; Lu, J.; Hu, X.; Hu, X.; Gao, X.; Zhou, J. PRMT5/FGFR3/AKT Signaling Axis Facilitates Lung Cancer Cell Metastasis. Technol. Cancer Res. Treat. 2023, 22, 15330338231161139. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.; Karkhanis, V.; Baiocchi, R.A.; Sif, S. Protein arginine methyltransferase 5 (PRMT5) promotes survival of lymphoma cells via activation of WNT/β-catenin and AKT/GSK3β proliferative signaling. J. Biol. Chem. 2019, 294, 7692–7710. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Zhang, H.; Zhu, K.; Jiang, C.S.; Zhang, X.; Chang, H.; Qiao, Y.; Sun, M.; Wang, J.; Wang, M.; et al. PRMT5 promotes ovarian cancer growth through enhancing Warburg effect by methylating ENO1. MedComm 2023, 4, e245. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.; Wang, H.; Xu, X.; Zhou, Z.; He, J.; Peng, W.; Du, F.; Zhang, Y.; Gong, A.; Xu, M. PRMT5 promotes epithelial-mesenchymal transition via EGFR-β-catenin axis in pancreatic cancer cells. J. Cell Mol. Med. 2020, 24, 1969–1979. [Google Scholar] [CrossRef]
- Yao, B.; Zhu, S.; Wei, X.; Chen, M.K.; Feng, Y.; Li, Z.; Xu, X.; Zhang, Y.; Wang, Y.; Zhou, J.; et al. The circSPON2/miR-331-3p axis regulates PRMT5, an epigenetic regulator of CAMK2N1 transcription and prostate cancer progression. Mol. Cancer 2022, 21, 119. [Google Scholar] [CrossRef]
- Deng, X.; Shao, G.; Zhang, H.T.; Li, C.; Zhang, D.; Cheng, L.; Elzey, B.D.; Pili, R.; Ratliff, T.L.; Huang, J.; et al. Protein arginine methyltransferase 5 functions as an epigenetic activator of the androgen receptor to promote prostate cancer cell growth. Oncogene 2017, 36, 1223–1231. [Google Scholar] [CrossRef]
- Andrade, M.A.; Perez-Iratxeta, C.; Ponting, C.P. Protein repeats: Structures, functions, and evolution. J. Struct. Biol. 2001, 134, 117–131. [Google Scholar] [CrossRef]
- Hadjikyriacou, A.A.-O.X.; Yang, Y.A.-O.; Espejo, A.A.-O.; Bedford, M.A.-O.; Clarke, S.A.-O. Unique Features of Human Protein Arginine Methyltransferase 9 (PRMT9) and Its Substrate RNA Splicing Factor SF3B2. J. Biol. Chem. 2015, 290, 16723–16743. [Google Scholar] [CrossRef]
- Yang, Y.; Hadjikyriacou, A.; Xia, Z.A.-O.; Gayatri, S.; Kim, D.; Zurita-Lopez, C.; Kelly, R.; Guo, A.; Li, W.; Clarke, S.G.; et al. PRMT9 is a type II methyltransferase that methylates the splicing factor SAP145. Nat. Commun. 2015, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Zhou, Z.; Jin, S.; Xu, K.; Zhang, H.; Xu, J.; Sun, Q.; Wang, J.; Xu, J. PRMT9 promotes hepatocellular carcinoma invasion and metastasis via activating PI3K/Akt/GSK-3β/Snail signaling. Cancer Sci. 2018, 109, 1414–1427. [Google Scholar] [CrossRef]
- Zhang, H.; Guo, X.; Feng, X.; Wang, T.; Hu, Z.; Que, X.; Tian, Q.; Zhu, T.; Guo, G.; Huang, W.; et al. MiRNA-543 promotes osteosarcoma cell proliferation and glycolysis by partially suppressing PRMT9 and stabilizing HIF-1α protein. Oncotarget 2017, 8, 2342–2355. [Google Scholar] [CrossRef] [PubMed]
- Santoro, A.; Di Micco, R. PRMT9 inhibition sparks immune responses in AML. Nat. Cancer 2024, 5, 539–541. [Google Scholar] [CrossRef] [PubMed]
- Miranda, T.B.; Miranda, M.; Frankel, A.; Clarke, S. PRMT7 is a member of the protein arginine methyltransferase family with a distinct substrate specificity. J. Biol. Chem. 2004, 279, 22902–22907. [Google Scholar] [CrossRef]
- Ferreira, T.R.; Dowle, A.A.; Parry, E.; Alves-Ferreira, E.V.C.; Hogg, K.; Kolokousi, F.; Larson, T.R.; Plevin, M.J.; Cruz, A.K.; Walrad, P.B. PRMT7 regulates RNA-binding capacity and protein stability in Leishmania parasites. Nucleic Acids Res. 2020, 48, 5511–5526. [Google Scholar] [CrossRef]
- Zhu, J.; Liu, X.; Cai, X.; Ouyang, G.; Fan, S.; Wang, J.; Xiao, W. Zebrafish prmt7 negatively regulates antiviral responses by suppressing the retinoic acid-inducible gene-I-like receptor signaling. Faseb J. 2020, 34, 988–1000. [Google Scholar] [CrossRef]
- Chen, M.; Wang, Y.; Lin, L.; Dong, F.; Wu, H.; Bao, S.; Gao, F. PRMT7 is involved in regulation of germ cell proliferation during embryonic stage. Biochem. Biophys. Res. Commun. 2020, 533, 938–944. [Google Scholar] [CrossRef]
- Jelinic, P.; Stehle, J.C.; Shaw, P. The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol. 2006, 4, e355. [Google Scholar] [CrossRef]
- Gao, H.; Zhang, M.; Guo, J.; Liu, Z.; Guo, F.; Wang, B.; Mu, Y. Prmt7 Downregulation in Mouse Spermatogonia Functions through miR-877-3p/Col6a3. Life 2022, 12, 1194. [Google Scholar] [CrossRef]
- Buhr, N.; Carapito, C.; Schaeffer, C.; Kieffer, E.; Van Dorsselaer, A.; Viville, S. Nuclear proteome analysis of undifferentiated mouse embryonic stem and germ cells. Electrophoresis 2008, 29, 2381–2390. [Google Scholar] [CrossRef]
- Vuong, T.A.; Jeong, H.J.; Lee, H.J.; Kim, B.G.; Leem, Y.E.; Cho, H.; Kang, J.S. PRMT7 methylates and suppresses GLI2 binding to SUFU thereby promoting its activation. Cell Death Differ. 2020, 27, 15–28. [Google Scholar] [CrossRef]
- Cali, E.; Suri, M.; Scala, M.; Ferla, M.P.; Alavi, S.; Faqeih, E.A.; Bijlsma, E.K.; Wigby, K.M.; Baralle, D.; Mehrjardi, M.Y.V.; et al. Biallelic PRMT7 pathogenic variants are associated with a recognizable syndromic neurodevelopmental disorder with short stature, obesity, and craniofacial and digital abnormalities. Genet. Med. 2023, 25, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Blanc, R.S.; Vogel, G.; Chen, T.; Crist, C.; Richard, S. PRMT7 Preserves Satellite Cell Regenerative Capacity. Cell Rep. 2016, 14, 1528–1539. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.J.; Lee, H.J.; Vuong, T.A.; Choi, K.S.; Choi, D.; Koo, S.H.; Cho, S.C.; Cho, H.; Kang, J.S. Prmt7 Deficiency Causes Reduced Skeletal Muscle Oxidative Metabolism and Age-Related Obesity. Diabetes 2016, 65, 1868–1882. [Google Scholar] [CrossRef] [PubMed]
- Leem, Y.E.; Bae, J.H.; Jeong, H.J.; Kang, J.S. PRMT7 deficiency enhances adipogenesis through modulation of C/EBP-β. Biochem. Biophys. Res. Commun. 2019, 517, 484–490. [Google Scholar] [CrossRef]
- Jeong, H.J.; Lee, S.J.; Lee, H.J.; Kim, H.B.; Anh Vuong, T.; Cho, H.; Bae, G.U.; Kang, J.S. Prmt7 promotes myoblast differentiation via methylation of p38MAPK on arginine residue 70. Cell Death Differ. 2020, 27, 573–586. [Google Scholar] [CrossRef]
- Liu, F.; Wan, L.; Zou, H.; Pan, Z.; Zhou, W.; Lu, X. PRMT7 promotes the growth of renal cell carcinoma through modulating the β-catenin/C-MYC axis. Int. J. Biochem. Cell Biol. 2020, 120, 105686. [Google Scholar] [CrossRef]
- Cheng, D.; He, Z.; Zheng, L.; Xie, D.; Dong, S.; Zhang, P. PRMT7 contributes to the metastasis phenotype in human non-small-cell lung cancer cells possibly through the interaction with HSPA5 and EEF2. Onco Targets Ther. 2018, 11, 4869–4876. [Google Scholar] [CrossRef]
- Li, W.J.; He, Y.H.; Yang, J.J.; Hu, G.S.; Lin, Y.A.; Ran, T.; Peng, B.L.; Xie, B.L.; Huang, M.F.; Gao, X.; et al. Profiling PRMT methylome reveals roles of hnRNPA1 arginine methylation in RNA splicing and cell growth. Nat. Commun. 2021, 12, 1946. [Google Scholar] [CrossRef] [PubMed]
- Geng, P.; Zhang, Y.; Liu, X.; Zhang, N.; Liu, Y.; Liu, X.; Lin, C.; Yan, X.; Li, Z.; Wang, G.; et al. Automethylation of protein arginine methyltransferase 7 and its impact on breast cancer progression. Faseb J. 2017, 31, 2287–2300. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Li, X.; Cai, X.; Zha, H.; Zhou, Z.; Sun, X.; Rong, F.; Tang, J.; Zhu, C.; Liu, X.; et al. Arginine monomethylation by PRMT7 controls MAVS-mediated antiviral innate immunity. Mol. Cell 2021, 81, 3171–3186.e3178. [Google Scholar] [CrossRef] [PubMed]
- Oksa, L.; Mäkinen, A.; Nikkilä, A.; Hyvärinen, N.; Laukkanen, S.; Rokka, A.; Haapaniemi, P.; Seki, M.; Takita, J.; Kauko, O.; et al. Arginine Methyltransferase PRMT7 Deregulates Expression of RUNX1 Target Genes in T-Cell Acute Lymphoblastic Leukemia. Cancers 2022, 14, 2169. [Google Scholar] [CrossRef]
- Srour, N.; Villarreal, O.D.; Hardikar, S.; Yu, Z.; Preston, S.; Miller, W.H., Jr.; Szewczyk, M.M.; Barsyte-Lovejoy, D.; Xu, H.; Chen, T.; et al. PRMT7 ablation stimulates anti-tumor immunity and sensitizes melanoma to immune checkpoint blockade. Cell Rep. 2022, 38, 110582. [Google Scholar] [CrossRef]
- Tran, T.T.V.; Jeong, Y.; Kim, S.; Yeom, J.E.; Lee, J.; Lee, W.; Bae, G.U.; Kang, J.S. PRMT1 Ablation in Endothelial Cells Causes Endothelial Dysfunction and Aggravates COPD Attributable to Dysregulated NF-κB Signaling. Adv. Sci. 2025, 12, e2411514. [Google Scholar] [CrossRef]
- Zhu, L.; He, X.; Dong, H.; Sun, J.; Wang, H.; Zhu, Y.; Huang, F.; Zou, J.; Chen, Z.; Zhao, X.; et al. Protein arginine methyltransferase 1 is required for maintenance of normal adult hematopoiesis. Int. J. Biol. Sci. 2019, 15, 2763–2773. [Google Scholar] [CrossRef]
- Waseem, S.; Kumar, S.; Lee, K.; Yoon, B.H.; Kim, M.; Kim, H.; Lee, K. Protein Arginine Methyltransferase 1 Is Essential for the Meiosis of Male Germ Cells. Int. J. Mol. Sci. 2021, 22, 7951. [Google Scholar] [CrossRef]
- Peng, Z.; Bao, L.; Iben, J.; Wang, S.; Shi, B.; Shi, Y.B. Protein arginine methyltransferase 1 regulates mouse enteroendocrine cell development and homeostasis. Cell Biosci. 2024, 14, 70. [Google Scholar] [CrossRef]
- Pyun, J.H.; Kim, H.J.; Jeong, M.H.; Ahn, B.Y.; Vuong, T.A.; Lee, D.I.; Choi, S.; Koo, S.H.; Cho, H.; Kang, J.S. Cardiac specific PRMT1 ablation causes heart failure through CaMKII dysregulation. Nat. Commun. 2018, 9, 5107. [Google Scholar] [CrossRef]
- Choi, S.; Jeong, H.J.; Kim, H.; Choi, D.; Cho, S.C.; Seong, J.K.; Koo, S.H.; Kang, J.S. Skeletal muscle-specific Prmt1 deletion causes muscle atrophy via deregulation of the PRMT6-FOXO3 axis. Autophagy 2019, 15, 1069–1081. [Google Scholar] [CrossRef] [PubMed]
- Bao, X.; Siprashvili, Z.; Zarnegar, B.J.; Shenoy, R.M.; Rios, E.J.; Nady, N.; Qu, K.; Mah, A.; Webster, D.E.; Rubin, A.J.; et al. CSNK1a1 Regulates PRMT1 to Maintain the Progenitor State in Self-Renewing Somatic Tissue. Dev. Cell 2017, 43, 227–239.e225. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Adams, A.; Roberts, B.; O’Neil, M.; Vittal, A.; Schmitt, T.; Kumer, S.; Cox, J.; Li, Z.; Weinman, S.A.; et al. Protein arginine methyl transferase 1- and Jumonji C domain-containing protein 6-dependent arginine methylation regulate hepatocyte nuclear factor 4 alpha expression and hepatocyte proliferation in mice. Hepatology 2018, 67, 1109–1126. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.H.; Yan, F.Z.; Xu, B.N.; Qian, H.; Hong, X.L.; Liu, S.Q.; Luo, Y.Y.; Wu, S.H.; Cai, L.Y.; Zhang, X.; et al. PRMT3 drives PD-L1-mediated immune escape through activating PDHK1-regulated glycolysis in hepatocellular carcinoma. Cell Death Dis. 2025, 16, 158. [Google Scholar] [CrossRef]
- O’Brien, K.B.; Alberich-Jordà, M.; Yadav, N.; Kocher, O.; Diruscio, A.; Ebralidze, A.; Levantini, E.; Sng, N.J.; Bhasin, M.; Caron, T.; et al. CARM1 is required for proper control of proliferation and differentiation of pulmonary epithelial cells. Development 2010, 137, 2147–2156. [Google Scholar] [CrossRef]
- Kim, K.H.; Oprescu, S.N.; Snyder, M.M.; Kim, A.; Jia, Z.; Yue, F.; Kuang, S. PRMT5 mediates FoxO1 methylation and subcellular localization to regulate lipophagy in myogenic progenitors. Cell Rep. 2023, 42, 113329. [Google Scholar] [CrossRef]
- Kumar, R.; Ranganathan, P. PRMT5: Splicing up tolerance. J. Clin. Investig. 2024, 134, e185701. [Google Scholar] [CrossRef]
- Kim, K.H.; Jia, Z.; Snyder, M.; Chen, J.; Qiu, J.; Oprescu, S.N.; Chen, X.; Syed, S.A.; Yue, F.; Roseguini, B.T.; et al. PRMT5 links lipid metabolism to contractile function of skeletal muscles. EMBO Rep. 2023, 24, e57306. [Google Scholar] [CrossRef]
- Ma, J.; He, X.; Cao, Y.; O’Dwyer, K.; Szigety, K.M.; Wu, Y.; Gurung, B.; Feng, Z.; Katona, B.W.; Hua, X. Islet-specific Prmt5 excision leads to reduced insulin expression and glucose intolerance in mice. J. Endocrinol. 2020, 244, 41–52. [Google Scholar] [CrossRef]
- Yang, J.; Yang, S.; Ge, X.; Yuan, L.; Qi, Y.; Huang, Z.; Yang, G.; Zhang, R. Protein arginine methyltransferase 5 in osteoblasts promotes the healing of extraction sockets. Oral. Dis. 2024, 30, 3951–3961. [Google Scholar] [CrossRef]
- Liu, F.; Cheng, G.; Hamard, P.J.; Greenblatt, S.; Wang, L.; Man, N.; Perna, F.; Xu, H.; Tadi, M.; Luciani, L.; et al. Arginine methyltransferase PRMT5 is essential for sustaining normal adult hematopoiesis. J. Clin. Investig. 2015, 125, 3532–3544. [Google Scholar] [CrossRef]
- Neault, M.; Mallette, F.A.; Vogel, G.; Michaud-Levesque, J.; Richard, S. Ablation of PRMT6 reveals a role as a negative transcriptional regulator of the p53 tumor suppressor. Nucleic Acids Res. 2012, 40, 9513–9521. [Google Scholar] [CrossRef]
- Tran, T.T.V.; Zhang, Y.; Wei, S.; Lee, J.; Jeong, Y.; Vuong, T.A.; Lee, S.J.; Ryu, D.; Bae, G.U.; Kang, J.S. Endothelial PRMT7 prevents dysfunction, promotes revascularization and enhances cardiac recovery post-myocardial infarction. Exp. Mol. Med. 2025, 57, 1759–1774. [Google Scholar] [CrossRef]
- Ahn, B.Y.; Jeong, M.H.; Pyun, J.H.; Jeong, H.J.; Vuong, T.A.; Bae, J.H.; An, S.; Kim, S.W.; Kim, Y.K.; Ryu, D.; et al. PRMT7 ablation in cardiomyocytes causes cardiac hypertrophy and fibrosis through β-catenin dysregulation. Cell Mol. Life Sci. 2022, 79, 99. [Google Scholar] [CrossRef]
- Shen, L.; Ma, X.; Wang, Y.; Wang, Z.; Zhang, Y.; Pham, H.Q.H.; Tao, X.; Cui, Y.; Wei, J.; Lin, D.; et al. Loss-of-function mutation in PRMT9 causes abnormal synapse development by dysregulation of RNA alternative splicing. Nat. Commun. 2024, 15, 2809. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, K.; Xia, Q.; Li, K.; Yan, W.; Wang, C. The Role of Protein Arginine Methylation as a Post-Translational Modification in Cellular Homeostasis and Disease. Biology 2025, 14, 1370. https://doi.org/10.3390/biology14101370
Li K, Xia Q, Li K, Yan W, Wang C. The Role of Protein Arginine Methylation as a Post-Translational Modification in Cellular Homeostasis and Disease. Biology. 2025; 14(10):1370. https://doi.org/10.3390/biology14101370
Chicago/Turabian StyleLi, Ke, Qing Xia, Kexin Li, Wenxin Yan, and Changshan Wang. 2025. "The Role of Protein Arginine Methylation as a Post-Translational Modification in Cellular Homeostasis and Disease" Biology 14, no. 10: 1370. https://doi.org/10.3390/biology14101370
APA StyleLi, K., Xia, Q., Li, K., Yan, W., & Wang, C. (2025). The Role of Protein Arginine Methylation as a Post-Translational Modification in Cellular Homeostasis and Disease. Biology, 14(10), 1370. https://doi.org/10.3390/biology14101370






