Roles of Toll-like Receptor Signaling in Inflammatory Bone Resorption
Abstract
Simple Summary
Abstract
1. Introduction
2. Prostaglandin E2 Is a Primary Mediator of Inflammatory Bone Resorption
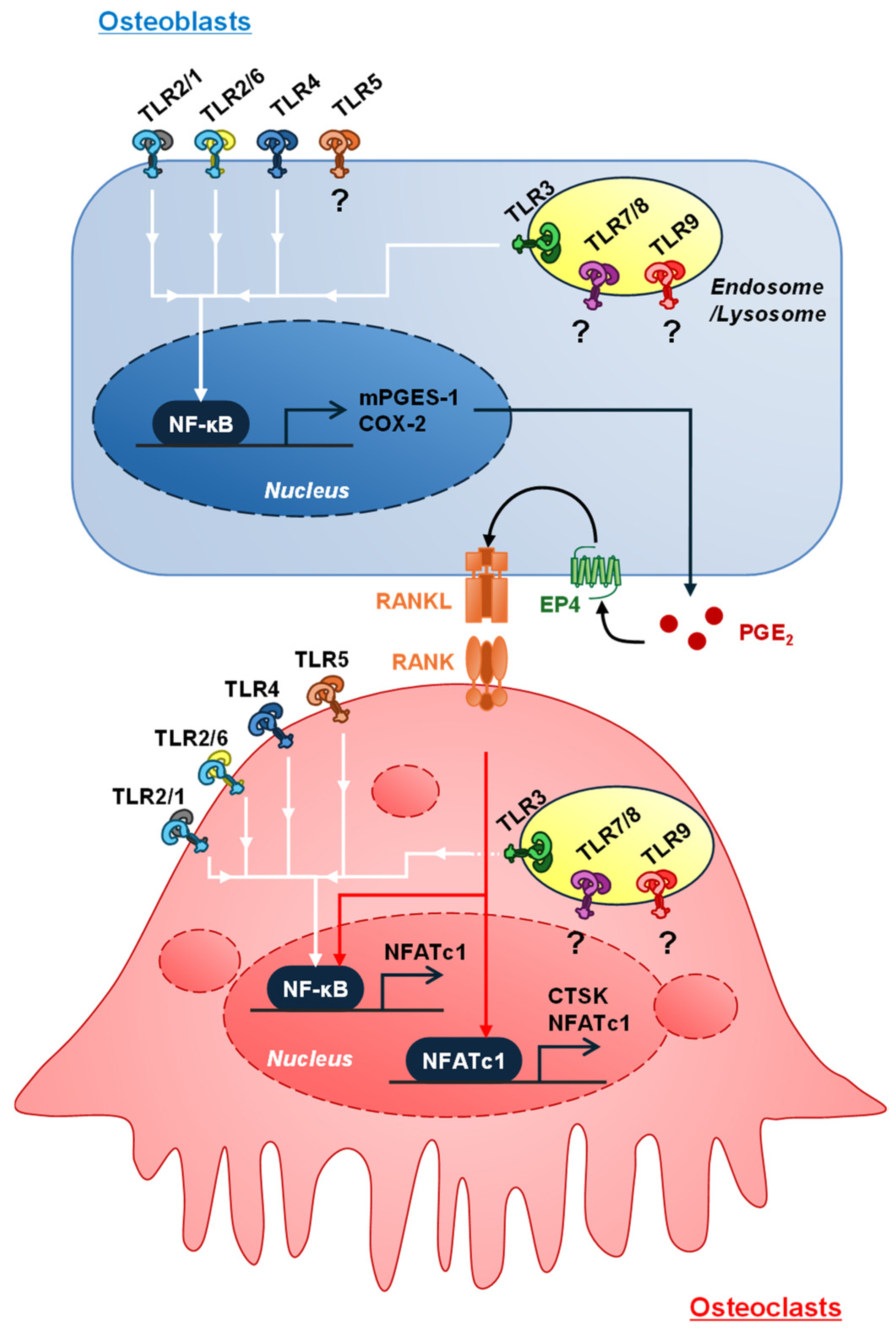
3. Roles of TLR Signaling in Osteoclast Differentiation
3.1. Exogenous and Endogenous Ligands for TLRs
3.2. Roles of Cell Surface and Intracellular TLR4 for LPS from Gram-Negative Bacteria
3.3. Cell Surface TLR2 Heterodimers as Receptors for Bacterial Cell-Wall Components from Gram-Positive and Gram-Negative Bacteria
3.4. Cell Surface TLR5, a Receptor for Flagellum from Gram-Positive and G-Negative Bacteria
3.5. Intracellular TLR3, 7, and 9 as Receptors for Nucleic Acids Including Endogenous and Exogenous RNA and DNA
3.6. TLR Ligands Bind TLRs and Coordinate the Inflammatory Signaling with Other Family of Cytosolic Sensor
3.7. Roles of Immune Cells Activated by TLR Ligands in Inflammatory Bone Resorption
4. Conclusions
5. Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Abbreviation or Acronym | Name |
| 1,25(OH)2D3 | 1,25-dihydroxyvitamin D3 |
| AA | Arachidonic acid |
| AGE | Advanced glycation end product |
| A. Actinomycetemcomitans | Aggregatibacter actinomycetemcomitans |
| Anti-MP | Antimicrobial peptide |
| AP-1 | Activator protein-1 |
| BMC | Bone marrow cell |
| BMD | Bone mineral density |
| CD | Cluster of differentiation |
| cGAS | Cyclic GMP-AMP synthase |
| COX | Cyclooxygenase |
| ODN | Oligodeoxynucleotide |
| DAMP | Damage-associated molecular pattern |
| DAP12 | DNAX activating protein 12 kDa |
| dsRNA | Double strand RNA |
| E. coli | Escherichia coli |
| EP | Prostaglandin E2 receptor |
| FLS | Fibroblast-like synoviocyte |
| FSL-1 | Fibroblast-stimulating lipopeptide |
| GEO | Gene Expression Omnibus |
| HMGB1 | High mobility group box 1 |
| HSP | Heat shock protein |
| IFN | Interferon |
| IκBα | Inhibitor of NF-κB α |
| IKK | IκB kinase |
| IL | Interleukin |
| IRAK | IL-1 receptor-associated kinase |
| IRF | Interferon regulatory factor |
| LDL | Low-density lipoprotein |
| LL37 | Cathelicidin-related antimicrobial peptide |
| LPS | Lipopolysaccharide |
| LTA | Lipoteichoic acid |
| MAMP | Microbe-associated molecular pattern |
| MALP2 | Macrophage-activating lipopeptide 2 |
| MAPK | Mitogen-activated protein kinase |
| M-CSF | Macrophage colony-stimulating factor |
| MD2 | Myeloid differentiation protein 2 |
| MDA5 | Melanoma differentiation-associated gene 5 |
| mPGES-1 | Membrane-bound PGE synthase |
| MyD88 | Myeloid differentiation factor 88 |
| NFATc1 | Nuclear factor of activated T cells, cytosolic 1 |
| NET | Neutrophil extracellular trap |
| NF-κB | Nuclear factor-κB |
| NLRC | NOD-like receptor family caspase recruitment domain-containing protein 4 |
| NLRP | NOD-like receptor protein 3 |
| NOD | Nucleotide oligomerization domain |
| NOX | NADPH oxidase |
| OMV | Bacterial outer membrane vesicle |
| OPG | Osteoprotegerin |
| PAMP | Pathogen-associated molecular pattern |
| PG | Prostaglandin |
| PI3K | Phosphatidylinositol-3 kinase |
| PLA2 | Phospholipase A2 |
| P. gingivalis | Porphyromonas gingivalis |
| PRR | Pattern recognition receptor |
| RA | Rheumatoid arthritis |
| RANK | Receptor activator of NF-κB |
| RANKL | Receptor activator of NF-κB ligand |
| RIG-I | Retinoic acid-inducible gene-I |
| RIP | Receptor interacting protein |
| snRNA | Small nuclear RNA |
| ssRNA | Single-strand RNA |
| STING | Stimulator of interferon genes |
| TIRAP | Toll/interleukin-1 receptor-domain-containing adapter protein |
| TLR | Toll-like receptor |
| TNF | Tumor necrosis factor |
| TRAF | TNF receptor-associated factor |
| TRAM | TRIF-related adaptor molecule |
| TRIF | Toll/interleukin-1 receptor-domain-containing adapter-inducing interferon-β |
References
- Duan, T.; Du, Y.; Xing, C.; Wang, H.Y.; Wang, R.F. Toll-Like Receptor Signaling and Its Role in Cell-Mediated Immunity. Front. Immunol. 2022, 13, 812774. [Google Scholar] [CrossRef]
- Fitzgerald, K.A.; Kagan, J.C. Toll-like Receptors and the Control of Immunity. Cell 2020, 180, 1044–1066. [Google Scholar] [CrossRef] [PubMed]
- Bolamperti, S.; Villa, I.; Rubinacci, A. Bone remodeling: An operational process ensuring survival and bone mechanical competence. Bone Res. 2022, 10, 48. [Google Scholar] [CrossRef]
- Roodman, G.D. Regulation of osteoclast differentiation. Ann. N. Y. Acad. Sci. 2006, 1068, 100–109. [Google Scholar] [CrossRef]
- Moriishi, T.; Fukuyama, R.; Ito, M.; Miyazaki, T.; Maeno, T.; Kawai, Y.; Komori, H.; Komori, T. Osteocyte network; a negative regulatory system for bone mass augmented by the induction of Rankl in osteoblasts and Sost in osteocytes at unloading. PLoS ONE 2012, 7, e40143. [Google Scholar] [CrossRef]
- Park, J.H.; Lee, N.K.; Lee, S.Y. Current Understanding of RANK Signaling in Osteoclast Differentiation and Maturation. Mol. Cells. 2017, 40, 706–713. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Wang, L.; Chen, S. Endogenous Toll-like Receptor Ligands and Their Biological Significance. J. Cell. Mol. Med. 2010, 14, 2592–2603. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Zhao, Y. Toll-like Receptors and Immune Regulation: Their Direct and Indirect Modulation on Regulatory CD4+ CD25+ T Cells. Immunology 2007, 122, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Reuter, S.; Herold, K.; Domroes, J.; Mrowka, R.; Schön, A. Toll-Like Receptor 5 as a Novel Receptor for Fungal Zymosan. Heliyon 2023. preprint. Available online: https://www.biorxiv.org/content/10.1101/2021.12.23.473960v1 (accessed on 29 July 2024). [CrossRef]
- McCoy, J.M.; Wicks, J.R.; Audoly, L.P. The Role of Prostaglandin E2 Receptors in the Pathogenesis of Rheumatoid Arthritis. J. Clin. Investig. 2002, 110, 651–658. [Google Scholar] [CrossRef]
- Inada, M.; Matsumoto, C.; Uematsu, S.; Akira, S.; Miyaura, C. Membrane-Bound Prostaglandin E Synthase-1-Mediated Prostaglandin E2 Production by Osteoblast Plays a Critical Role in Lipopolysaccharide-Induced Bone Loss Associated with Inflammation. J. Immunol. 2006, 177, 1879–1885. [Google Scholar] [CrossRef]
- Jiang, W.; Jin, Y.; Zhang, S.; Ding, Y.; Huo, K.; Yang, J.; Zhao, L.; Nian, B.; Zhong, T.P.; Lu, W.; et al. PGE2 Activates EP4 in Subchondral Bone Osteoclasts to Regulate Osteoarthritis. Bone Res. 2022, 10, 27. [Google Scholar] [CrossRef]
- Båge, T.; Kats, A.; Lopez, B.S.; Morgan, G.; Nilsson, G.; Burt, I.; Korotkova, M.; Corbett, L.; Knox, A.J.; Pino, L.; et al. Expression of Prostaglandin E Synthases in Periodontitis Immunolocalization and Cellular Regulation. Am. J. Pathol. 2011, 178, 1676–1688. [Google Scholar] [CrossRef] [PubMed]
- Suzawa, T.; Miyaura, C.; Inada, M.; Maruyama, T.; Sugimoto, Y.; Ushikubi, F.; Ichikawa, A.; Narumiya, S.; Suda, T. The Role of Prostaglandin E Receptor Subtypes (EP1, EP2, EP3, and EP4) in Bone Resorption: An Analysis Using Specific Agonists for the Respective EPs. Endocrinology 2000, 141, 1554–1559. [Google Scholar] [CrossRef] [PubMed]
- Miyaura, C.; Inada, M.; Suzawa, T.; Sugimoto, Y.; Ushikubi, F.; Ichikawa, A.; Narumiya, S.; Suda, T. Impaired Bone Resorption to Prostaglandin E2 in Prostaglandin E Receptor EP4-Knockout Mice. J. Biol. Chem. 2000, 275, 19819–19823. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Mizoguchi, T.; Take, I.; Kurihara, S.; Udagawa, N.; Takahashi, N. Prostaglandin E2 Enhances Osteoclastic Differentiation of Precursor Cells through Protein Kinase A-Dependent Phosphorylation of TAK1. J. Biol. Chem. 2005, 280, 11395–11403. [Google Scholar] [CrossRef] [PubMed]
- Kaji, H.; Sugimoto, T.; Kanatani, M.; Fukase, M.; Kumegawa, M.; Chihara, K. Prostaglandin E2 Stimulates Osteoclast-like Cell Formation and Bone-resorbing Activity via Osteoblasts: Role of CAMP-dependent Protein Kinase. J. Bone Miner. Res. 1996, 11, 62–71. [Google Scholar] [CrossRef]
- Nyman, S.; Schroeder, H.E.; Lindhe, J. Suppression of Inflammation and Bone Resorption by Indomethacin During Experimental Periodontitis in Dogs. J. Periodontol. 1979, 50, 450–461. [Google Scholar] [CrossRef]
- Zubery, Y.; Dunstan, C.R.; Story, B.M.; Kesavalu, L.; Ebersole, J.L.; Holt, S.C.; Boyce, B.F. Bone Resorption Caused by Three Periodontal Pathogens In Vivo in Mice Is Mediated in Part by Prostaglandin. Infect. Immun. 1998, 66, 4158–4162. [Google Scholar] [CrossRef]
- Matsumoto, C.; Oda, T.; Yokoyama, S.; Tominari, T.; Hirata, M.; Miyaura, C.; Inada, M. Toll-like Receptor 2 Heterodimers, TLR2/6 and TLR2/1 Induce Prostaglandin E Production by Osteoblasts, Osteoclast Formation and Inflammatory Periodontitis. Biochem. Biophys. Res. Commun. 2012, 428, 110–115. [Google Scholar] [CrossRef]
- Tominari, T.; Sanada, A.; Ichimaru, R.; Matsumoto, C.; Hirata, M.; Itoh, Y.; Numabe, Y.; Miyaura, C.; Inada, M. Gram-Positive Bacteria Cell Wall-Derived Lipoteichoic Acid Induces Inflammatory Alveolar Bone Loss through Prostaglandin E Production in Osteoblasts. Sci. Rep. 2021, 11, 13353. [Google Scholar] [CrossRef] [PubMed]
- Tominari, T.; Akita, M.; Matsumoto, C.; Hirata, M.; Yoshinouchi, S.; Tanaka, Y.; Karouji, K.; Itoh, Y.; Maruyama, T.; Miyaura, C.; et al. Endosomal TLR3 Signaling in Stromal Osteoblasts Induces Prostaglandin E2–Mediated Inflammatory Periodontal Bone Resorption. J. Biol. Chem. 2022, 298, 101603. [Google Scholar] [CrossRef]
- Luo, G.; Li, F.; Li, X.; Wang, Z.G.; Zhang, B. TNF-α and RANKL Promote Osteoclastogenesis by Upregulating RANK via the NF-ΚB Pathway. Mol. Med. Rep. 2018, 17, 6605–6611. [Google Scholar] [CrossRef]
- Liao, R.; Feng, Z.; Li, W.; Liu, R.; Xu, X.; Yao, S.; Tian, J. Interleukin-1 Induces Receptor Activator of Nuclear Factor-ΚB Ligand-Independent Osteoclast Differentiation in RAW264.7 Cells. Exp. Ther. Med. 2021, 21, 640. [Google Scholar] [CrossRef]
- Takayanagi, H.; Kim, S.; Taniguchi, T. Signaling Crosstalk between RANKL and Interferons in Osteoclast Differentiation. Arthritis Res. Ther. 2002, 4, S227. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.K.; Kim, S.J.; Rah, S.H.; Kang, J.I.; Jung, H.E.; Lee, D.; Lee, H.K.; Lee, J.O.; Park, B.S.; Yoon, T.Y.; et al. Reconstruction of LPS Transfer Cascade Reveals Structural Determinants within LBP, CD14, and TLR4-MD2 for Efficient LPS Recognition and Transfer. Immunity 2017, 46, 38–50. [Google Scholar] [CrossRef]
- Marongiu, L.; Gornati, L.; Artuso, I.; Zanoni, I.; Granucci, F. Below the Surface: The Inner Lives of TLR4 and TLR9. J. Leukoc. Biol. 2019, 106, 147–160. [Google Scholar] [CrossRef]
- Miyaura, C.; Inada, M.; Matsumoto, C.; Ohshiba, T.; Uozumi, N.; Shimizu, T.; Ito, A. An Essential Role of Cytosolic Phospholipase A2α in Prostaglandin E2–Mediated Bone Resorption Associated with Inflammation. J. Exp. Med. 2003, 197, 1303–1310. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, C.; Ashida, N.; Yokoyama, S.; Tominari, T.; Hirata, M.; Ogawa, K.; Sugihara, M.; Yano, M.; Inada, M.; Miyaura, C. The Protective Effects of β-Cryptoxanthin on Inflammatory Bone Resorption in a Mouse Experimental Model of Periodontitis. Biosci. Biotechnol. Biochem. 2013, 77, 860–862. [Google Scholar] [CrossRef]
- Hirata, N.; Ichimaru, R.; Tominari, T.; Matsumoto, C.; Watanabe, K.; Taniguchi, K.; Hirata, M.; Ma, S.; Suzuki, K.; Grundler, F.M.W.; et al. Beta-Cryptoxanthin Inhibits Lipopolysaccharide-Induced Osteoclast Differentiation and Bone Resorption via the Suppression of Inhibitor of NF-ΚB Kinase Activity. Nutrients 2019, 11, 368. [Google Scholar] [CrossRef]
- Takeda, H.; Tominari, T.; Ichimaru, R.; Matsumoto, C.; Watanabe, K.; Hirata, M.; Inada, M.; Miyaura, C. Lutein, a Carotenoid, Inhibits Lipopolysaccharide-Induced Alveolar Bone Loss Associated with Inflammation in a Mouse Model of Periodontitis. Curr. Top. Biochem. Res. 2016, 17, 71–76. [Google Scholar]
- Tominari, T.; Matsumoto, C.; Watanabe, K.; Hirata, M.; Grundler, F.M.W.; Miyaura, C.; Inada, M. Epigallocatechin Gallate (EGCG) Suppresses Lipopolysaccharide-induced Inflammatory Bone Resorption, and Protects against Alveolar Bone Loss in Mice. FEBS Open Bio 2015, 5, 522–527. [Google Scholar] [CrossRef]
- Tominari, T.; Ichimaru, R.; Yoshinouchi, S.; Matsumoto, C.; Watanabe, K.; Hirata, M.; Grundler, F.M.W.; Inada, M.; Miyaura, C. Effects of O-methylated (−)-epigallocatechin Gallate (EGCG) on LPS-induced Osteoclastogenesis, Bone Resorption, and Alveolar Bone Loss in Mice. FEBS Open Bio 2017, 7, 1972–1981. [Google Scholar] [CrossRef]
- Tominari, T.; Hirata, M.; Matsumoto, C.; Inada, M.; Miyaura, C. Polymethoxy Flavonoids, Nobiletin and Tangeretin, Prevent Lipopolysaccharide-Induced Inflammatory Bone Loss in an Experimental Model for Periodontitis. J. Pharmacol. Sci. 2012, 119, 390–394. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, S.; Tominari, T.; Matsumoto, C.; Yoshinouchi, S.; Ichimaru, R.; Watanabe, K.; Hirata, M.; Grundler, F.M.W.; Miyaura, C.; Inada, M. Effects of Polymethoxyflavonoids on Bone Loss Induced by Estrogen Deficiency and by LPS-Dependent Inflammation in Mice. Pharmaceuticals 2018, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.; Hassan, F.; Tumurkhuu, G.; Dagvadorj, J.; Koide, N.; Naiki, Y.; Mori, I.; Yoshida, T.; Yokochi, T. Bacterial Lipopolysaccharide Induces Osteoclast Formation in RAW 264.7 Macrophage Cells. Biochem. Biophys. Res. Commun. 2007, 360, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Hou, G.Q.; Guo, C.; Song, G.H.; Fang, N.; Fan, W.J.; Chen, X.D.; Yuan, L.; Wang, Z.Q. Lipopolysaccharide (LPS) Promotes Osteoclast Differentiation and Activation by Enhancing the MAPK Pathway and COX-2 Expression in RAW264.7 Cells. Int. J. Mol. Med. 2013, 32, 503–510. [Google Scholar] [CrossRef]
- AlQranei, M.S.; Senbanjo, L.T.; Aljohani, H.; Hamza, T.; Chellaiah, M.A. Lipopolysaccharide- TLR-4 Axis Regulates Osteoclastogenesis Independent of RANKL/RANK Signaling. BMC Immunol. 2021, 22, 23. [Google Scholar] [CrossRef]
- Zou, W.; Bar-Shavit, Z. Dual Modulation of Osteoclast Differentiation by Lipopolysaccharide. J. Bone Miner. Res. 2002, 17, 1211–1218. [Google Scholar] [CrossRef]
- Takami, M.; Kim, N.; Rho, J.; Choi, Y. Stimulation by Toll-Like Receptors Inhibits Osteoclast Differentiation. J. Immunol. 2002, 169, 1516–1523. [Google Scholar] [CrossRef]
- Itoh, K.; Udagawa, N.; Kobayashi, K.; Suda, K.; Li, X.; Takami, M.; Okahashi, N.; Nishihara, T.; Takahashi, N. Lipopolysaccharide Promotes the Survival of Osteoclasts Via Toll-Like Receptor 4, but Cytokine Production of Osteoclasts in Response to Lipopolysaccharide Is Different from That of Macrophages. J. Immunol. 2003, 170, 3688–3695. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Han, J.; Xi, C.; Xie, J.; Feng, X.; Wang, C.; Mei, L.; Xiong, W. HMGB1 Regulates RANKL-Induced Osteoclastogenesis in a Manner Dependent on RAGE1. J. Bone Miner. Res. 2009, 23, 1084–1096. [Google Scholar] [CrossRef] [PubMed]
- Nishida, M.; Saegusa, J.; Tanaka, S.; Morinobu, A. S100A12 Facilitates Osteoclast Differentiation from Human Monocytes. PLoS ONE 2018, 13, e0204140. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, Y.; Okui, T.; Yoneda, T.; Ryumon, S.; Nakamura, T.; Kawai, H.; Kunisada, Y.; Ibaragi, S.; Masui, M.; Ono, K.; et al. High-Mobility Group Box 1 Induces Bone Destruction Associated with Advanced Oral Squamous Cancer via RAGE and TLR4. Biochem. Biophys. Res. Commun. 2020, 531, 422–430. [Google Scholar] [CrossRef]
- Das, N.; Dewan, V.; Grace, P.M.; Gunn, R.J.; Tamura, R.; Tzarum, N.; Watkins, L.R.; Wilson, I.A.; Yin, H. HMGB1 Activates Proinflammatory Signaling via TLR5 Leading to Allodynia. Cell Rep. 2016, 17, 1128–1140. [Google Scholar] [CrossRef]
- Buwitt-Beckmann, U.; Heine, H.; Wiesmüller, K.-H.; Jung, G.; Brock, R.; Akira, S.; Ulmer, A.J. TLR1- and TLR6-Independent Recognition of Bacterial Lipopeptides. J. Biol. Chem. 2006, 281, 9049–9057. [Google Scholar] [CrossRef] [PubMed]
- Yano, A.; Suzuki, K.; Yamamoto, M.; Yamada, S. Pam3CSK4, a TLR2 Agonist, Induces Osteoclastogenesis in RAW 264.7 Cells. Dent. Med. Res. 2012, 32, 181–188. [Google Scholar] [CrossRef]
- Dou, C.; Zhen, G.; Dan, Y.; Wan, M.; Limjunyawong, N.; Cao, X. Sialylation of TLR2 Initiates Osteoclast Fusion. Bone Res. 2022, 10, 24. [Google Scholar] [CrossRef]
- Ha, H.; Lee, J.H.; Kim, H.N.; Kwak, H.B.; Kim, H.M.; Lee, S.E.; Rhee, J.H.; Kim, H.H.; Lee, Z.H. Stimulation by TLR5 Modulates Osteoclast Differentiation through STAT1/IFN-β. J. Immunol. 2008, 180, 1382–1389. [Google Scholar] [CrossRef]
- Kassem, A.; Henning, P.; Kindlund, B.; Lindholm, C.; Lerner, U.H. TLR5, a Novel Mediator of Innate Immunity-induced Osteoclastogenesis and Bone Loss. FASEB J. 2015, 29, 4449–4460. [Google Scholar] [CrossRef]
- Chamberlain, N.D.; Vila, O.M.; Volin, M.V.; Volkov, S.; Pope, R.M.; Swedler, W.; Mandelin, A.M.; Shahrara, S. TLR5, a Novel and Unidentified Inflammatory Mediator in Rheumatoid Arthritis That Correlates with Disease Activity Score and Joint TNF-α Levels. J. Immunol. 2012, 189, 475–483. [Google Scholar] [CrossRef]
- Kim, S.; Chen, Z.; Chamberlain, N.D.; Essani, A.B.; Volin, M.V.; Amin, M.A.; Volkov, S.; Gravallese, E.M.; Arami, S.; Swedler, W.; et al. Ligation of TLR5 Promotes Myeloid Cell Infiltration and Differentiation into Mature Osteoclasts in Rheumatoid Arthritis and Experimental Arthritis. J. Immunol. 2014, 193, 3902–3913. [Google Scholar] [CrossRef]
- Demaria, O.; Pagni, P.P.; Traub, S.; de Gassart, A.; Branzk, N.; Murphy, A.J.; Valenzuela, D.M.; Yancopoulos, G.D.; Flavell, R.A.; Alexopoulou, L. TLR8 Deficiency Leads to Autoimmunity in Mice. J. Clin. Investig. 2010, 120, 3651–3662. [Google Scholar] [CrossRef]
- Kim, K.W.; Cho, M.L.; Oh, H.J.; Kim, H.R.; Kang, C.M.; Heo, Y.M.; Lee, S.H.; Kim, H.Y. TLR-3 Enhances Osteoclastogenesis through Upregulation of RANKL Expression from Fibroblast-like Synoviocytes in Patients with Rheumatoid Arthritis. Immunol. Lett. 2009, 124, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Karikó, K.; Ni, H.; Capodici, J.; Lamphier, M.; Weissman, D. MRNA Is an Endogenous Ligand for Toll-like Receptor 3. J. Biol. Chem. 2004, 279, 12542–12550. [Google Scholar] [CrossRef] [PubMed]
- Cavassani, K.A.; Ishii, M.; Wen, H.; Schaller, M.A.; Lincoln, P.M.; Lukacs, N.W.; Hogaboam, C.M.; Kunkel, S.L. TLR3 Is an Endogenous Sensor of Tissue Necrosis during Acute Inflammatory Events. J. Exp. Med. 2008, 205, 2609–2621. [Google Scholar] [CrossRef]
- Lee, J.; Shim, Y.; Seo, W.; Kim, M.; Choi, W.; Kim, H.; Kim, Y.E.; Yang, K.; Ryu, T.; Jeong, J.M.; et al. Mitochondrial Double-Stranded RNA in Exosome Promotes Interleukin-17 Production Through Toll-Like Receptor 3 in Alcohol-associated Liver Injury. Hepatology 2020, 72, 609–625. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, A.; Takami, M.; Matsumoto, A.; Mochizuki, A.; Yamada, T.; Tachi, K.; Shibuya, I.; Nakamachi, T.; Shioda, S.; Baba, K.; et al. R848, a Toll-like Receptor 7 Agonist, Inhibits Osteoclast Differentiation but Not Survival or Bone-Resorbing Function of Mature Osteoclasts. Cytotechnology 2012, 64, 331–339. [Google Scholar] [CrossRef]
- Suzuki, H.; Mochizuki, A.; Yoshimura, K.; Miyamoto, Y.; Kaneko, K.; Inoue, T.; Chikazu, D.; Takami, M.; Kamijo, R. Bropirimine Inhibits Osteoclast Differentiation through Production of Interferon-β. Biochem. Biophys. Res. Commun. 2015, 467, 146–151. [Google Scholar] [CrossRef]
- Alzabin, S.; Kong, P.; Medghalchi, M.; Palfreeman, A.; Williams, R.; Sacre, S. Investigation of the Role of Endosomal Toll-like Receptors in Murine Collagen-Induced Arthritis Reveals a Potential Role for TLR7 in Disease Maintenance. Arthritis Res. Ther. 2012, 14, R142. [Google Scholar] [CrossRef]
- Kim, K.W.; Kim, B.M.; Won, J.Y.; Lee, K.A.; Kim, H.R.; Lee, S.H. Toll-like Receptor 7 Regulates Osteoclastogenesis in Rheumatoid Arthritis. J. Biochem. 2019, 166, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Hegewald, A.B.; Breitwieser, K.; Ottinger, S.M.; Mobarrez, F.; Korotkova, M.; Rethi, B.; Jakobsson, P.-J.; Catrina, A.I.; Wähämaa, H.; Saul, M.J. Extracellular MiR-574-5p Induces Osteoclast Differentiation via TLR 7/8 in Rheumatoid Arthritis. Front. Immunol. 2020, 11, 585282. [Google Scholar] [CrossRef]
- Amcheslavsky, A.; Bar-Shavit, Z. Toll-Like Receptor 9 Ligand Blocks Osteoclast Differentiation Through Induction of Phosphatase. J. Bone Miner. Res. 2007, 22, 1301–1310. [Google Scholar] [CrossRef]
- Zou, W.; Schwartz, H.; Endres, S.; Hartmann, G.; Bar-Shavit, Z. CpG Oligonucleotides: Novel Regulators of Osteoclast Differentiation. FASEB J. 2002, 16, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Ding, P.; Tan, Q.; Wei, Z.; Chen, Q.; Wang, C.; Qi, L.; Wen, L.; Zhang, C.; Yao, C. Toll-like Receptor 9 Deficiency Induces Osteoclastic Bone Loss via Gut Microbiota-Associated Systemic Chronic Inflammation. Bone Res. 2022, 10, 42. [Google Scholar] [CrossRef] [PubMed]
- Kim, P.D.; Xia-Juan, X.; Crump, K.E.; Abe, T.; Hajishengallis, G.; Sahingur, S.E. Toll-Like Receptor 9-Mediated Inflammation Triggers Alveolar Bone Loss in Experimental Murine Periodontitis. Infect. Immun. 2015, 83, 2992–3002. [Google Scholar] [CrossRef]
- Vanaja, S.K.; Russo, A.J.; Behl, B.; Banerjee, I.; Yankova, M.; Deshmukh, S.D.; Rathinam, V.A.K. Bacterial Outer Membrane Vesicles Mediate Cytosolic Localization of LPS and Caspase-11 Activation. Cell 2016, 165, 1106–1119. [Google Scholar] [CrossRef]
- Hagar, J.A.; Powell, D.A.; Aachoui, Y.; Ernst, R.K.; Miao, E.A. Cytoplasmic LPS Activates Caspase-11: Implications in TLR4-Independent Endotoxic Shock. Science 2013, 341, 1250–1253. [Google Scholar] [CrossRef]
- Miao, E.A.; Mao, D.P.; Yudkovsky, N.; Bonneau, R.; Lorang, C.G.; Warren, S.E.; Leaf, I.A.; Aderem, A. Innate Immune Detection of the Type III Secretion Apparatus through the NLRC4 Inflammasome. Proc. Natl. Acad. Sci. USA 2010, 107, 3076–3080. [Google Scholar] [CrossRef]
- Zhao, Y.; Yang, J.; Shi, J.; Gong, Y.N.; Lu, Q.; Xu, H.; Liu, L.; Shao, F. The NLRC4 Inflammasome Receptors for Bacterial Flagellin and Type III Secretion Apparatus. Nature 2011, 477, 596–600. [Google Scholar] [CrossRef]
- Alippe, Y.; Wang, C.; Ricci, B.; Xiao, J.; Qu, C.; Zou, W.; Novack, D.V.; Abu-Amer, Y.; Civitelli, R.; Mbalaviele, G. Bone Matrix Components Activate the NLRP3 Inflammasome and Promote Osteoclast Differentiation. Sci. Rep. 2017, 7, 6630. [Google Scholar] [CrossRef]
- Alippe, Y.; Kress, D.; Ricci, B.; Sun, K.; Yang, T.; Wang, C.; Xiao, J.; Abu-Amer, Y.; Mbalaviele, G. Actions of the NLRP3 and NLRC4 Inflammasomes Overlap in Bone Resorption. FASEB J. 2021, 35, e21837. [Google Scholar] [CrossRef] [PubMed]
- Reikine, S.; Nguyen, J.B.; Modis, Y. Pattern Recognition and Signaling Mechanisms of RIG-I and MDA5. Front. Immunol. 2014, 5, 342. [Google Scholar] [CrossRef]
- Wang, X.; Lin, M.; Zhu, L.; Ye, Z. GAS-STING: A Classical DNA Recognition Pathways to Tumor Therapy. Front. Immunol. 2023, 14, 1200245. [Google Scholar] [CrossRef] [PubMed]
- MacLauchlan, S.; Kushwaha, P.; Tai, A.; Chen, S.; Manning, C.; Swarnkar, G.; Abu-Amer, Y.; Fitzgerald, K.A.; Sharma, S.; Gravallese, E.M. STING-Dependent Interferon Signatures Restrict Osteoclast Differentiation and Bone Loss in Mice. Proc. Natl. Acad. Sci. USA 2023, 120, e2210409120. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, Z.; Wang, Z.M. Differential immune cell infiltrations between healthy periodontal and chronic periodontitis tissues. BMC Oral Health 2020, 20, 293. [Google Scholar] [CrossRef] [PubMed]
- Chakravarti, A.; Raquil, M.A.; Tessier, P.; Poubelle, P.E. Surface RANKL of Toll-like receptor 4-stimulated human neutrophils activates osteoclastic bone resorption. Blood 2009, 114, 1633–1644. [Google Scholar] [CrossRef]
- Poli, C.; Martin, J.C.; Braudeau, C.; Bériou, G.; Hémont, C.; Charrier, C.; Guérin, S.; Heslan, M.; Josien, R. Receptor activating NF-κB ligand (RANKL) is a constitutive intracellular protein in resting human basophils and is strongly induced on their surface by interleukin 3. Immunobiology 2015, 220, 692–700. [Google Scholar] [CrossRef]
- Loser, K.; Mehling, A.; Loeser, S.; Apelt, J.; Kuhn, A.; Grabbe, S.; Schwarz, T.; Penninger, J.M.; Beissert, S. Epidermal RANKL controls regulatory T-cell numbers via activation of dendritic cells. Nat. Med. 2006, 12, 1372–1379. [Google Scholar] [CrossRef]
- Schmiedel, B.J.; Nuebling, T.; Steinbacher, J.; Malinovska, A.; Wende, C.M.; Azuma, M.; Schneider, P.; Grosse-Hovest, L.; Salih, H.R. Receptor activator for NF-κB ligand in acute myeloid leukemia: Expression, function, and modulation of NK cell immunosurveillance. J. Immunol. 2013, 190, 821–831. [Google Scholar] [CrossRef]
- Hienz, S.A.; Paliwal, S.; Ivanovski, S. Mechanisms of Bone Resorption in Periodontitis. J. Immunol. Res. 2015, 2015, 615486. [Google Scholar] [CrossRef]
- Ando, Y.; Tsukasaki, M.; Huynh, N.C.; Zang, S.; Yan, M.; Muro, R.; Nakamura, K.; Komagamine, M.; Komatsu, N.; Okamoto, K.; et al. The neutrophil-osteogenic cell axis promotes bone destruction in periodontitis. Int. J. Oral Sci. 2024, 16, 18. [Google Scholar] [CrossRef] [PubMed]
- Makkawi, H.; Hoch, S.; Burns, E.; Hosur, K.; Hajishengallis, G.; Kirschning, C.J.; Nussbaum, G. Porphyromonas gingivalis Stimulates TLR2-PI3K Signaling to Escape Immune Clearance and Induce Bone Resorption Independently of MyD88. Front. Cell Infect. Microbiol. 2017, 7, 359. [Google Scholar] [CrossRef] [PubMed]
- Guilherme Neto, J.L.; Rodrigues Venturini, L.G.; Schneider, A.H.; Taira, T.M.; Duffles Rodrigues, L.F.; Veras, F.P.; Oliveira, S.R.; da Silva, T.A.; Cunha, F.Q.; Fukada, S.Y. Neutrophil Extracellular Traps Aggravate Apical Periodontitis by Stimulating Osteoclast Formation. J. Endod. 2023, 49, 1514–1521. [Google Scholar] [CrossRef]
- Kim, T.S.; Silva, L.M.; Theofilou, V.I.; Greenwell-Wild, T.; Li, L.; Williams, D.W.; Ikeuchi, T.; Brenchley, L. NIDCD/NIDCR Genomics and Computational Biology Core; Bugge, T.H.; et al. Neutrophil extracellular traps and extracellular histones potentiate IL-17 inflammation in periodontitis. J. Exp. Med. 2023, 220, e20221751. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Li, Y.; Sun, R.; Hu, H.; Liu, Y.; Herrmann, M.; Zhao, Y.; Muñoz, L.E. Receptor-Mediated NETosis on Neutrophils. Front. Immunol. 2021, 12, 775267. [Google Scholar] [CrossRef]
- Han, Y.K.; Jin, Y.; Miao, Y.B.; Shi, T.; Lin, X.P. CD8+ Foxp3+ T Cells Affect Alveolar Bone Homeostasis via Modulating Tregs/Th17 During Induced Periodontitis: An Adoptive Transfer Experiment. Inflammation 2018, 41, 1791–1803. [Google Scholar] [CrossRef]
- Li, S.; Su, L.; Luan, Q.; Liu, G.; Zeng, W.; Yu, X. Regulatory B cells induced by interleukin-35 inhibit inflammation and alveolar bone resorption in ligature-induced periodontitis. J. Periodontol. 2023, 94, 1376–1388. [Google Scholar] [CrossRef]
- Luo, C.Y.; Wang, L.; Sun, C.; Li, D.J. Estrogen enhances the functions of CD4(+)CD25(+)Foxp3(+) regulatory T cells that suppress osteoclast differentiation and bone resorption in vitro. Cell Mol. Immunol. 2011, 8, 50–58. [Google Scholar] [CrossRef]


| TLR | Location | Exogenous Ligands (MAMPs) | Endogenous Ligands (DAMPs) |
|---|---|---|---|
| TLR2/1 and/or TLR2/6 | Cell surface (Cell membrane) | Peptide glycan | Amyloid, HSPs, HMGB1, Hyaluronan |
| TLR2/1 | Cell surface (Cell membrane) | Triacylated lipopeptide | - |
| TLR2/6 | Cell surface (Cell membrane) | Diacylated lipopeptide, LTA, Fungal zymosan | - |
| TLR3 | Endosome/lysosome | dsRNA | snRNA, Mitochondrial dsRNA, RNA from dead cells |
| TLR4 | Cell surface (Cell membrane) | LPS, Virus envelope protein | Oxidized LDL, AGE-LDL, HSPs, HMGB1, Hyaluronan, S100 proteins, Fibrinogen, Anti-MPs |
| TLR5 | Cell surface (Cell membrane) | Flagellin Fungal zymosan | Lectin, HSPs |
| TLR7 | Endosome/lysosome | ssRNA | RNA from dead cells Anti-MPs |
| TLR8 | Endosome/lysosome | ssRNA | RNA from dead cells Anti-MPs |
| TLR9 | Endosome/lysosome | Unmethylated CpG DNA | HMGB1, Mitochondria DNA, Anti-MPs |
| TLR10 | Endosome/lysosome | - | - |
| TLR11 | Cell surface (Cell membrane) | Profilin-like protein | - |
| TLR12 | Endosome/lysosome | Profilin | - |
| TLR13 | Endosome/lysosome | 23S rRNA | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tominari, T.; Matsumoto, C.; Tanaka, Y.; Shimizu, K.; Takatoya, M.; Sugasaki, M.; Karouji, K.; Kasuga, U.; Miyaura, C.; Miyata, S.; et al. Roles of Toll-like Receptor Signaling in Inflammatory Bone Resorption. Biology 2024, 13, 692. https://doi.org/10.3390/biology13090692
Tominari T, Matsumoto C, Tanaka Y, Shimizu K, Takatoya M, Sugasaki M, Karouji K, Kasuga U, Miyaura C, Miyata S, et al. Roles of Toll-like Receptor Signaling in Inflammatory Bone Resorption. Biology. 2024; 13(9):692. https://doi.org/10.3390/biology13090692
Chicago/Turabian StyleTominari, Tsukasa, Chiho Matsumoto, Yuki Tanaka, Kensuke Shimizu, Masaru Takatoya, Moe Sugasaki, Kento Karouji, Urara Kasuga, Chisato Miyaura, Shinji Miyata, and et al. 2024. "Roles of Toll-like Receptor Signaling in Inflammatory Bone Resorption" Biology 13, no. 9: 692. https://doi.org/10.3390/biology13090692
APA StyleTominari, T., Matsumoto, C., Tanaka, Y., Shimizu, K., Takatoya, M., Sugasaki, M., Karouji, K., Kasuga, U., Miyaura, C., Miyata, S., Itoh, Y., Hirata, M., & Inada, M. (2024). Roles of Toll-like Receptor Signaling in Inflammatory Bone Resorption. Biology, 13(9), 692. https://doi.org/10.3390/biology13090692









