Liver Progenitor Cells: Cellular Origins, Plasticity, and Signaling Pathways in Liver Regeneration
Abstract
Simple Summary
Abstract
1. Introduction
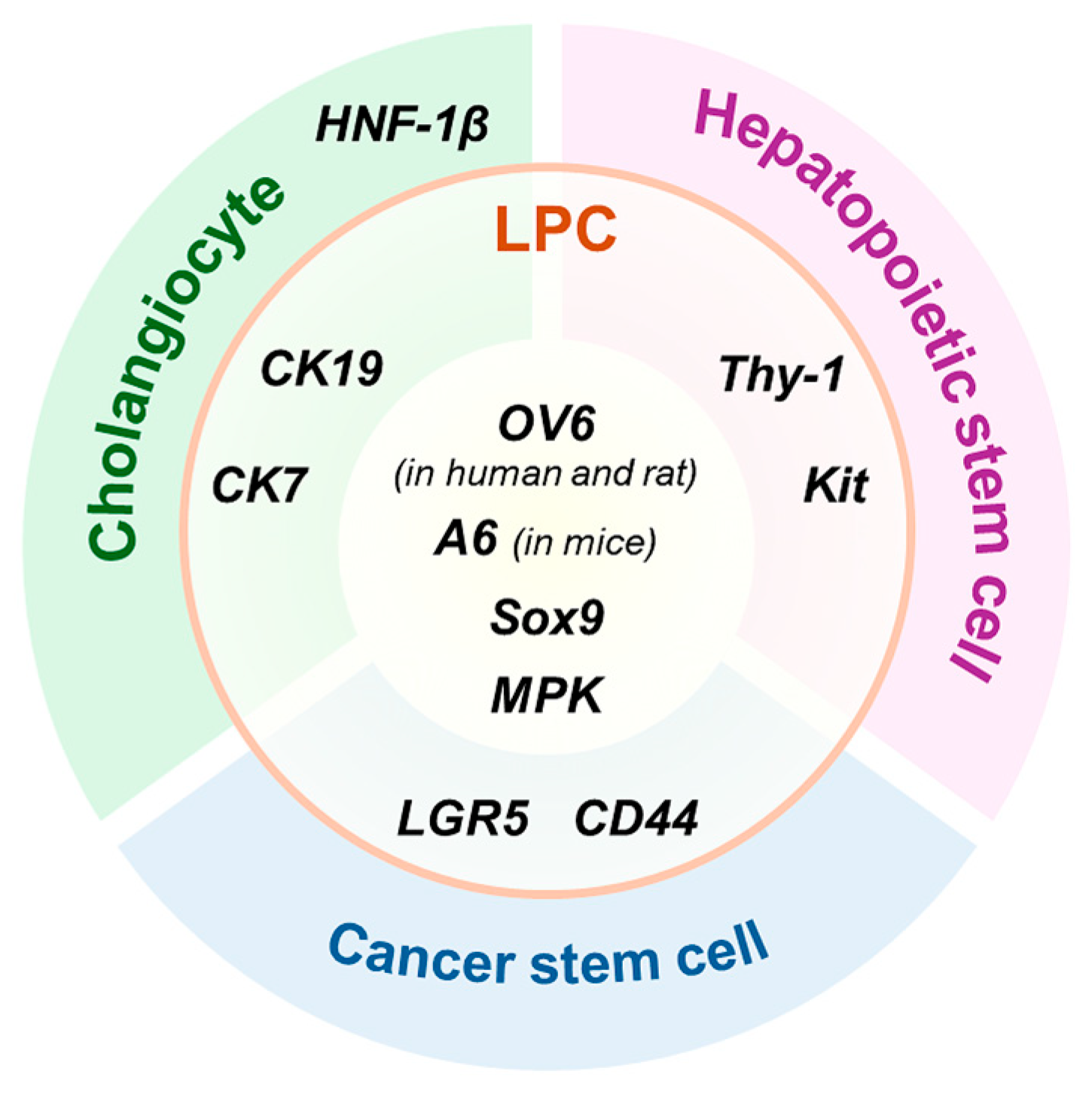
2. General Information on Liver Progenitor Cells
3. Origin of Liver Progenitor Cells in Liver Regeneration
3.1. Dedifferentiation of Hepatocytes into Liver Progenitor Cells
3.2. Conversion of Cholangiocytes into Liver Progenitor Cells
3.3. Transdifferentiation of Hepatic Stellate Cells into Liver Progenitor Cells
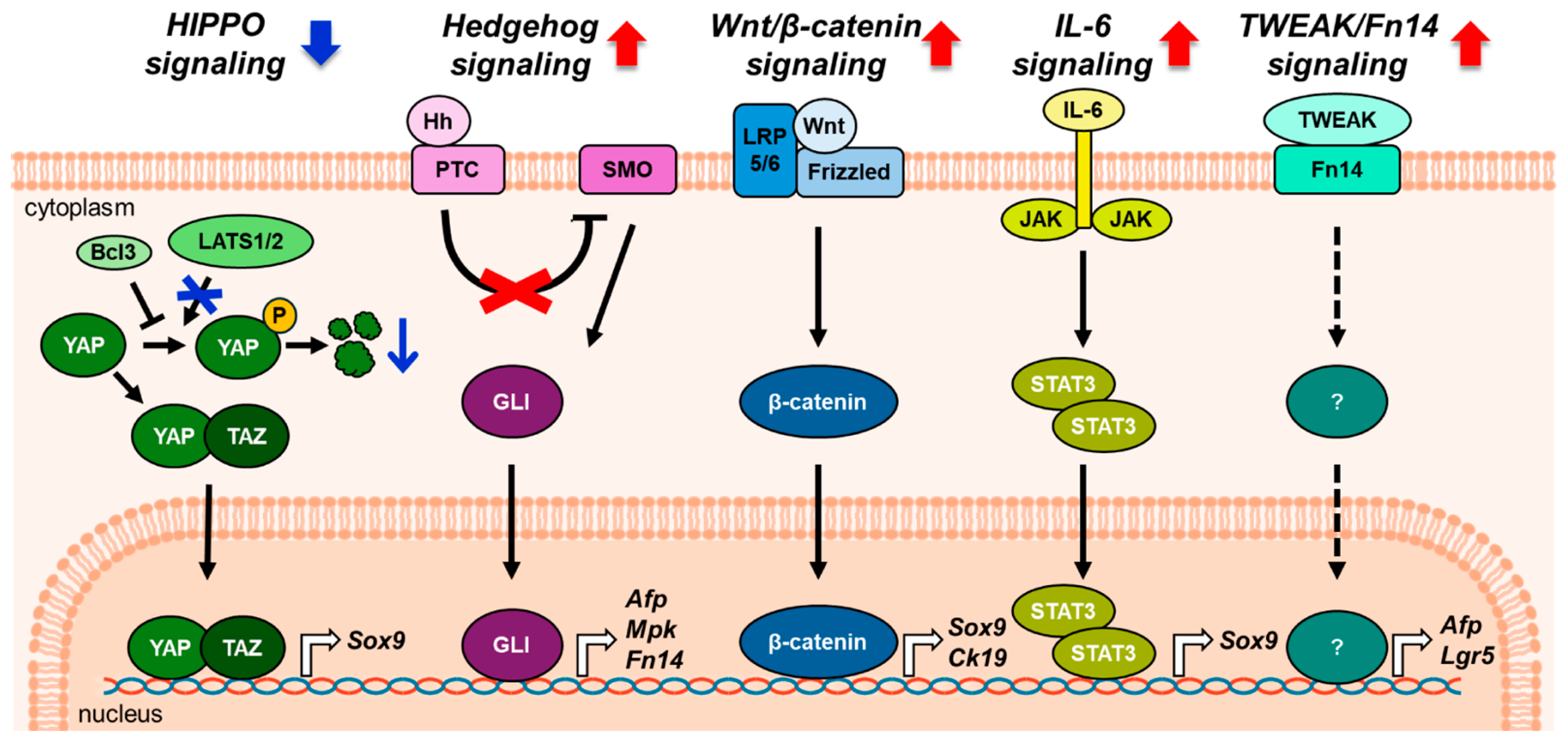
4. Liver Progenitor Cells Differentiation into Mature Hepatic Cells During the Repair Process
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 2-AAF | 2-acetamidofluorene |
| ABIC score | Age, serum bilirubin, international normalized ratio, and serum creatinine score |
| AFP | Alpha fetoprotein |
| AKT | Protein kinase B |
| Bcl3 | B-cell lymphoma 3 |
| CCl4 | Carbon tetrachloride |
| CD44 | Cluster of differentiation 44 |
| CDE | Choline-deficient, ethionine-supplemented |
| CK19 | Cytokeratin 19 |
| CK7 | Cytokeratin 7 |
| c-MET | Tyrosine kinase mesenchymal–epithelial transition factor |
| DAPM | 4,4′-diaminodiphenylmethane |
| DDC | 3,5-diethoxycarbonyl-1,4-dihydrocollidine |
| DR | Ductular reaction |
| EGF | Epidermal growth factor |
| EGFR | Epidermal growth factor receptor |
| Fah | Fumarylacetoacetase |
| Fn14 | Fibroblast growth factor-inducible 14 |
| GLI | Glioma-associated oncogene homolog |
| HGF | Hepatocyte growth factor |
| Hh | Hedgehog |
| HNF1α | Hepatocyte nuclear factor 1α |
| HNF1β | Hepatocyte nuclear factor 1β |
| HNF4α | Hepatocyte nuclear factor 4α |
| HSC | Hepatic stellate cell |
| Ihh | Indian hedgehog |
| IL-6 | Interleukin-6 |
| Kit | Receptor tyrosine kinase Kit |
| KO | Knockout |
| LATS | Large tumor suppressor kinase |
| LGR5 | Leucine-rich repeat-containing G protein-coupled receptor 5 |
| LPC | Liver progenitor cell |
| LRP5/6 | Low-density lipoprotein-related receptors 5 and 6 |
| MCD diet | methionine- and choline-deficient diet |
| MPK | Muscle pyruvate kinase |
| NICD | Notch intracellular domain |
| NTR-Mtx system | nitroreductase–metronidazole system |
| OV6 | Oval cell marker |
| PHx | Partial hepatectomy |
| PI3K | Phosphoinositide 3-kinase |
| PTC | Patched |
| RBPJ | Recombination signal binding protein for immunoglobulin kappa J region |
| Shh | Sonic hedgehog |
| SMO | Smoothened |
| Sox9 | SRY-box transcription factor 9 |
| STAT3 | Signal transducer and activator of transcription 3 |
| TAZ | Transcriptional coactivator with PDZ-binding motif |
| TGF-β | Transforming growth factor-β |
| Thy-1 | Thymocyte differentiation antigen 1 |
| TWEAK | Tumor necrosis factor-like weak inducer of apoptosis |
| Wnt | Wingless-type MMTV integration site family |
| YAP | Yes-associated protein |
References
- Yagi, S.; Hirata, M.; Miyachi, Y.; Uemoto, S. Liver Regeneration after Hepatectomy and Partial Liver Transplantation. Int. J. Mol. Sci. 2020, 21, 8414. [Google Scholar] [CrossRef] [PubMed]
- Michalopoulos, G.K.; Bhushan, B. Liver regeneration: Biological and pathological mechanisms and implications. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 40–55. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.A.; Glorioso, J.M.; Nyberg, S.L. Liver regeneration. Transl. Res. 2014, 163, 352–362. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.; Lee, C.; Lee, M.J.; Jung, Y. Therapeutic strategies to improve liver regeneration after hepatectomy. Exp. Biol. Med. 2023, 248, 1313–1318. [Google Scholar] [CrossRef] [PubMed]
- Haga, J.; Shimazu, M.; Wakabayashi, G.; Tanabe, M.; Kawachi, S.; Fuchimoto, Y.; Hoshino, K.; Morikawa, Y.; Kitajima, M.; Kitagawa, Y. Liver regeneration in donors and adult recipients after living donor liver transplantation. Liver. Transpl. 2008, 14, 1718–1724. [Google Scholar] [CrossRef] [PubMed]
- Korchilava, B.; Khachidze, T.; Megrelishvili, N.; Svanadze, L.; Kakabadze, M.; Tsomaia, K.; Jintcharadze, M.; Kordzaia, D. Liver regeneration after partial hepatectomy: Triggers and mechanisms. World J. Hepatol. 2025, 17, 107378. [Google Scholar] [CrossRef] [PubMed]
- Riehle, K.J.; Dan, Y.Y.; Campbell, J.S.; Fausto, N. New concepts in liver regeneration. J. Gastroenterol. Hepatol. 2011, 26 (Suppl. 1), 203–212. [Google Scholar] [CrossRef]
- Stanger, B.Z. Cellular homeostasis and repair in the mammalian liver. Annu. Rev. Physiol. 2015, 77, 179–200. [Google Scholar] [CrossRef]
- Dubuquoy, L.; Louvet, A.; Lassailly, G.; Truant, S.; Boleslawski, E.; Artru, F.; Maggiotto, F.; Gantier, E.; Buob, D.; Leteurtre, E.; et al. Progenitor cell expansion and impaired hepatocyte regeneration in explanted livers from alcoholic hepatitis. Gut 2015, 64, 1949–1960. [Google Scholar] [CrossRef] [PubMed]
- Best, J.; Manka, P.; Syn, W.K.; Dollé, L.; van Grunsven, L.A.; Canbay, A. Role of liver progenitors in liver regeneration. Hepatobiliary Surg. Nutr. 2015, 4, 48–58. [Google Scholar] [CrossRef]
- Kholodenko, I.V.; Yarygin, K.N. Cellular Mechanisms of Liver Regeneration and Cell-Based Therapies of Liver Diseases. BioMed Res. Int. 2017, 2017, 8910821. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Miyajima, A. Liver regeneration and fibrosis after inflammation. Inflamm. Regen. 2016, 36, 19. [Google Scholar] [CrossRef]
- Chen, Y.; Wong, P.P.; Sjeklocha, L.; Steer, C.J.; Sahin, M.B. Mature hepatocytes exhibit unexpected plasticity by direct dedifferentiation into liver progenitor cells in culture. Hepatology 2012, 55, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Tarlow, B.D.; Pelz, C.; Naugler, W.E.; Wakefield, L.; Wilson, E.M.; Finegold, M.J.; Grompe, M. Bipotential adult liver progenitors are derived from chronically injured mature hepatocytes. Cell Stem Cell 2014, 15, 605–618. [Google Scholar] [CrossRef] [PubMed]
- Rodrigo-Torres, D.; Affò, S.; Coll, M.; Morales-Ibanez, O.; Millán, C.; Blaya, D.; Alvarez-Guaita, A.; Rentero, C.; Lozano, J.J.; Maestro, M.A.; et al. The biliary epithelium gives rise to liver progenitor cells. Hepatology 2014, 60, 1367–1377. [Google Scholar] [CrossRef]
- Raven, A.; Lu, W.Y.; Man, T.Y.; Ferreira-Gonzalez, S.; O’Duibhir, E.; Dwyer, B.J.; Thomson, J.P.; Meehan, R.R.; Bogorad, R.; Koteliansky, V.; et al. Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature 2017, 547, 350–354. [Google Scholar] [CrossRef]
- Kordes, C.; Sawitza, I.; Götze, S.; Herebian, D.; Häussinger, D. Hepatic stellate cells contribute to progenitor cells and liver regeneration. J. Clin. Investig. 2014, 124, 5503–5515. [Google Scholar] [CrossRef]
- Swiderska-Syn, M.; Syn, W.K.; Xie, G.; Krüger, L.; Machado, M.V.; Karaca, G.; Michelotti, G.A.; Choi, S.S.; Premont, R.T.; Diehl, A.M. Myofibroblastic cells function as progenitors to regenerate murine livers after partial hepatectomy. Gut 2014, 63, 1333–1344. [Google Scholar] [CrossRef]
- Boulter, L.; Lu, W.Y.; Forbes, S.J. Differentiation of progenitors in the liver: A matter of local choice. J. Clin. Investig. 2013, 123, 1867–1873. [Google Scholar] [CrossRef]
- Oh, S.H.; Hatch, H.M.; Petersen, B.E. Hepatic oval ‘stem’ cell in liver regeneration. Semin. Cell Dev. Biol. 2002, 13, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Foster, M.; Al-Dhalimy, M.; Lagasse, E.; Finegold, M.; Grompe, M. The origin and liver repopulating capacity of murine oval cells. Proc. Natl. Acad. Sci. USA 2003, 100 (Suppl. 1), 11881–11888. [Google Scholar] [CrossRef] [PubMed]
- So, J.; Kim, A.; Lee, S.H.; Shin, D. Liver progenitor cell-driven liver regeneration. Exp. Mol. Med. 2020, 52, 1230–1238. [Google Scholar] [CrossRef] [PubMed]
- Sell, S. Comparison of liver progenitor cells in human atypical ductular reactions with those seen in experimental models of liver injury. Hepatology 1998, 27, 317–331. [Google Scholar] [CrossRef] [PubMed]
- Van Haele, M.; Roskams, T. Hepatic Progenitor Cells: An Update. Gastroenterol. Clin. N. Am. 2017, 46, 409–420. [Google Scholar] [CrossRef]
- Gilgenkrantz, H.; Collin de l’Hortet, A. Understanding Liver Regeneration: From Mechanisms to Regenerative Medicine. Am. J. Pathol. 2018, 188, 1316–1327. [Google Scholar] [CrossRef]
- Kordes, C.; Häussinger, D. Hepatic stem cell niches. J. Clin. Investig. 2013, 123, 1874–1880. [Google Scholar] [CrossRef]
- Roskams, T.A.; Theise, N.D.; Balabaud, C.; Bhagat, G.; Bhathal, P.S.; Bioulac-Sage, P.; Brunt, E.M.; Crawford, J.M.; Crosby, H.A.; Desmet, V.; et al. Nomenclature of the finer branches of the biliary tree: Canals, ductules, and ductular reactions in human livers. Hepatology 2004, 39, 1739–1745. [Google Scholar] [CrossRef] [PubMed]
- Saxena, R.; Theise, N. Canals of Hering: Recent insights and current knowledge. Semin. Liver Dis. 2004, 24, 43–48. [Google Scholar] [CrossRef]
- Miyajima, A.; Tanaka, M.; Itoh, T. Stem/progenitor cells in liver development, homeostasis, regeneration, and reprogramming. Cell Stem Cell 2014, 14, 561–574. [Google Scholar] [CrossRef]
- Yovchev, M.I.; Grozdanov, P.N.; Zhou, H.; Racherla, H.; Guha, C.; Dabeva, M.D. Identification of adult hepatic progenitor cells capable of repopulating injured rat liver. Hepatology 2008, 47, 636–647. [Google Scholar] [CrossRef] [PubMed]
- Yovchev, M.I.; Grozdanov, P.N.; Joseph, B.; Gupta, S.; Dabeva, M.D. Novel hepatic progenitor cell surface markers in the adult rat liver. Hepatology 2007, 45, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Petersen, B.E.; Goff, J.P.; Greenberger, J.S.; Michalopoulos, G.K. Hepatic oval cells express the hematopoietic stem cell marker Thy-1 in the rat. Hepatology 1998, 27, 433–445. [Google Scholar] [CrossRef]
- Crosby, H.A.; Kelly, D.A.; Strain, A.J. Human hepatic stem-like cells isolated using c-kit or CD34 can differentiate into biliary epithelium. Gastroenterology 2001, 120, 534–544. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Chen, K.; Bolkestein, M.; Yin, Y.; Verstegen, M.M.A.; Bijvelds, M.J.C.; Wang, W.; Tuysuz, N.; Ten Berge, D.; Sprengers, D.; et al. Dynamics of Proliferative and Quiescent Stem Cells in Liver Homeostasis and Injury. Gastroenterology 2017, 153, 1133–1147. [Google Scholar] [CrossRef]
- Kon, J.; Ooe, H.; Oshima, H.; Kikkawa, Y.; Mitaka, T. Expression of CD44 in rat hepatic progenitor cells. J. Hepatol. 2006, 45, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Furuyama, K.; Kawaguchi, Y.; Akiyama, H.; Horiguchi, M.; Kodama, S.; Kuhara, T.; Hosokawa, S.; Elbahrawy, A.; Soeda, T.; Koizumi, M.; et al. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat. Genet. 2011, 43, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Jelnes, P.; Santoni-Rugiu, E.; Rasmussen, M.; Friis, S.L.; Nielsen, J.H.; Tygstrup, N.; Bisgaard, H.C. Remarkable heterogeneity displayed by oval cells in rat and mouse models of stem cell-mediated liver regeneration. Hepatology 2007, 45, 1462–1470. [Google Scholar] [CrossRef] [PubMed]
- Crosby, H.A.; Hubscher, S.G.; Joplin, R.E.; Kelly, D.A.; Strain, A.J. Immunolocalization of OV-6, a putative progenitor cell marker in human fetal and diseased pediatric liver. Hepatology 1998, 28, 980–985. [Google Scholar] [CrossRef]
- Petersen, B.E.; Grossbard, B.; Hatch, H.; Pi, L.; Deng, J.; Scott, E.W. Mouse A6-positive hepatic oval cells also express several hematopoietic stem cell markers. Hepatology 2003, 37, 632–640. [Google Scholar] [CrossRef]
- Petersen, B.E.; Bowen, W.C.; Patrene, K.D.; Mars, W.M.; Sullivan, A.K.; Murase, N.; Boggs, S.S.; Greenberger, J.S.; Goff, J.P. Bone marrow as a potential source of hepatic oval cells. Science 1999, 284, 1168–1170. [Google Scholar] [CrossRef]
- Bae, S.H.; Choi, J.Y.; Yoon, S.K.; Oh, I.H.; Yoon, K.H.; Park, S.T.; Kim, G.D.; Oh, S.H.; Petersen, B.E. Thy1-positive bone marrow stem cells express liver-specific genes in vitro and can mature into hepatocytes in vivo. Hepatol. Int. 2008, 2, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Menthena, A.; Deb, N.; Oertel, M.; Grozdanov, P.N.; Sandhu, J.; Shah, S.; Guha, C.; Shafritz, D.A.; Dabeva, M.D. Bone marrow progenitors are not the source of expanding oval cells in injured liver. Stem Cells 2004, 22, 1049–1061. [Google Scholar] [CrossRef]
- Yanger, K.; Knigin, D.; Zong, Y.; Maggs, L.; Gu, G.; Akiyama, H.; Pikarsky, E.; Stanger, B.Z. Adult hepatocytes are generated by self-duplication rather than stem cell differentiation. Cell Stem Cell 2014, 15, 340–349. [Google Scholar] [CrossRef]
- Malato, Y.; Naqvi, S.; Schürmann, N.; Ng, R.; Wang, B.; Zape, J.; Kay, M.A.; Grimm, D.; Willenbring, H. Fate tracing of mature hepatocytes in mouse liver homeostasis and regeneration. J. Clin. Investig. 2011, 121, 4850–4860. [Google Scholar] [CrossRef] [PubMed]
- Avril, A.; Pichard, V.; Bralet, M.P.; Ferry, N. Mature hepatocytes are the source of small hepatocyte-like progenitor cells in the retrorsine model of liver injury. J. Hepatol. 2004, 41, 737–743. [Google Scholar] [CrossRef] [PubMed]
- Camargo, F.D.; Gokhale, S.; Johnnidis, J.B.; Fu, D.; Bell, G.W.; Jaenisch, R.; Brummelkamp, T.R. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 2007, 17, 2054–2060. [Google Scholar] [CrossRef]
- Dong, J.; Feldmann, G.; Huang, J.; Wu, S.; Zhang, N.; Comerford, S.A.; Gayyed, M.F.; Anders, R.A.; Maitra, A.; Pan, D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 2007, 130, 1120–1133. [Google Scholar] [CrossRef]
- Fu, M.; Hu, Y.; Lan, T.; Guan, K.L.; Luo, T.; Luo, M. The Hippo signalling pathway and its implications in human health and diseases. Signal Transduct. Target Ther. 2022, 7, 376. [Google Scholar] [CrossRef] [PubMed]
- Moya, I.M.; Halder, G. Hippo-YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat. Rev. Mol. Cell Biol. 2019, 20, 211–226. [Google Scholar] [CrossRef] [PubMed]
- Yimlamai, D.; Christodoulou, C.; Galli, G.G.; Yanger, K.; Pepe-Mooney, B.; Gurung, B.; Shrestha, K.; Cahan, P.; Stanger, B.Z.; Camargo, F.D. Hippo pathway activity influences liver cell fate. Cell 2014, 157, 1324–1338. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Lu, L.; Yanger, K.; Wang, W.; Sohn, B.H.; Stanger, B.Z.; Zhang, M.; Martin, J.F.; Ajani, J.A.; Chen, J.; et al. Large tumor suppressor homologs 1 and 2 regulate mouse liver progenitor cell proliferation and maturation through antagonism of the coactivators YAP and TAZ. Hepatology 2016, 64, 1757–1772. [Google Scholar] [CrossRef] [PubMed]
- Shao, C.; Jing, Y.; Zhao, S.; Yang, X.; Hu, Y.; Meng, Y.; Huang, Y.; Ye, F.; Gao, L.; Liu, W.; et al. LPS/Bcl3/YAP1 signaling promotes Sox9(+)HNF4α(+) hepatocyte-mediated liver regeneration after hepatectomy. Cell Death Dis. 2022, 13, 277. [Google Scholar] [CrossRef]
- Zhang, Y.; Beachy, P.A. Cellular and molecular mechanisms of Hedgehog signalling. Nat. Rev. Mol. Cell Biol. 2023, 24, 668–687. [Google Scholar] [CrossRef]
- Briscoe, J.; Thérond, P.P. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 2013, 14, 416–429. [Google Scholar] [CrossRef] [PubMed]
- Rangwala, F.; Guy, C.D.; Lu, J.; Suzuki, A.; Burchette, J.L.; Abdelmalek, M.F.; Chen, W.; Diehl, A.M. Increased production of sonic hedgehog by ballooned hepatocytes. J. Pathol. 2011, 224, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, T. Hedgehog pathway, cell cycle, and primary cilium. Cell Death Discov. 2025, 11, 302. [Google Scholar] [CrossRef]
- Sicklick, J.K.; Li, Y.X.; Melhem, A.; Schmelzer, E.; Zdanowicz, M.; Huang, J.; Caballero, M.; Fair, J.H.; Ludlow, J.W.; McClelland, R.E.; et al. Hedgehog signaling maintains resident hepatic progenitors throughout life. Am. J. Physiol. Gastrointest. Liver. Physiol. 2006, 290, G859–G870. [Google Scholar] [CrossRef]
- Jung, Y.; McCall, S.J.; Li, Y.X.; Diehl, A.M. Bile ductules and stromal cells express hedgehog ligands and/or hedgehog target genes in primary biliary cirrhosis. Hepatology 2007, 45, 1091–1096. [Google Scholar] [CrossRef] [PubMed]
- Fleig, S.V.; Choi, S.S.; Yang, L.; Jung, Y.; Omenetti, A.; VanDongen, H.M.; Huang, J.; Sicklick, J.K.; Diehl, A.M. Hepatic accumulation of Hedgehog-reactive progenitors increases with severity of fatty liver damage in mice. Lab Investig. 2007, 87, 1227–1239. [Google Scholar] [CrossRef]
- Ochoa, B.; Syn, W.K.; Delgado, I.; Karaca, G.F.; Jung, Y.; Wang, J.; Zubiaga, A.M.; Fresnedo, O.; Omenetti, A.; Zdanowicz, M.; et al. Hedgehog signaling is critical for normal liver regeneration after partial hepatectomy in mice. Hepatology 2010, 51, 1712–1723. [Google Scholar] [CrossRef]
- Swiderska-Syn, M.; Xie, G.; Michelotti, G.A.; Jewell, M.L.; Premont, R.T.; Syn, W.K.; Diehl, A.M. Hedgehog regulates yes-associated protein 1 in regenerating mouse liver. Hepatology 2016, 64, 232–244. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Chu, Q.; Shi, Q.; Zeng, Y.; Lu, J.; Li, L. Wnt signaling pathways in biology and disease: Mechanisms and therapeutic advances. Signal Transduct. Target Ther. 2025, 10, 106. [Google Scholar] [CrossRef]
- Liu, J.; Xiao, Q.; Xiao, J.; Niu, C.; Li, Y.; Zhang, X.; Zhou, Z.; Shu, G.; Yin, G. Wnt/β-catenin signalling: Function, biological mechanisms, and therapeutic opportunities. Signal Transduct. Target Ther. 2022, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Nejak-Bowen, K.; Monga, S.P. Wnt-β-catenin in hepatobiliary homeostasis, injury, and repair. Hepatology 2023, 78, 1907–1921. [Google Scholar] [CrossRef] [PubMed]
- Monga, S.P.; Pediaditakis, P.; Mule, K.; Stolz, D.B.; Michalopoulos, G.K. Changes in WNT/beta-catenin pathway during regulated growth in rat liver regeneration. Hepatology 2001, 33, 1098–1109. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.D.; Awuah, P.; Singh, S.; Monga, S.P. Disparate cellular basis of improved liver repair in beta-catenin-overexpressing mice after long-term exposure to 3,5-diethoxycarbonyl-1,4-dihydrocollidine. Am. J. Pathol. 2010, 177, 1812–1822. [Google Scholar] [CrossRef] [PubMed]
- Okabe, H.; Yang, J.; Sylakowski, K.; Yovchev, M.; Miyagawa, Y.; Nagarajan, S.; Chikina, M.; Thompson, M.; Oertel, M.; Baba, H.; et al. Wnt signaling regulates hepatobiliary repair following cholestatic liver injury in mice. Hepatology 2016, 64, 1652–1666. [Google Scholar] [CrossRef]
- Murakami, M.; Kamimura, D.; Hirano, T. Pleiotropy and Specificity: Insights from the Interleukin 6 Family of Cytokines. Immunity 2019, 50, 812–831. [Google Scholar] [CrossRef]
- Guo, R.; Jiang, M.; Wang, G.; Li, B.; Jia, X.; Ai, Y.; Chen, S.; Tang, P.; Liu, A.; Yuan, Q.; et al. IL6 supports long-term expansion of hepatocytes in vitro. Nat. Commun. 2022, 13, 7345. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Cui, L.; Lin, P.; Liu, Z.; Bao, S.; Ma, X.; Nan, H.; Zhu, W.; Cen, J.; Mao, Y.; et al. Kupffer-cell-derived IL-6 is repurposed for hepatocyte dedifferentiation via activating progenitor genes from injury-specific enhancers. Cell Stem Cell 2023, 30, 283–299.e289. [Google Scholar] [CrossRef]
- Wang, Z.; Du, K.; Jin, N.; Tang, B.; Zhang, W. Macrophage in liver Fibrosis: Identities and mechanisms. Int. Immunopharmacol. 2023, 120, 110357. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, A.; Shepherd, E.L.; Amatucci, A.; Munir, M.; Reynolds, G.; Humphreys, E.; Resheq, Y.; Adams, D.H.; Hübscher, S.; Burkly, L.C.; et al. Interaction of TWEAK with Fn14 leads to the progression of fibrotic liver disease by directly modulating hepatic stellate cell proliferation. J. Pathol. 2016, 239, 109–121. [Google Scholar] [CrossRef]
- Karaca, G.; Swiderska-Syn, M.; Xie, G.; Syn, W.K.; Krüger, L.; Machado, M.V.; Garman, K.; Choi, S.S.; Michelotti, G.A.; Burkly, L.C.; et al. TWEAK/Fn14 signaling is required for liver regeneration after partial hepatectomy in mice. PLoS ONE 2014, 9, e83987. [Google Scholar] [CrossRef]
- Affò, S.; Dominguez, M.; Lozano, J.J.; Sancho-Bru, P.; Rodrigo-Torres, D.; Morales-Ibanez, O.; Moreno, M.; Millán, C.; Loaeza-del-Castillo, A.; Altamirano, J.; et al. Transcriptome analysis identifies TNF superfamily receptors as potential therapeutic targets in alcoholic hepatitis. Gut 2013, 62, 452–460. [Google Scholar] [CrossRef]
- Banales, J.M.; Huebert, R.C.; Karlsen, T.; Strazzabosco, M.; LaRusso, N.F.; Gores, G.J. Cholangiocyte pathobiology. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 269–281. [Google Scholar] [CrossRef]
- Lou, C.; Lan, T.; Xu, S.; Hu, X.; Li, J.; Xiang, Z.; Lin, S.; Fan, X.; Chen, J.; Xu, X. Heterogeneity and plasticity of cholangiocytes in liver injury: A journey from pathophysiology to therapeutic utility. Gut 2025. [Google Scholar] [CrossRef]
- Maroni, L.; Haibo, B.; Ray, D.; Zhou, T.; Wan, Y.; Meng, F.; Marzioni, M.; Alpini, G. Functional and structural features of cholangiocytes in health and disease. Cell Mol. Gastroenterol. Hepatol. 2015, 1, 368–380. [Google Scholar] [CrossRef]
- Petersen, B.E.; Zajac, V.F.; Michalopoulos, G.K. Bile ductular damage induced by methylene dianiline inhibits oval cell activation. Am. J. Pathol. 1997, 151, 905–909. [Google Scholar] [PubMed]
- Tarlow, B.D.; Finegold, M.J.; Grompe, M. Clonal tracing of Sox9+ liver progenitors in mouse oval cell injury. Hepatology 2014, 60, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Pu, W.; Zhu, H.; Zhang, M.; Pikiolek, M.; Ercan, C.; Li, J.; Huang, X.; Han, X.; Zhang, Z.; Lv, Z.; et al. Bipotent transitional liver progenitor cells contribute to liver regeneration. Nat. Genet. 2023, 55, 651–664. [Google Scholar] [CrossRef] [PubMed]
- Choi, T.Y.; Ninov, N.; Stainier, D.Y.; Shin, D. Extensive conversion of hepatic biliary epithelial cells to hepatocytes after near total loss of hepatocytes in zebrafish. Gastroenterology 2014, 146, 776–788. [Google Scholar] [CrossRef] [PubMed]
- Eski, S.E.; Mi, J.; Pozo-Morales, M.; Hovhannisyan, G.G.; Perazzolo, C.; Manco, R.; Ez-Zammoury, I.; Barbhaya, D.; Lefort, A.; Libert, F.; et al. Cholangiocytes contribute to hepatocyte regeneration after partial liver injury during growth spurt in zebrafish. Nat. Commun. 2025, 16, 5260. [Google Scholar] [CrossRef]
- He, J.; Lu, H.; Zou, Q.; Luo, L. Regeneration of liver after extreme hepatocyte loss occurs mainly via biliary transdifferentiation in zebrafish. Gastroenterology 2014, 146, 789–800.e788. [Google Scholar] [CrossRef] [PubMed]
- Cai, P.; Ni, R.; Lv, M.; Liu, H.; Zhao, J.; He, J.; Luo, L. VEGF signaling governs the initiation of biliary-mediated liver regeneration through the PI3K-mTORC1 axis. Cell Rep. 2023, 42, 113028. [Google Scholar] [CrossRef] [PubMed]
- Schwabe, R.F.; Brenner, D.A. Hepatic stellate cells: Balancing homeostasis, hepatoprotection and fibrogenesis in health and disease. Nat. Rev. Gastroenterol. Hepatol. 2025, 22, 481–499. [Google Scholar] [CrossRef]
- Trivedi, P.; Wang, S.; Friedman, S.L. The Power of Plasticity-Metabolic Regulation of Hepatic Stellate Cells. Cell Metab. 2021, 33, 242–257. [Google Scholar] [CrossRef]
- Tsuchida, T.; Friedman, S.L. Mechanisms of hepatic stellate cell activation. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 397–411. [Google Scholar] [CrossRef]
- Hou, W.; Syn, W.K. Role of Metabolism in Hepatic Stellate Cell Activation and Fibrogenesis. Front. Cell Dev. Biol. 2018, 6, 150. [Google Scholar] [CrossRef] [PubMed]
- De Smet, V.; Eysackers, N.; Merens, V.; Kazemzadeh Dastjerd, M.; Halder, G.; Verhulst, S.; Mannaerts, I.; van Grunsven, L.A. Initiation of hepatic stellate cell activation extends into chronic liver disease. Cell Death Dis. 2021, 12, 1110. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wan, L.; Jin, X.; Wang, W.; Li, D. Transforming growth factor-β signaling confers hepatic stellate cells progenitor features after partial hepatectomy. J. Cell Physiol. 2020, 235, 2655–2667. [Google Scholar] [CrossRef]
- Peng, D.; Fu, M.; Wang, M.; Wei, Y.; Wei, X. Targeting TGF-β signal transduction for fibrosis and cancer therapy. Mol. Cancer 2022, 21, 104. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Lin, W.; Long, Y.; Yang, Y.; Zhang, H.; Wu, K.; Chu, Q. Notch signaling pathway: Architecture, disease, and therapeutics. Signal Transduct. Target Ther. 2022, 7, 95. [Google Scholar] [CrossRef] [PubMed]
- Gallenstein, N.; Tichy, L.; Weigand, M.A.; Schenz, J. Notch Signaling in Acute Inflammation and Sepsis. Int. J. Mol. Sci. 2023, 24, 3458. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Zhou, Y.; Hu, T.; Zhang, H.; Shen, M.; Cheng, P.; Dai, W.; Wang, F.; Chen, K.; Zhang, Y.; et al. Notch Signaling Coordinates Progenitor Cell-Mediated Biliary Regeneration Following Partial Hepatectomy. Sci. Rep. 2016, 6, 22754. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Chen, Y.; Fu, G.B.; Yuan, T.J.; Huang, W.J.; Wang, Z.Y.; Li, W.J.; Jiao, Y.F.; Yu, W.F.; Yan, H.X. EpCAM inhibits differentiation of human liver progenitor cells into hepatocytes in vitro by activating Notch1 signaling. Biochem. Biophys. Res. Commun. 2020, 525, 238–243. [Google Scholar] [CrossRef] [PubMed]
- Kitade, M.; Factor, V.M.; Andersen, J.B.; Tomokuni, A.; Kaji, K.; Akita, H.; Holczbauer, A.; Seo, D.; Marquardt, J.U.; Conner, E.A.; et al. Specific fate decisions in adult hepatic progenitor cells driven by MET and EGFR signaling. Genes Dev. 2013, 27, 1706–1717. [Google Scholar] [CrossRef]
- Boulter, L.; Govaere, O.; Bird, T.G.; Radulescu, S.; Ramachandran, P.; Pellicoro, A.; Ridgway, R.A.; Seo, S.S.; Spee, B.; Van Rooijen, N.; et al. Macrophage-derived Wnt opposes Notch signaling to specify hepatic progenitor cell fate in chronic liver disease. Nat. Med. 2012, 18, 572–579. [Google Scholar] [CrossRef]
- Williams, J.M.; Oh, S.H.; Jorgensen, M.; Steiger, N.; Darwiche, H.; Shupe, T.; Petersen, B.E. The role of the Wnt family of secreted proteins in rat oval “stem” cell-based liver regeneration: Wnt1 drives differentiation. Am. J. Pathol. 2010, 176, 2732–2742. [Google Scholar] [CrossRef]
- Ishikawa, T.; Factor, V.M.; Marquardt, J.U.; Raggi, C.; Seo, D.; Kitade, M.; Conner, E.A.; Thorgeirsson, S.S. Hepatocyte growth factor/c-met signaling is required for stem-cell-mediated liver regeneration in mice. Hepatology 2012, 55, 1215–1226. [Google Scholar] [CrossRef]
- Organ, S.L.; Tsao, M.S. An overview of the c-MET signaling pathway. Ther. Adv. Med. Oncol. 2011, 3, S7–S19. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Su, X.; Li, Z.; Deng, L.; Liu, X.; Feng, X.; Peng, J. HGF/c-MET pathway in cancer: From molecular characterization to clinical evidence. Oncogene 2021, 40, 4625–4651. [Google Scholar] [CrossRef]
- Hasuike, S.; Ido, A.; Uto, H.; Moriuchi, A.; Tahara, Y.; Numata, M.; Nagata, K.; Hori, T.; Hayashi, K.; Tsubouchi, H. Hepatocyte growth factor accelerates the proliferation of hepatic oval cells and possibly promotes the differentiation in a 2-acetylaminofluorene/partial hepatectomy model in rats. J. Gastroenterol. Hepatol. 2005, 20, 1753–1761. [Google Scholar] [CrossRef]
- Wang, P.; Liu, T.; Cong, M.; Wu, X.; Bai, Y.; Yin, C.; An, W.; Wang, B.; Jia, J.; You, H. Expression of extracellular matrix genes in cultured hepatic oval cells: An origin of hepatic stellate cells through transforming growth factor beta? Liver. Int. 2009, 29, 575–584. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhang, X.; Xu, Y.; Li, X.; Ren, S.; Zhou, Y.; Duan, Y.; Zern, M.; Zhang, H.; Chen, G.; et al. Hepatic Progenitor Cells Contribute to the Progression of 2-Acetylaminofluorene/Carbon Tetrachloride-Induced Cirrhosis via the Non-Canonical Wnt Pathway. PLoS ONE 2015, 10, e0130310. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.R.; Jing, Y.Y.; Liu, W.T.; Han, Z.P.; Li, R.; Yang, Y.; Zhu, J.N.; Li, X.Y.; Li, P.P.; Wei, L.X. Lipopolysaccharide induces the differentiation of hepatic progenitor cells into myofibroblasts via activation of the Hedgehog signaling pathway. Cell Cycle 2017, 16, 1357–1365. [Google Scholar] [CrossRef] [PubMed]
- Passino, M.A.; Adams, R.A.; Sikorski, S.L.; Akassoglou, K. Regulation of hepatic stellate cell differentiation by the neurotrophin receptor p75NTR. Science 2007, 315, 1853–1856. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.; Russell, J.O.; Molina, L.M.; Monga, S.P. Liver Progenitors and Adult Cell Plasticity in Hepatic Injury and Repair: Knowns and Unknowns. Annu. Rev. Pathol. 2020, 15, 23–50. [Google Scholar] [CrossRef] [PubMed]
- Forbes, S.J.; Newsome, P.N. Liver regeneration—Mechanisms and models to clinical application. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 473–485. [Google Scholar] [CrossRef]
- Liu, C.; Wang, K.; Mei, J.; Zhao, R.; Shen, J.; Zhang, W.; Li, L.; Roy, B.; Fang, X. Integrative single-cell and spatial transcriptome analysis reveals heterogeneity of human liver progenitor cells. Hepatol. Commun. 2025, 9, e0662. [Google Scholar] [CrossRef] [PubMed]
- Holland, C.H.; Ramirez Flores, R.O.; Myllys, M.; Hassan, R.; Edlund, K.; Hofmann, U.; Marchan, R.; Cadenas, C.; Reinders, J.; Hoehme, S.; et al. Transcriptomic Cross-Species Analysis of Chronic Liver Disease Reveals Consistent Regulation Between Humans and Mice. Hepatol. Commun. 2022, 6, 161–177. [Google Scholar] [CrossRef]
- Matchett, K.P.; Wilson-Kanamori, J.R.; Portman, J.R.; Kapourani, C.A.; Fercoq, F.; May, S.; Zajdel, E.; Beltran, M.; Sutherland, E.F.; Mackey, J.B.G.; et al. Multimodal decoding of human liver regeneration. Nature 2024, 630, 158–165. [Google Scholar] [CrossRef] [PubMed]

| Source | Species | Injury | Timing | Signaling | Validation Method |
|---|---|---|---|---|---|
| Hepatocyte | Human | Alcoholic hepatitis | ABIC score ≥ 6.71 points | TWEAK/Fn14 | Liver biopsy |
| Mouse | 70% PHx | 24, 48 h after PHx | TWEAK/Fn14 | Systemic Fn14 deletion | |
| 3 h after PHx | Hippo | Systemic Bcl3 deletion | |||
| 12, 24, 48 h after PHx | Hedgehog | Immunohistochemistry and Western blot | |||
| 12, 24, 48 h after PHx | Hedgehog | Isolated pHeps from HSC-specific Smo deleted mice | |||
| 0.1% DDC | 150 days of DDC diet | Wnt/β-catenin | Hepatocyte-specific β-catenin overexpression | ||
| 2 wks or 1 month of DDC diet | Wnt/β-catenin | Liver-specific Wls or Lrp5/6 deletion | |||
| 1 wks of DDC diet | IL-6 | Hepatocyte-specific IL-6 deletion | |||
| Cholangiocyte | Mouse | 70% PHx | 7 days after PHx | - | HNF1β lineage tracing |
| MCD and 50% PHx | 7–14 days of MCD diet; 48 h after PHx | - | CK19 lineage tracing and hepatocyte-specific p21 overexpression | ||
| 0.1% DDC | 14 days for recovery after 12 days of DDC diet | - | CK19 lineage tracing and hepatocyte-specific β1 integrin deletion | ||
| 3 wks of DDC diet | Notch | CK19 lineage tracing and systemic Fah deletion | |||
| 4 wks of DDC diet | - | SOX9 lineage tracing | |||
| CDE | 2 wks for recovery after 3 wks of CDE diet | - | SOX9 lineage tracing | ||
| CCl4 | 1 wks for recovery after 5 wks of CCl4 | - | SOX9 lineage tracing | ||
| Zebrafish | Hepatocyte ablation | 8 and 24 h after hepatocyte ablation | - | NTR-Mtz system | |
| 24 h after hepatocyte ablation | - | NTR-Mtz system | |||
| HSC | Mouse | 70% PHx | 24, 48, 72 h after PHx | Hedgehog | SMO lineage tracing |
| 1, 3 days after PHx | TGF-β | Immunohistochemistry |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, J.; Sung, A.; Jeong, H.; Jung, Y. Liver Progenitor Cells: Cellular Origins, Plasticity, and Signaling Pathways in Liver Regeneration. Biology 2025, 14, 1361. https://doi.org/10.3390/biology14101361
Han J, Sung A, Jeong H, Jung Y. Liver Progenitor Cells: Cellular Origins, Plasticity, and Signaling Pathways in Liver Regeneration. Biology. 2025; 14(10):1361. https://doi.org/10.3390/biology14101361
Chicago/Turabian StyleHan, Jinsol, Ahyeon Sung, Hayeong Jeong, and Youngmi Jung. 2025. "Liver Progenitor Cells: Cellular Origins, Plasticity, and Signaling Pathways in Liver Regeneration" Biology 14, no. 10: 1361. https://doi.org/10.3390/biology14101361
APA StyleHan, J., Sung, A., Jeong, H., & Jung, Y. (2025). Liver Progenitor Cells: Cellular Origins, Plasticity, and Signaling Pathways in Liver Regeneration. Biology, 14(10), 1361. https://doi.org/10.3390/biology14101361






