Simple Summary
The current review article focused on the complex interaction between plant hormones and transcription factors in the response to salt stress, a pressing global issue that has a considerable effect on agricultural productivity. The study unveils the effect of salt stress on the ion equilibrium and triggers a cascade of molecular reactions that have a response to plant growth and development. Transcription factors are essential in regulating gene expression under salt stress. The transcription factors function in collaboration with hormones to regulate environmental stress responses. The study sheds light on the underlying regulatory networks, thus providing crucial knowledge for cultivating salt-tolerant crops through selective breeding and genetic engineering. This review significantly contributes to the establishment of sustainable agricultural techniques, which are essential in addressing the increasing problem of soil salinization and securing global food security.
Abstract
The negative impacts of soil salinization on ion homeostasis provide a significant global barrier to agricultural production and development. Plant physiology and biochemistry are severely affected by primary and secondary NaCl stress impacts, which damage cellular integrity, impair water uptake, and trigger physiological drought. Determining how transcriptional factors (TFs) and hormone networks are regulated in plants in response to salt stress is necessary for developing crops that tolerate salt. This study investigates the complex mechanisms of several significant TF families that influence plant responses to salt stress, involving AP2/ERF, bZIP, NAC, MYB, and WRKY. It demonstrates how these transcription factors (TFs) help plants respond to the detrimental effects of salinity by modulating gene expression through mechanisms including hormone signaling, osmotic stress pathway activation, and ion homeostasis. Additionally, it explores the hormonal imbalances triggered by salt stress, which entail complex interactions among phytohormones like jasmonic acid (JA), salicylic acid (SA), and abscisic acid (ABA) within the hormonal regulatory networks. This review highlights the regulatory role of key transcription factors in salt-stress response, and their interaction with plant hormones is crucial for developing genome-edited crops that can enhance agricultural sustainability and address global food security challenges.
1. Introduction
1.1. Overview of Salt Stress in Plants
Plant breeding is severely hampered by soil salinization, which is also harmful to agricultural productivity, yield, and crop growth worldwide [1]. The main challenge in producing plants in highly salty soils is the disturbance of the water cycle balance. The two terms “primary salt harm” and “secondary salt harm” indicate the negative impacts that salt-stress effects on plants. Salt damage occurs by the direct influence of salt ions, which may lead the cell membrane to show significant harm. Secondary salt damage is the term for the indirect impacts of salt ions that produce osmotic stress, which inhibits the plant from absorbing water [2]. The concentration of the soil solution is induced as the concentration of salt in the soil rises. Osmotic stress induction can significantly reduce a plant’s capability to absorb water [3]. The plant observes that it is difficult to consume water from the ground when the concentration of salt in the soil increases too much. The soil solution has a greater osmotic pressure than that of the plant cells. This condition may lead to plant roots being dehydrated, producing a situation similar to a physiological drought and, in extreme situations, possibly resulting in the plant’s death [4]. Due to the alteration of the lipid bilayer organization and the structure of membrane proteins, salt stress negatively impacts cell membranes, resulting in increased membrane lipid permeability, leading to lipid peroxidation [5,6]. Consequently, these changes affect the natural physiological functions of the membrane [7]. Under salt stress, plants lose water from cells, leading to alterations in cell turgor and osmotic potential. The destruction of both the cell membrane and plasma membrane results in a reduction or loss of selective permeability in the cell membrane. Consequently, this causes the leakage of beneficial ions like Ca2+ and K2+ from the cell [8]. Simultaneously, toxic ions such as Na+ and Cl− accumulate within the cell, causing an ion imbalance and further compromising cellular integrity. Cell membrane, organelle membrane, organelle structure, chlorophyll content, and photosynthetic rate lead to decreased and enhanced ribonuclease activities [9]. This leads to stress damage, weakening the plant’s resistance [10,11].
1.2. Importance of Understanding Plant Responses to Salt Stress
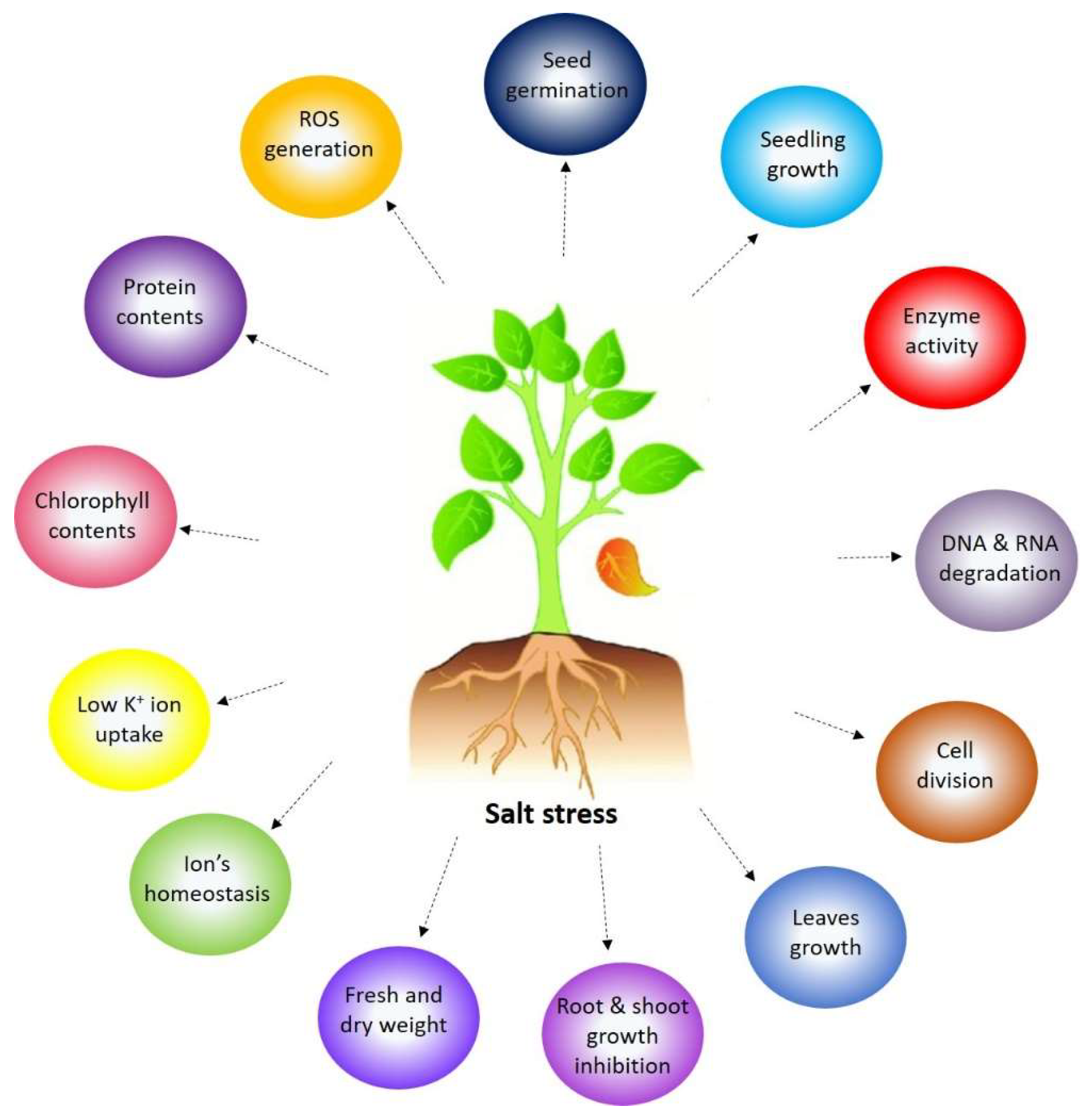
During salt stress, plants show different responses efficiently expressing physiological attributes that adapt to or alleviate the impact of salt stress [12,13]. Figure 1 shows the effect of salt stress on plant growth and development. At low salinity concentrations, the development of the root is generally lower influenced than shoot growth, resulting in an increased root-to-shoot ratio [14]. In contrast, higher salt concentration decreases the root growth [15]. With increased salinity stress, the plant experiences a significant decrease in the accumulation of dry biomass due to a reduction in shoot and root growth [16]. It has been previously reported that inhibition of growth may occur due to reduced rates of new cell formation [17]. Salinity-induced changes in the cell wall structure may have contributed to the cell wall’s increased stiffness and decreased dry weight accumulation. Osmotic stress, which affects salt stress in the root zone, impairs cell ion homeostasis by causing both an increase in the accumulation of Na+ and Cl− and an inhibition of the consumption of vital nutrients like K+ [18]. Increased uptake of Na+ competes with the absorption of other essential nutrient ions, particularly K+, resulting in a deficiency of potassium, which causes a lower K+/Na+ ratio in plants under salt stress [18]. Plants under salt stress exhibit notable alterations in their physiological and biochemical characteristics, including reduced levels of chlorophyll content in their leaves, a decline in protein synthesis, an increase in reactive oxygen species (ROS) accumulation, an increased accumulation of compatible solutes like proline, and modifications in antioxidant enzyme activities. Therefore, the total changes in plant growth and productivity are caused by the combination of all the morphological, physiological, and biochemical modifications in plants exposed to salt stress.
Figure 1.
The morphological, physiological, and biochemical response of a plant under salinity stress.
1.3. Significance of Transcriptional Factors and Hormonal Networks in Salt-Stress Response
It has recently been discovered that NaCl stress has a significant impact on the hormonal balance and the expression of specific transcription factors. Phytohormones, including abscisic acid (ABA), salicylic acid (SA), and jasmonic acid (JA), were shown to increase, whereas the levels of gibberellic acid (GA) decreased. Additionally, there was an initial increase and then a decrease in trans-zeatin (tZ) and indoleacetic acid (IAA) [19]. RNA-seq was used to completely investigate the expression patterns of genes associated with hormone production and signal transduction. As a result, a coextrusion network involving hormones and a genome-wide co-expression network were constructed using weighted gene co-expression network analysis (WGCNA). The study suggested a transcriptional regulatory model including six different hormone types and identified 20 hormone-related candidate genes connected with salt stress. This provides a significant understanding of the molecular mechanisms and insights into tomatoes’ abilities to tolerate salt [19]. Modulating gene expression in response to salt stress is mostly dependent on transcription factors. In combination with hormones, biotic and abiotic factors, symbiotic relationships, cell differentiation, and stress-signaling pathways, they intricately control gene expression. Additionally, by regulating the expression of downstream target genes, transcription factors (TFs) contribute significantly to the development of plant salt tolerance. For instance, when ABA binds to PYL, it reduces negative regulation, activating AREB/ABF transcription factors, which are bZIP proteins that bind to ABA response elements in the promoter regions of target genes [20]. It has been studied that endogenous ABA levels increase rapidly after oxidative stress, acting as a signaling molecule that regulates stress-responsive genes, such as DREB proteins and bZIP, to enhance plant productivity through drought stress tolerance mechanisms [21]. The crosstalk between transcription factors and hormones under saline conditions involves the coordination of stress-responsive transcription factors, such as those from the AP2/ERF, MYB, and bZIP families, with plant hormones like ABA and ethylene. This interaction regulates the expression of antioxidant genes, helping to maintain reactive oxygen species (ROS) homeostasis and enhancing plant tolerance to salt stress. Furthermore, primary osmotic stress, ion toxicity, secondary oxidative stress, and nutritional stress are all addressed by this control, which can help reduce the adverse effects that salt stress causes to plants [22,23]. Plant responses to salt stress are mostly mediated by the hormonal networks, particularly those involving ethylene. The formation of ethylene during salt stress suggests it is important for the salt response. In addition to regulating proteins at the post-translational level, salt stress stimulates specific genes at the transcriptional level. This complicated mechanism controls the complex interaction between gene expression and hormone signaling in salt-stressed environments [24]. An investigation of the association between hormones and transcription factors under salt stress was conducted on tomato plants. A complex transcriptional regulatory network has been revealed by a detailed examination of the expression patterns of transcription factors (TFs) and candidate genes connected with hormones under salt stress. The complex interactions between gene expression and signaling pathways play a significant part in the ability of plants to respond to salt stress [19].
2. Transcriptional Factors in Salt-Stress Regulation
2.1. Role of TFs in Gene Expression Regulation
The regulation of plant responses to the growing worldwide problem of soil salinization is mainly reliant on transcription factors (TFs). A comprehensive study into their function and the several families of transcription factors activated in the regulation of salt stress [25]. There are significantly more connections between the regulation of salinity stress and transcription factor families such as AP2/ERF, bZIP, NAC, MYB, and WRKY [26]. Two more important TF families that are linked to the ability to deal with abiotic stress like salt stress are ABF and DREB [27]. Specific DNA sequences are linked by transcription factors (TFs) that regulate the rate at which genetic information is transcriptionally transformed from DNA to messenger RNA. Thus, the regulatory process facilitates the expression of genes that are essential for salt-stress tolerance and adaptation in plants [28]. Transcription factors (TFs) have a major impact on signal-transduction networks that respond to salt stress in plants. They are a part of the mechanisms that alert the plant’s genetic machinery to the presence of salt stress, which causes stress-responsive genes to express themselves [23]. TFs contribute to salt tolerance in crops by signaling and activating genetic responses, which are crucial for ion homeostasis, osmotic stress pathway activation, and plant hormone signaling, enabling plant survival under salt stress [28]. To illustrate the specificity and critical role certain TF families play in responding to salt stress, bZIP Transcription Factors are specifically linked to the regulation of plant salt stress [29]. Soil salinity has a major impact on crop development and yield. It is imperative to understand and manipulate TFs to advance plant breeding and agricultural advancements [29].
2.2. Key Transcriptional Factors Involved in Salt-Stress Response
TFs are proteins that bind to DNA regulatory sequences, found upstream in the 5′ region of target genes, and control gene transcription. Consequently, TFs play a crucial role in activating and deactivating specific genes by binding to other DNA sequences and regulating gene transcription and protein synthesis, leading to modifications in cellular function within plant tissues [27]. TFs are highly conserved in the plant genome, altering gene expression and endowing plants with resilience to salinity. Some TFs are generally found in all cells of a plant.
There are various cell types and stages of development for which a few transcription factors are specialized. These transcription factors are essential for controlling the expression of genes, which affects development. It is crucial for understanding the transcription mechanism because it regulates changes in gene transcription. In all eukaryotes, the enzyme in charge of transcription is called RNA Polymerase II, or RNAP II. However, RNAP II cannot function on its initiative. Trans-acting factors, which are transcription factors, and cis-regulatory regions, which are certain DNA sequences within or closer to a gene, control its activity. Transcription factors are necessary for RNAP II to be able to transcribe DNA throughout this process. Particular DNA sequences known as “cis-acting elements” are essential for regulating transcription [30]. The TF families bZIP, MYB, NAC, and WRKY are associated with stress tolerance [31].
2.3. Role of Stress-Responsive Transcription Factors in Enhancing Abiotic Stress Tolerance in Plants
Stress-responsive transcription factors are significant for the development of stress tolerance as well as abiotic stress responses [32]. Therefore, targeting stress-responsive transcription factors is essential for developing crops that are more resilient to abiotic stress, ensuring enhanced tolerance [33]. Genetic engineering is recognized as an alternative strategy for enhancing stress tolerance in crops by significantly changing their agronomic characteristics. Abiotic stress response has been identified in numerous genes, including functional proteins and proteins involved in signal-transduction pathways [34]. Also known as trans-acting factors, they are essential components in the signal-transduction pathways for abiotic stress. Transcription factors bind to different cis-elements and regulate the expression of a family of related genes, which is essential to the mechanism of plant resistance. The transcriptional regulation domain, nuclear localization signal (NLS), oligomerization site, and DNA-binding domain are the four fundamental parts of transcription factors. The combined action of these structural domains determines the time, space, and mode of action of the regulatory functions of transcription factors [35]. Transcription factors can be divided into several categories according to the characteristics of DNA-binding regions. In these responses, WRKY, bZIP, MYB, and NAC. Transcription factors play key roles in various physiological processes in various plants shown in Table 1, Table 2, Table 3 and Table 4. Under salt stress, the dynamic changes in hormones had a significant effect on the normal growth and development of plants [24]. Salt stress may be reduced by the buildup of Abscisic acid, which can regulate stomatal closure, ion homeostasis, gene expression, and metabolic changes in response to salt stress [36]. When plants were exposed to salt stress, the accumulated Indole acetic acid and Cytokinin in root tips could confer augmented resistance [24]. Previous research has shown that under salt stress, the expression of the IAA-related genes SAUR32, SAUR36, and ARF5, as well as the gene related to cell division, IPT5, could be significantly induced in the roots of apple rootstocks. These genes could improve salt tolerance by increasing the IAA and CK content in the apple rootstocks [37]. Jasmonic acid activation of antioxidant enzymes can delay flowering, reduce plant growth and root elongation, and increase plant viability under salt stress [38]. Additionally, Salicylic acid and Gibberellins were also crucial in salt stress [39].

Table 1.
WRKY Transcription Factors in Response to Salt Stress.

Table 2.
bZIP Transcription Factors in Response to Salt Stress.

Table 3.
MYB Transcription Factors in Response to Salt Stress.

Table 4.
NAC transcription factors in response to salt stress.
3. Major TF Families
In this study, we focused on four TF families implicated in abiotic stress tolerance, and the significance of these transcription factors in salt-stress signaling is described in Table 5.

Table 5.
The importance of transcription factors in salt-stress signaling and those involved in four TF families responsible for abiotic stress tolerance.
4. Mechanisms of TF Activation during Salt-Stress
Complex networks characterize plant abiotic stress signal-transduction processes. One environmental stimulus can cause several secondary stress signals in plant cells, and each of these secondary stress signals might be transmitted through a various signaling pathway [127]. This shows a challenge for developing a thorough understanding of TF functions in plant signaling pathways. Nevertheless, these pathways may eventually regulate the same target genes or exhibit crosstalk at certain nodes during the signal-transduction cascades [128]. Abiotic stress-signaling pathways may exhibit crosstalk toward single or multiple stressors. To address this, molecular analyses of stress-induced genes using RNA-seq, oligo arrays, or full-length cDNA microarrays are several ways to investigate [129]. Such strategies have demonstrated that plants express crosstalk variously in response to drought and salt stress compared to their response to drought and cold stress. Most drought-inducible genes were also triggered by high salinity and ABA treatments, according to the recent finding that 10% of drought-inducible genes could be activated by cold [130]. Abiotic stress responses in plants can be regulated by TFs from different families, either independently of ABA (CaNAC05 and CaNAC41, for example) [131] or dependently on this hormone (such as AtAREB1 and GhWRKY17) [132]. Alternatively, they can function in both ABA-dependent and ABA-independent pathways (such as TaMYB19, MbDREB1, and DREB2A) [133]. Target gene promoters contain significant cis-elements that can interact indirectly with TFs to produce crosstalk. The abiotic-stress-responsive RD29A promoter, for example, has both DRE/CRT and ABRE cis-elements that can be bound by TFs that bind DRE/CRT (AtDREB1 and AtDREB2) and ABRE (AtAREB1 and AtAREB2), respectively. Transactivation studies in Arabidopsis have demonstrated that these TFs work together to cooperatively regulate the target gene’s expression [134]. This investigation further shows that the degree of RD29A transcriptional activity depends on a combination of DREB/AREB regulators simultaneously to the promoter’s cis-elements. Different studies on TF binding to the Arabidopsis RD29A promoter indicated that this promoter also contains the binding site (named NACRS) for ANAC096, and TF was able to bind with AtAREB1 and AtAREB2 in proximity and NACRS and ABRE regions [135]. Furthermore, studies performed by Oh et al. suggested that the target gene might be more probable to be highly regulated by AREBs or DREBs if there were more DRE or ABRE sequences in the promoter region, which contains both cis-elements [136]. Various transcription factors have been assigned as multi-functional regulatory proteins that engage in both biotic and abiotic stress pathways. This has been demonstrated by the NACs from Arabidopsis [ATAF1 (Arabidopsis transcription activation factor 1) and ATAF2] and rice (OsNAC6) [137], and by MYBs from wheat (TaPIMP1) and rice (OsMYB4) [138], and WRKY TFs from rice (OsWRKY45) and grapevine (VvWRKY11) [139].
5. Hormonal Networks in Salt-Stress Response
5.1. Overview of Plant Hormones Involved in Salt-Stress Response
Plant hormones play a crucial role in how plants respond to salt stress, which is a significant environmental stressor that limits growth and productivity. There are nine well-characterized plant hormones, and among these, abscisic acid (ABA), ethylene, salicylic acid (SA), and jasmonic acid (JA) are specifically recognized for their roles in stress response. Meanwhile, others like auxin, gibberellin (GA), cytokinins (CKs), brassinosteroids (BRs), and strigolactones (SLs) are primarily associated with growth promotion [24]. Hormonal regulation is part of a sophisticated network of biological mechanisms that plants employ to adapt to salt stress, involving osmoregulation, redox, and ionic homeostasis, as well as adjustments in growth through hormone or light signaling-mediated pathways [140]. Interaction between these hormones contributes to a complex regulatory system that allows plants to sense, signal, and respond to the harsh conditions imposed by high salinity levels [141]. The balance and interaction between these hormones and their receptors play a critical role in determining the plant’s response to salt stress. For instance, the dominance of receptor signaling over ethylene might make the plant more susceptible to salt stress [142]. This crosstalk among hormones underlines the importance of both stress and growth hormones in plant adaptation to salt stress [24]. Specifically, one significant phytohormone that is crucial for responding to various stress signals is ABA. When ABA is applied to plants, it simulates the effects of stress. The expression patterns of stress genes and ABA treatment overlap because numerous abiotic stimuli lead to cell desiccation and osmotic imbalance. It suggests that to maintain cellular homeostasis, numerous stress signals and ABA might have similar parts in the signaling pathway that communicate with one another [143]. Abscisic acid (ABA) is essential for many physiological functions, such as reducing the germination of seeds and causing dormancy, controlling seed growth, encouraging stomatal closure, controlling embryo morphogenesis, facilitating the synthesis of lipids and storage proteins, accelerating leaf senescence, and promoting defense mechanisms against infections [144]. The main function of ABA seems to be the regulation of plant water balance and osmotic stress tolerance. Several ABA-deficient mutants namely aba1, aba2, and aba3 have been noted in Arabidopsis [145].
5.2. Interplay between ABA, Salicylic Acid (SA), Jasmonic Acid (JA) and Other Hormones
A well-organized mechanism that involves the plant’s capacity to adapt to changing environmental conditions determines the way plants respond to abiotic stressors. A signal-transduction cascade’s crosstalk is the point at which several hormones interact with one another. Salicylic acid, JA, and ABA are essential phytohormones in stress signaling. For stomatal closure, ABA always triggers JA by a Ca2+ influx, which mediates the CDPK signaling cascade. Studies further demonstrated that PYL6 (RCAR9) and an ABA receptor with the corresponding transcription factor MYC2 influenced the expression of JAZ6 and JAZ8, indicating a possible association between ABA and JA in stress tolerance [146]. Cytokinin hormone is also involved in delayed senescence, cell growth, and leaf expansion. ABA and CK, however, have been found to be negatively correlated, which eventually results in stomatal closure and reduced water loss [147]. The CK receptor kinase, which significantly regulates ABA levels, has also been found to be responsible for a relationship between Arabidopsis histidine phosphotransferase proteins; however, stress tolerance is additionally observed in CK-deficient mutants [148]. On the other hand, by inhibiting ABA in response to dryness, ethylene has demonstrated a negative correlation between gas exchange and the development of leaves and roots [149]. Increasing ABA levels caused ABI5 to be triggered, which controls LEA genes that serve as an osmoprotectant for the seed under stress [150]. Additionally, ABA and GA showed an antagonistic interaction, and their balance regulated the dormancy and germination of seeds. The interplay between ABA and GA signaling under abiotic stress conditions is significantly influenced by DELLA proteins. The RINGH2 factor, which encodes XERICO and mediates ABA signaling, ABA accumulation, and ABI5 activity to disrupt GA levels, has been observed to be induced by RGL2, a protein related to the DELLA protein family [151]. Thus, by inactivating through the 26S proteasome pathway and increasing the GA level, RGL2 has been identified to be a crucial factor in breaking seed dormancy [152]. Because RGL2 regulated MFT for phosphatidylethanolamine binding protein, which showed an adverse relationship with ABI5 but a positive correlation with ABI3, MFT was additionally identified in ABA and GA signaling [153]. In phytohormone-mediated agricultural plant growth and development under environmental stress, ABA primarily functions via interacting with other related plant hormones.
5.3. Hormonal Crosstalk in Salt-Stress Adaptation
Physiological drought is caused by salt stress, which leads to degraded protein and photosynthesis [154]. Reactive oxygen species (ROS) formation, excessive amounts of sodium ions (Na+), and changes in intracellular Ca2+ levels are examples of signals that trigger the salt-stress response [155]. Ethylene is the primary hormone in salt-stress response [156]. The amounts of ET and its precursor ACC (1-aminocyclopropane-1-carboxylate) increase under NaCl stress [157]. Production of ET synthesis may induce salt sensitivity [154]. Five ethylene receptors (ETR1, ERS1, ETR2, EIN4, and ERS2), a protein kinase known as CTR1 (Constitutive Triple Response 1) functioning as a negative regulator, and an essential positive regulator known as EIN2 are all involved in the intricate process of ethylene signaling. Associated with several downstream ethylene response factors, this signaling route further activates main transcription factors like EIN3, EIL1 (Ethylene Insensitive Like 1), and EIL2. Osmotic stress, which is produced by several abiotic variables, including salinity, prevents ETR1 (Ethylene Response 1) expression [154]. Cotton showed upregulation of ethylene receptor genes (ETR1, ETR2, and EIN4), signaling genes (CTR1, EIN3, ERF1, and ERF2), and MAPK cascade genes (MEKK1-MKK2-MPK4/6) in response to both short- and long-term salt stress [158]. Ethylene and salt stress both affect the many Ethylene-Responsive Element Binding Factor (ERF) genes, particularly ESE1–ESE3. EIN3 accumulation and transcriptional activity are enhanced, and EBF1/EBF2 is degraded by salt stress [158]. When exposed to salt stress, 1-aminocyclopropane-1-carboxylic acid synthases (ACSs) are significantly expressed [159]. Salt stress tolerance in Arabidopsis seedlings increases with ACC pretreatment [160]. Salt causes the tobacco to induce ACS1 transcripts [161]. Four ACS genes were upregulated in both salt and non-acclimated plants under salt stress [162]. Stress-triggered (MAPK) cascades phosphorylate ACSs during the post-transcriptional regulation of synthases (ACSs), preventing the 26S proteasome from degrading ACSs detrimentally [156]. The effects of salt acclimatization are reduced by the decrease in MAPK6 function [162]. Stabilization of ACSs apparently needs MPK6 to maintain high ethylene levels. ACSs are also stabilized by CDPKs (Calcium-Dependent Protein Kinases) in tomatoes and ACC content and activity of 1-aminocyclopropane-1-carboxylic acid oxidase (ACO) is increased under salt stress in Cicer arietinum roots [156]. ABA modulates several genes that are sensitive to stress [163]. ABA and ET collaborate to mediate salt stress. Several genes involved in ABA production, including ZEP, AAO, and MCSU, are activated during exposure to salt via downstream signaling pathways and Ca2+ dependent phosphorylation processes [164]. It has been found that certain plants, including Oryza sativa [165] Brassica [166], and Zea mays [167], demonstrated significant ABA levels. Increased levels of ABA lead to stomatal closure and assist in the accumulation of proteins for osmotic modification. Salt-stress tolerance is enhanced by high accumulation of ABA due to ectopic expression of drought-responsive genes in rice, particularly OsCam1–1 (Oryza Sativa Calmodulin1–1) and OsDSM2 (Drought-Hypersensitive Mutant 2) [163]. Many MAPKs are upregulated in response to salt stress and ABA treatment, and plants that express MAPKs are more tolerant of salt stress [163]. Salt stress is influenced by ABA-regulated Ca2+-dependent kinases and SnRks, as well as phosphorylate ABA-related transcription factors, and affects gene expression [168]. Several regulatory sequences, including DRE/CRT, ABRE, MYC recognition sequence (MYCRS), and MYB recognition sequence (MYBRS), have been identified in the promoters of stress-responsive genes. ABA-dependent transcription factors (ABFs), MYCs, and MYBs directly bind to specific regions on the promoters of salt-stress-responsive genes and promote their activation. All LEA genes have ABRE motifs in their promoters, which bind ABF. Dihydroorotate Dehydrogenase1 is a drought-inducible gene that is essential to salt and drought stress responses, and it is regulated by ABFs and DREB2 [163]. The mutation in ACS7 reduces tolerance to heat, osmotic, and salt stress due to the crosstalk between ethylene (ET) and (ABA) during stress conditions [156]. There is limited information reported on the salt-stress mechanism via auxins [163]. Induced hypersensitivity to salt stress leads to increased auxin production caused by the UCCA3 gene, which is involved in auxin biosynthesis [169]. Changes in the root architecture are caused by auxin accumulation and redistribution in response to salt stress [170]. In previous studies, we reported that auxin levels significantly decreased in tomato under salt stress [171]. Salt stress leads to decreasing growth, an indication of changes in both IAA biosynthesis levels and distribution, and it decreases CK production in wheat [172]. The Na+ transporter-encoding HKT1-1 gene was expressed more often in mutants with lower CK levels [173].
6. Crosstalk between Transcriptional Factors and Hormonal Networks
6.1. Interaction between TFs and Hormone Signaling Pathways
The complex interactions between hormone signaling pathways and gene regulation are largely dependent on transcription factors (TFs) (Figure 2). They act as key regulators of essential cellular processes, including differentiation, development, and the cellular response to external signals [174]. Hormone signals often first impact transcription factors, which then determine how biological pathways will develop, such as during somatic embryogenesis [175]. In plants, for instance, the cross-regulation between different hormone signaling pathways can influence the activity of TFs. Researchers have examined this cross-regulation, identifying network components that might be responsible for such interactions. There is a large-scale protein–protein interaction network that provides a comprehensive analysis of this cross-regulation in plant hormone signaling [176]. Furthermore, the activity of transcription factors in response to plant hormones has been extensively characterized, and dynamic transcriptional regulatory models for several hormones, including abscisic acid, brassinosteroid, ethylene, jasmonic acid, salicylic acid, and strigolactone/Karrikin, have been reconstructed to understand these relationships better [39]. A specific instance of TFs interacting with hormone signaling pathways can be seen in the Arabidopsis transcription factor LONG HYPOCOTYL 5 (HY5). This factor plays a significant role in photomorphogenic development by promoting the expression of genes that are negative regulators of auxin signaling, therefore linking hormone and light signaling pathways. Mutations in HY5 disrupt this balance, leading to altered hormone signaling and plant development phenotypes [177]. Moreover, in the context of stress responses, WRKY transcription factors in plants have been identified as key players in regulating hormone signal-transduction pathways. These TFs are integral to plant processes responding to both biotic and abiotic stress, suggesting their pivotal role in hormone-mediated stress-response mechanisms [178].
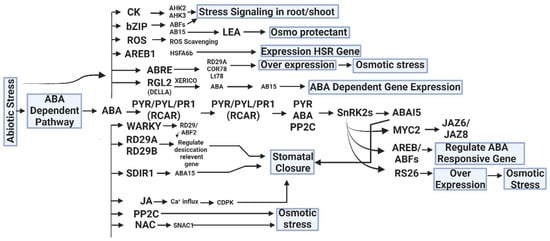
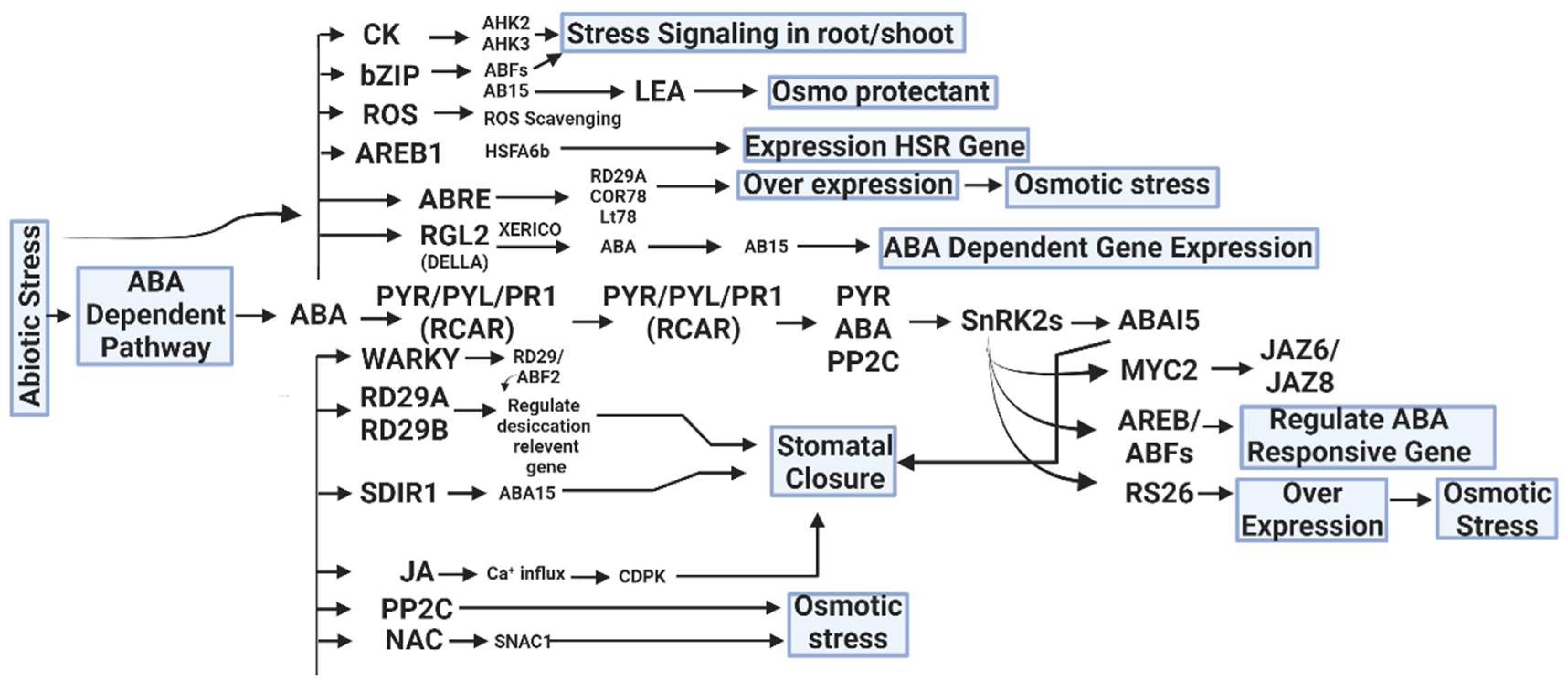
Figure 2.
Hormonal crosstalk and abiotic stress tolerance are caused CK (Cytokinin), ROS (Reactive oxygen species), LEA (Late Embryogenesis Abundant), AREB1(Abscisic Acid Response Element Binding Protein 1), RGL2 (Repressor of GA LIKE 2), PYR (Pyrabactin Resistance Gene), RD29A (Response-to-Dehydration 29A), SDIR1 (Salt and Drought-Induced Ring Finger), JA (Jasmonic Acid), PP2C (Protein Phosphate 2C), JAZ (Jasmonate-ZIM domain), and SnRK2s (Sucrose Non-Fermenting 1-Related Protein Kinase 2s) by an ABA-dependent and independent pathway [150].
6.2. Feedback Mechanisms Regulating TFs and Hormonal Responses
The cross-regulation of hormone signaling pathways plays a major role in plant growth and development [179]. Plants utilize this mechanism to analyze several types of environmental and internal signals from their surroundings, absorb and interpret the data, and then initiate appropriate responses. This enables plants to respond to stressors, show plastic growth, and adapt to their local environment [180]. The growth defense trade-off is a well-known example of when plants, under pathogen attack, prioritize the allocation of resources to defense mechanisms [181]. Nevertheless, this trade-off can be dependent on specific conditions, as plants thriving in nutrient-rich environments may not necessarily be prioritized to favor one response over the other [181]. Each plant hormone has a recognized, distinct signaling pathway [182]. Signal transduction and transcription regulation include cross-regulation of these pathways [183]. Transcription factors shared between pathways and independent TFs regulating target genes shared by both pathways can cause cross-regulation of transcription. The transcriptional responses to various hormones share a small number of genes [39]. The DELLA and JASMONATE-ZIM DOMAIN (JAZ) proteins and NONEXPRESSER OF PR GENES 1 (NPR1) are classic examples of hormone cross-regulation. Each of these examples involves proteins that are mostly controlled by a single hormone and that act on that hormone’s route, but they also have an impact on other hormone signaling pathways [184]. A hormone stimulation causes thousands of genes to change in expression [185]. Gene expression changes dynamically and demonstrates an extensive amount of variance throughout time, showing a range of gene expression patterns [186]. Different TFs dynamically regulate genes at different times in response to stress. Jasmonic acid (JA), ethylene (ET), and abscisic acid (ABA) are the three hormones that control the expression of tens to hundreds of TFs; all hormones probably function similarly [187]. Several TFs may target a single gene, and individual TFs may target hundreds to thousands of genes. This enables dynamic and complex expression patterns but presents a substantial issue in determining which TFs regulate these patterns [184]. The primary JA signaling pathway is significantly regulated by the JAZ repressor JAZ10, which encodes both the dominant negative and active forms of the protein [188].
6.3. Coordinated Regulation of Gene Expression during Salt Stress
Saline soil causes decreased seed germination rates and plant growth. One of the primary adverse factors impacting food security and agricultural productivity is soil salinization [189]. Plants have developed complex signaling pathways to adapt to changing environmental conditions, typically consisting of signal transducers, secondary signals, plant hormones, and receptors [190]. Transcription factors (TFs), also observed as trans-acting factors, are significant components of abiotic stress signal-transduction pathways. Plant resistance is a result of transcription factors that bind to certain cis-elements and control the expression of a gene family associated with them. The DNA-binding domain, transcriptional regulatory domain, nuclear localization signal (NLS), and oligomerization site are the four basic components of typical transcription factors. In Table 6, the TFs and their domains associated with salinity stress are summarized. The combined action of these structural domains influences the time, space, and mode of action of the regulatory functions of transcription factors [35]. Many transcription factors act as major regulators to select genes, controlling the determination of cell type, development pattern, and specific pathway control [191]. Plants produce and exchange several signals that activate transcription factors in response to biotic and abiotic stressors, including salt, drought, extremes in temperature, and pathogens. Transcription factors bind with corresponding cis-acting elements to switch on RNA polymerase and transcribe complexes, therefore creating the transcription and expression of specific genes. Products eventually begin responding to the signal [192].

Table 6.
A summary of the transcription factors associated with salt stress.
7. Regulation of Salt-Stress-Related Genes
7.1. Molecular Mechanisms Controlling the Expression of Salt-Responsive Genes
Plants develop a response through the perception and transduction of osmotic and ionic signals, resulting in modifications to cellular characteristics under salt stress. To date, no salinity sensor/receptor information has been reported in plants [226]. On the other hand, the Arabidopsis signaling pathway includes salt excessively sensitive (SOS) and calcineurin B-like (CBL)/CBL-interacting kinases. Higher cytosolic Ca2+ concentrations activate the Ca2+-dependent protein kinase complex (SOS2-SOS3), which phosphorylates SOS1 and starts its activity. Furthermore, a novel pathway for the control of vacuolar Na+ sequestration modulated by the CBL10-CIPK24 complex was revealed. Furthermore, TFs are dependent on both ABA-dependent and ABA-independent pathways, and salinity stress promotes the increased formation of ABA that triggers signaling (Figure 3). Through phosphorylation, it activates sucrose non-fermenting 1-related protein kinase 2 (SnRK2). Plants show inhibition of snRK2 under control conditions, but in reaction to stress, they bind to PYRABACTIN RESISTANCE 1 (PYR1), a receptor protein. Several downstream TFs are activated or inhibited when PP2C is released from SnRK2 and self-phosphorylates, namely the ABA-responsive factors (ABF) and the abscisic acid (ABA)-responsive element (ABRE)-binding protein (AREB) [227]. The mitogen-activated protein kinases (MAPKs) are another possible option for salt-stress sensing. It is responsible for mediating the homeostasis of secondary stimuli, including oxidative, osmotic, and ionic stresses. It is classified into three distinct classes, determined by the phosphorylation and activation of elements, i.e., MAPK, MAPKK, and MAPKKK.
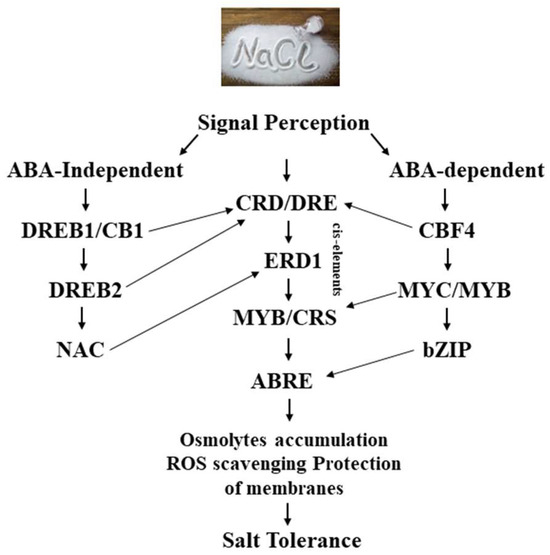
Figure 3.
Schematic representation of the salinity stress signal perception and gene expression.
7.2. Epigenetic Modifications and Chromatin Remodeling
Widespread salinity and sodality have adversely impacted large areas of arable land, limiting crop development and lowering land production per person. Understanding the molecular mechanisms underlying each stress and being able to control them to feed the world’s population has become crucial for scientists. In this situation, modifying key histone proteins through methylation, acetylation, and phosphorylation resulted in epigenetic solutions that have been of unknown benefit in affecting plant responses to salt-induced stress [228]. Scientific understanding of the fundamental epigenetic process regulating blooming, germination, fruit ripening, vernalization of fruiting photoperiodism, etc., has been made easier by advances in biotechnology. To assist in the identification of the configurational changes that the genome undergoes during cell differentiation. Studies investigating plant-heritable epigenetic marks have been performed to find out more about natural selection processes and other adaptive responses of plants to their surroundings [229]. Studies on various abiotic stress responses have clearly shown that some abiotic stressors trigger somatic memory through mitotic cell divisions, which can persist for a long period [230]. Memory quickly resets to baseline levels when normalcy can thrive. However, in meiotic cells, some memories influence chromatin. They are heritable and thus have the potential to pass on from parent plants to the stress-free offspring of the generation, which is known as a transgenerational epigenetic inheritance [231]. DNA methylation and histone modifications may have a synergistic effect on stress-induced genes because salinity stress affects the expression of various transcription factors in soybeans [232]. Hypomethylation might be associated with changes in the expression of DNA demethylases under salt stress [233]. When salinity stress was applied, contrasting variations in cytosine methylation patterns were observed in the progenitor and salinity-tolerant wheat cultivar SR3. The responses of contrasting wheat genotypes under NaCl stress could be attributed to the altered expression levels of high-affinity potassium transporters (HKTs) regulated through genetic and epigenetic mechanisms [234]. The hypersensitivity of Arabidopsis thaliana to NaCl stress is attributed to the transcriptional adaptor ADA2b, which is responsible for histone acetyltransferase activity. On the other hand, plant responses are more complicated due to the histone modifications and crosstalk between histone acetylation and cytosine methylation [235]. Both genome-wide DNA methylation and histone modifications are affected by salt stress, and both mechanisms interact to provide a synchronized response to salt stress [236]. Plant responses discussed above under various stress conditions, salinity stress also triggers common reactions including modifications in histones and changes in DNA methylation. The reactions to stress stimuli result in a noticeable shift in chromatin organization and dynamics, facilitating locus-specific gene expression in plants [237]. Researchers observed that the expression of genes, including SUVH2/5/8, ROS1, MSH6, APUM3, MOS6, and DRB2 was down-regulated in a few filial generations of saline-stressed Arabidopsis thaliana cultures. The reduction of H3K9ac in the promoter region of the coding sites, the amplification of H3K9me2, and DNA hypermethylation are the causes of this phenomenon [238]. The regulatory agents for the salinity-responsive genes Glyma11g0200, Glyma08g41450, and Glyma20g30840 in soybean (Glycine max) are increased amounts of H3K4me3, H3K9ac, and reduced levels of H3K9me2 in combination with DNA hypermethylation [232].
7.3. Post-Transcriptional and Post-Translational Modifications of Proteins
Gene expression is regulated through two major modes, transcriptional and post-transcriptional regulation mechanisms, for the initiation of transcription, determining whether to activate or repress the process and consequently inflecting the number of proteins synthesized during translation [232]. Plants under stress exhibit a network of regulatory mechanisms involving the reprogramming of the expression of several key genes at both transcriptional and post-transcriptional levels [239]. These regulatory processes are vital for plants to recover and regenerate cellular homeostasis during both the stress period and the subsequent recovery phase. Understanding these regulatory mechanisms of salinity stress response and tolerance at transcriptional and post-transcriptional levels has been considerably clearer by high-throughput sequencing techniques, readily available databases, and in silico tools. Current studies have demonstrated the presence of several significant functions at both the transcriptional and post-transcriptional levels, including regulatory noncoding RNA species, small and micro-RNAs, and others that play crucial regulatory roles in identifying and regulating the effects of salinity stress on important crops [239]. These regulatory elements are considered key roles in enhancing salinity tolerance in crucial crops through genetic engineering. The formation of a Special Issue aims to highlight diverse transcriptional and post-transcriptional regulatory mechanisms involved in sensing, signaling, and responding to salinity stress in significant crops and model plants using a wide variety of biological, analytical, and computational tools. Many researchers have thoroughly explored the physiological, molecular, and biochemical responses to salinity stress. Various strategies, such as melatonin, play a significant role in regulating plant growth, development, and responses to stress [240]. Various genomic and non-genomic strategies are currently undergoing validation to assess their potential to enhance crop salt tolerance. These strategies encompass stress-inducible promoters, protein post-translational modification, halotolerant microbiomes, and the gene resources of halophytes. The development of novel genetic resources for enhancing salt tolerance has been made easier by cutting-edge techniques in plant phenotyping, next-generation sequencing, and molecularly assisted breeding [241]. The draft genome and transcriptome analysis of Oryza coarctata, halophylic rice, and a wild relative of cultivated rice have been conducted to provide a potential resource for factors related to salinity and submergence stress response [242]. There is a clear varietal difference between indica and japonica rice cultivars related to their varying salinity tolerance capacities, as shown by the transcript profiling of stress-responsive genes and alter in metabolism during salinity [243]. De novo transcriptome assembly, sequencing, and gene expression profiling of Sosola drummondii, a salt-stressed halophyte from a saline environment [244]. Chickpea plant response to NaCl stress was examined with DNA polymorphisms identified by whole-genome sequencing and their potential functional implications [245].
8. Conclusions
Enhancing NaCl stress tolerance in plants has far-reaching implications that extend beyond agriculture and social aspects. The development and adoption of salt-tolerant crops show a significant increase in more sustainable global food. Research has identified several key TFs concerned with salt-stress response, including members of the AP2/ERF, MYB, and bZIP families. These TFs play crucial roles in regulating the expression of stress-responsive genes. TFs regulate the expression of downstream target genes by binding to specific cis-elements in their promoters. In this review paper, we highlighted the role of TF networks, indicating crosstalk and interaction between plant hormones under salt stress. Further research aims to understand the regulatory mechanisms of transcription factors in plant morphogenesis, therefore facilitating the development of genome-edited plants that hold potential for biotechnological applications.
Author Contributions
M.A. and S.A. collected the data and wrote the manuscript draft. All authors have read and agreed to the published version of the manuscript.
Funding
This work was carried out with the support of “Cooperative Research Program for Agriculture Science and Technology Development (Project No. RS-2024-00322408)” Rural Development Administration, Republic of Korea.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Wani, S.H.; Kumar, V.; Khare, T.; Guddimalli, R.; Parveda, M.; Solymosi, K.; Suprasanna, P.; Kavi Kishor, P. Engineering salinity tolerance in plants: Progress and prospects. Planta 2020, 251, 76. [Google Scholar] [CrossRef]
- Wang, G.; Shen, W.; Zhang, Z.; Guo, S.; Hu, J.; Feng, R.; Zhao, Q.; Du, J.; Du, Y. The Effect of Neutral Salt and Alkaline Stress with the Same Na+ Concentration on Root Growth of Soybean (Glycine max (L.) Merr.) Seedlings. Agronomy 2022, 12, 2708. [Google Scholar] [CrossRef]
- Antunes, J.; Paz, A.M.; Castanheira, N.; Gonçalves, M.C.; Cortez, N. Study of the influence of the standing time in the electrical conductivity of the saturated soil paste in three soils. Rev. De Ciências Agrárias 2023, 46, 93–98. [Google Scholar]
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D. Interplay between membrane proteins and membrane protein-lipid pertaining to plant salinity stress. Cell Biochem. Funct. 2023, 41, 399–412. [Google Scholar] [CrossRef]
- Hussain, N.; Sohail, Y.; Shakeel, N.; Javed, M.; Bano, H.; Gul, H.S.; Zafar, Z.U.; Frahat Zaky Hassan, I.; Ghaffar, A.; Athar, H.-u.-R. Role of mineral nutrients, antioxidants, osmotic adjustment and PSII stability in salt tolerance of contrasting wheat genotypes. Sci. Rep. 2022, 12, 12677. [Google Scholar] [CrossRef] [PubMed]
- Balasubramaniam, T.; Shen, G.; Esmaeili, N.; Zhang, H. Plants’ Response Mechanisms to Salinity Stress. Plants 2023, 12, 2253. [Google Scholar] [CrossRef]
- Hualpa-Ramirez, E.; Carrasco-Lozano, E.C.; Madrid-Espinoza, J.; Tejos, R.; Ruiz-Lara, S.; Stange, C.; Norambuena, L. Stress salinity in plants: New strategies to cope with in the foreseeable scenario. Plant Physiol. Biochem. 2024, 108507. [Google Scholar] [CrossRef] [PubMed]
- Flowers, T.J.; Munns, R.; Colmer, T.D. Sodium chloride toxicity and the cellular basis of salt tolerance in halophytes. Ann. Bot. 2015, 115, 419–431. [Google Scholar] [CrossRef]
- Wu, H.; Fan, S.; Gong, H.; Guo, J. Roles of salicylic acid in selenium-enhanced salt tolerance in tomato plants. Plant Soil 2023, 484, 569–588. [Google Scholar] [CrossRef]
- Slama, I.; Abdelly, C.; Bouchereau, A.; Flowers, T.; Savouré, A. Diversity, distribution and roles of osmoprotective compounds accumulated in halophytes under abiotic stress. Ann. Bot. 2015, 115, 433–447. [Google Scholar] [CrossRef]
- Acosta-Motos, J.R.; Ortuño, M.F.; Bernal-Vicente, A.; Diaz-Vivancos, P.; Sanchez-Blanco, M.J.; Hernandez, J.A. Plant responses to salt stress: Adaptive mechanisms. Agronomy 2017, 7, 18. [Google Scholar] [CrossRef]
- Çulha, Ş.; Çakirlar, H. Tuzluluğun bitkiler üzerine etkileri ve tuz tolerans mekanizmaları. Afyon Kocatepe Üniversitesi Fen Ve Mühendislik Bilimleri Derg. 2011, 11, 11–34. [Google Scholar]
- Suo, J.; Zhao, Q.; David, L.; Chen, S.; Dai, S. Salinity response in chloroplasts: Insights from gene characterization. Int. J. Mol. Sci. 2017, 18, 1011. [Google Scholar] [CrossRef] [PubMed]
- Shelden, M.C.; Munns, R. Crop root system plasticity for improved yields in saline soils. Front. Plant Sci. 2023, 14. [Google Scholar] [CrossRef]
- Bouhraoua, S.; Ferioun, M.; Nassira, S.; Boussakouran, A.; Akhazzane, M.; Belahcen, D.; Hammani, K.; Louahlia, S. Biomass partitioning and physiological responses of four Moroccan barley varieties subjected to salt stress in a hydroponic system. J. Plant Biotechnol. 2023, 50, 115–126. [Google Scholar] [CrossRef]
- Dabravolski, S.A.; Isayenkov, S.V. The regulation of plant cell wall organisation under salt stress. Front. Plant Sci. 2023, 14, 1118313. [Google Scholar] [CrossRef]
- Abbas, A.; Mansha, S.; Waheed, H.; Siddiq, Z.; Hayyat, M.U.; Zhang, Y.-J.; Alwutayd, K. NaCl stress, tissue specific Na+ and K+ up-take and their effect on growth and physiology of Helianthus annuus L. and Solanum lycopersicum L. Sci. Hortic. 2024, 326, 112454. [Google Scholar] [CrossRef]
- Wang, B.; Wang, J.; Yang, T.; Wang, J.; Dai, Q.; Zhang, F.; Xi, R.; Yu, Q.; Li, N. The transcriptional regulatory network of hormones and genes under salt stress in tomato plants (Solanum lycopersicum L.). Front. Plant Sci. 2023, 14, 1115593. [Google Scholar] [CrossRef]
- Shaffique, S.; Kang, S.-M.; Hoque, M.I.U.; Imran, M.; Aaqil khan, M.; Lee, I.-J. Research progress in soybean by phytohormone modulation and metal chelation over the past decade. Agriculture 2023, 13, 1325. [Google Scholar] [CrossRef]
- Shaffique, S.; Hussain, S.; Kang, S.-M.; Imran, M.; Injamum-Ul-Hoque, M.; Khan, M.A.; Lee, I.-J. Phytohormonal modulation of the drought stress in soybean: Outlook, research progress, and cross-talk. Front. Plant Sci. 2023, 14, 1237295. [Google Scholar] [CrossRef]
- Kumar, P.; Kumar, P.; Suniti; Kumar, U.; Avni; Mann, A. Transcriptional Regulatory Network Involved in Drought and Salt Stress Response in Rice. In Salinity and Drought Tolerance in Plants: Physiological Perspectives; Springer: Berlin/Heidelberg, Germany, 2023; pp. 237–274. [Google Scholar]
- Pan, L.; Ma, J.; Li, J.; Yin, B.; Fu, C. Advances of salt stress-responsive transcription factors in plants. Sheng Wu Gong Cheng Xue Bao = Chin. J. Biotechnol. 2022, 38, 50–65. [Google Scholar]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How plant hormones mediate salt stress responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef]
- Lan Thi Hoang, X.; Du Nhi, N.H.; Binh Anh Thu, N.; Phuong Thao, N.; Phan Tran, L.-S. Transcription factors and their roles in signal transduction in plants under abiotic stresses. Curr. Genom. 2017, 18, 483–497. [Google Scholar]
- Kumar, J.; Singh, S.; Singh, M.; Srivastava, P.K.; Mishra, R.K.; Singh, V.P.; Prasad, S.M. Transcriptional regulation of salinity stress in plants: A short review. Plant Gene 2017, 11, 160–169. [Google Scholar] [CrossRef]
- Fernando, V.D. Major transcription factor families involved in salinity stress tolerance in plants. Transcr. Factors Abiotic Stress Toler. Plants 2020, 99–109. [Google Scholar]
- Zhang, T.; Zhou, Y. Plant transcription factors and salt stress. In Plant Transcription Factors; Elsevier: Amsterdam, The Netherlands, 2023; pp. 369–381. [Google Scholar]
- Liu, H.; Tang, X.; Zhang, N.; Li, S.; Si, H. Role of bZIP Transcription Factors in Plant Salt Stress. Int. J. Mol. Sci. 2023, 24, 7893. [Google Scholar] [CrossRef] [PubMed]
- Ray-Jones, H.; Spivakov, M. Transcriptional enhancers and their communication with gene promoters. Cell. Mol. Life Sci. 2021, 78, 6453–6485. [Google Scholar] [CrossRef]
- Hussain, Q.; Asim, M.; Zhang, R.; Khan, R.; Farooq, S.; Wu, J. Transcription factors interact with ABA through gene expression and signaling pathways to mitigate drought and salinity stress. Biomolecules 2021, 11, 1159. [Google Scholar] [CrossRef]
- Chauhan, D.; Singh, D.; Pandey, H.; Khan, Z.; Srivastava, R.; Dhiman, V.K.; Dhiman, V.K. Chapter 13 - Impact of transcription factors in plant abiotic stress: A recent advancement for crop improvement. In Plant Transcription Factors; Srivastava, V., Mishra, S., Mehrotra, S., Upadhyay, S.K., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 271–286. [Google Scholar]
- Kimotho, R.N.; Baillo, E.H.; Zhang, Z. Transcription factors involved in abiotic stress responses in Maize (Zea mays L.) and their roles in enhanced productivity in the post genomics era. PeerJ 2019, 7, e7211. [Google Scholar] [CrossRef]
- Cohen, I.; Zandalinas, S.I.; Huck, C.; Fritschi, F.B.; Mittler, R. Meta-analysis of drought and heat stress combination impact on crop yield and yield components. Physiol. Plant. 2021, 171, 66–76. [Google Scholar]
- Jia, J.; Zhao, P.; Cheng, L.; Yuan, G.; Yang, W.; Liu, S.; Chen, S.; Qi, D.; Liu, G.; Li, X. MADS-box family genes in sheepgrass and their involvement in abiotic stress responses. BMC Plant Biol. 2018, 18, 42. [Google Scholar] [CrossRef]
- Sarkar, B.; Bandyopadhyay, P.; Das, A.; Pal, S.; Hasanuzzaman, M.; Adak, M.K. Abscisic acid priming confers salt tolerance in maize seedlings by modulating osmotic adjustment, bond energies, ROS homeostasis, and organic acid metabolism. Plant Physiol. Biochem. 2023, 202, 107980. [Google Scholar] [CrossRef] [PubMed]
- Quint, M.; Gray, W.M. Auxin signaling. Curr. Opin. Plant Biol. 2006, 9, 448–453. [Google Scholar] [CrossRef]
- Ali, M.S.; Baek, K.-H. Jasmonic acid signaling pathway in response to abiotic stresses in plants. Int. J. Mol. Sci. 2020, 21, 621. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Zander, M.; Huang, S.-s.C.; Xie, M.; Song, L.; Guzmán, J.P.S.; Hann, E.; Shanbhag, B.K.; Ng, S.; Jain, S.; et al. Transcription Factor Dynamics in Cross-Regulation of Plant Hormone Signaling Pathways. bioRxiv 2023. [Google Scholar] [CrossRef]
- Wang, H.; Hao, J.; Chen, X.; Hao, Z.; Wang, X.; Lou, Y.; Peng, Y.; Guo, Z. Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol. Biol. 2007, 65, 799–815. [Google Scholar] [CrossRef]
- Yan, H.; Jia, H.; Chen, X.; Hao, L.; An, H.; Guo, X. The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic Nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production. Plant Cell Physiol. 2014, 55, 2060–2076. [Google Scholar] [CrossRef] [PubMed]
- Cai, R.; Dai, W.; Zhang, C.; Wang, Y.; Wu, M.; Zhao, Y.; Ma, Q.; Xiang, Y.; Cheng, B. The maize WRKY transcription factor ZmWRKY17 negatively regulates salt stress tolerance in transgenic Arabidopsis plants. Planta 2017, 246, 1215–1231. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Chi, X.; Guo, F.; Jin, X.; Luo, H.; Hawar, A.; Chen, Y.; Feng, K.; Wang, B.; Qi, J. SbWRKY30 enhances the drought tolerance of plants and regulates a drought stress-responsive gene, SbRD19, in sorghum. J. Plant Physiol. 2020, 246, 153142. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Li, C.; He, X.; Zhang, X.; Zhu, L. ABA signaling is negatively regulated by GbWRKY1 through JAZ1 and ABI1 to affect salt and drought tolerance. Plant Cell Rep. 2020, 39, 181–194. [Google Scholar] [CrossRef]
- Qin, Z.; Hou, F.; Li, A.; Dong, S.; Wang, Q.; Zhang, L. Transcriptome-wide identification of WRKY transcription factor and their expression profiles under salt stress in sweetpotato (Ipomoea batatas L.). Plant Biotechnol. Rep. 2020, 14, 599–611. [Google Scholar] [CrossRef]
- Chanwala, J.; Satpati, S.; Dixit, A.; Parida, A.; Giri, M.K.; Dey, N. Genome-wide identification and expression analysis of WRKY transcription factors in pearl millet (Pennisetum glaucum) under dehydration and salinity stress. BMC Genom. 2020, 21, 231. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Li, J.; Sui, Y.; Han, G.; Zhang, Y.; Guo, S.; Sui, N. The sweet sorghum SbWRKY50 is negatively involved in salt response by regulating ion homeostasis. Plant Mol. Biol. 2020, 102, 603–614. [Google Scholar] [CrossRef]
- Li, W.; Pang, S.; Lu, Z.; Jin, B. Function and mechanism of WRKY transcription factors in abiotic stress responses of plants. Plants 2020, 9, 1515. [Google Scholar] [CrossRef]
- Han, D.; Hou, Y.; Wang, Y.; Ni, B.; Li, Z.; Yang, G. Overexpression of a Malus baccata WRKY transcription factor gene (MbWRKY5) increases drought and salt tolerance in transgenic tobacco. Can. J. Plant Sci. 2018, 99, 173–183. [Google Scholar] [CrossRef]
- Bankaji, I.; Sleimi, N.; Vives-Peris, V.; Gomez-Cadenas, A.; Perez-Clemente, R.M. Identification and expression of the Cucurbita WRKY transcription factors in response to water deficit and salt stress. Sci. Hortic. 2019, 256, 108562. [Google Scholar] [CrossRef]
- Lin, L.; Yuan, K.; Huang, Y.; Dong, H.; Qiao, Q.; Xing, C.; Huang, X.; Zhang, S. A WRKY transcription factor PbWRKY40 from Pyrus betulaefolia functions positively in salt tolerance and modulating organic acid accumulation by regulating PbVHA-B1 expression. Environ. Exp. Bot. 2022, 196, 104782. [Google Scholar] [CrossRef]
- Zhu, L.; Li, S.; Ouyang, M.; Yang, L.; Sun, S.; Wang, Y.; Cai, X.; Wu, G.; Li, Y. Overexpression of watermelon ClWRKY20 in transgenic Arabidopsis improves salt and low-temperature tolerance. Sci. Hortic. 2022, 295, 110848. [Google Scholar] [CrossRef]
- Han, D.; Xu, T.; Han, J.; Liu, W.; Wang, Y.; Li, X.; Sun, X.; Wang, X.; Li, T.; Yang, G. Overexpression of MxWRKY53 increased iron and high salinity stress tolerance in Arabidopsis thaliana. Vitr. Cell. Dev. Biol.-Plant 2021, 266–278. [Google Scholar] [CrossRef]
- Han, D.; Han, J.; Xu, T.; Li, T.; Yao, C.; Wang, Y.; Luo, D.; Yang, G. Isolation and preliminary functional characterization of MxWRKY64, a new WRKY transcription factor gene from Malus xiaojinensis Cheng et Jiang. Vitr. Cell. Dev. Biol.-Plant 2021, 57, 202–213. [Google Scholar] [CrossRef]
- Zhu, H.; Jiang, Y.; Guo, Y.; Huang, J.; Zhou, M.; Tang, Y.; Sui, J.; Wang, J.; Qiao, L. A novel salt inducible WRKY transcription factor gene, AhWRKY75, confers salt tolerance in transgenic peanut. Plant Physiol. Biochem. 2021, 160, 175–183. [Google Scholar] [CrossRef]
- Xiang, X.-Y.; Chen, J.; Xu, W.-X.; Qiu, J.-R.; Song, L.; Wang, J.-T.; Tang, R.; Chen, D.; Jiang, C.-Z.; Huang, Z. Dehydration-induced WRKY transcriptional factor MfWRKY70 of Myrothamnus flabellifolia enhanced drought and salinity tolerance in Arabidopsis. Biomolecules 2021, 11, 327. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.-T.; Yu, D.-Q. Expression profiles of AtWRKY25, AtWRKY26 and AtWRKY33 under abiotic stresses. Yi Chuan = Hered. 2010, 32, 848–856. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Wang, N.-N.; Gong, S.-Y.; Lu, R.; Li, Y.; Li, X.-B. Overexpression of a cotton (Gossypium hirsutum) WRKY gene, GhWRKY34, in Arabidopsis enhances salt-tolerance of the transgenic plants. Plant Physiol. Biochem. 2015, 96, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Shen, Z.; Zhang, Y.; Wu, X.; Wang, J.; Sa, G.; Zhang, Y.; Zhang, H.; Deng, C.; Liu, J. Populus euphratica WRKY1 binds the promoter of H+-ATPase gene to enhance gene expression and salt tolerance. J. Exp. Bot. 2020, 71, 1527–1539. [Google Scholar] [CrossRef]
- Singh, D.; Debnath, P.; Sane, A.P.; Sane, V.A. Tomato (Solanum lycopersicum) WRKY23 enhances salt and osmotic stress tolerance by modulating the ethylene and auxin pathways in transgenic Arabidopsis. Plant Physiol. Biochem. 2023, 195, 330–340. [Google Scholar] [CrossRef]
- Liu, J.X.; Srivastava, R.; Che, P.; Howell, S.H. Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J. 2007, 51, 897–909. [Google Scholar] [CrossRef]
- Huang, C.; Zhou, J.; Jie, Y.; Xing, H.; Zhong, Y.; Yu, W.; She, W.; Ma, Y.; Liu, Z.; Zhang, Y. A ramie bZIP transcription factor BnbZIP2 is involved in drought, salt, and heavy metal stress response. DNA Cell Biol. 2016, 35, 776–786. [Google Scholar] [CrossRef]
- Lee, S.C.; Choi, H.W.; Hwang, I.S.; Choi, D.S.; Hwang, B.K. Functional roles of the pepper pathogen-induced bZIP transcription factor, CAbZIP1, in enhanced resistance to pathogen infection and environmental stresses. Planta 2006, 224, 1209–1225. [Google Scholar] [CrossRef]
- Gai, W.-X.; Ma, X.; Qiao, Y.-M.; Shi, B.-H.; ul Haq, S.; Li, Q.-H.; Wei, A.-M.; Liu, K.-K.; Gong, Z.-H. Characterization of the bZIP transcription factor family in pepper (Capsicum annuum L.): CabZIP25 positively modulates the salt tolerance. Front. Plant Sci. 2020, 11, 139. [Google Scholar] [CrossRef]
- Yang, Y.; Yu, T.-F.; Ma, J.; Chen, J.; Zhou, Y.-B.; Chen, M.; Ma, Y.-Z.; Wei, W.-L.; Xu, Z.-S. The soybean bZIP transcription factor gene GmbZIP2 confers drought and salt resistances in transgenic plants. Int. J. Mol. Sci. 2020, 21, 670. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, H.; Ohmiya, K.; Hattori, T. A rice bZIP protein, designated OSBZ8, is rapidly induced by abscisic acid. Plant J. 1996, 9, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xie, J.; Wang, L.; Si, L.; Zheng, S.; Yang, Y.; Yang, H.; Tian, S. Wheat TabZIP8, 9, 13 participate in ABA biosynthesis in NaCl-stressed roots regulated by TaCDPK9-1. Plant Physiol. Biochem. 2020, 151, 650–658. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, L.; Xia, C.; Gao, L.; Hao, C.; Zhao, G.; Jia, J.; Kong, X. A novel wheat C-bZIP gene, TabZIP14-B, participates in salt and freezing tolerance in transgenic plants. Front. Plant Sci. 2017, 8, 710. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Liu, G.; Liu, Y.; Zheng, L.; Nie, X.; Wang, Y. The bZIP protein from Tamarix hispida, ThbZIP1, is ACGT elements binding factor that enhances abiotic stress signaling in transgenic Arabidopsis. BMC Plant Biol. 2013, 13, 151. [Google Scholar] [CrossRef]
- Bi, C.; Yu, Y.; Dong, C.; Yang, Y.; Zhai, Y.; Du, F.; Xia, C.; Ni, Z.; Kong, X.; Zhang, L. The bZIP transcription factor TabZIP15 improves salt stress tolerance in wheat. Plant Biotechnol. J. 2021, 19, 209. [Google Scholar] [CrossRef]
- Wang, Z.; Zhu, J.; Yuan, W.; Wang, Y.; Hu, P.; Jiao, C.; Xia, H.; Wang, D.; Cai, Q.; Li, J. Genome-wide characterization of bZIP transcription factors and their expression patterns in response to drought and salinity stress in Jatropha curcas. Int. J. Biol. Macromol. 2021, 181, 1207–1223. [Google Scholar] [CrossRef]
- Ma, H.; Liu, C.; Li, Z.; Ran, Q.; Xie, G.; Wang, B.; Fang, S.; Chu, J.; Zhang, J. ZmbZIP4 contributes to stress resistance in maize by regulating ABA synthesis and root development. Plant Physiol. 2018, 178, 753–770. [Google Scholar] [CrossRef]
- Orellana, S.; Yanez, M.; Espinoza, A.; Verdugo, I.; Gonzalez, E.; RUIZ-LARA, S.; Casaretto, J.A. The transcription factor SlAREB1 confers drought, salt stress tolerance and regulates biotic and abiotic stress-related genes in tomato. Plant Cell Environ. 2010, 33, 2191–2208. [Google Scholar] [CrossRef]
- Zhao, P.; Ye, M.; Wang, R.; Wang, D.; Chen, Q. Systematic identification and functional analysis of potato (Solanum tuberosum L.) bZIP transcription factors and overexpression of potato bZIP transcription factor StbZIP-65 enhances salt tolerance. Int. J. Biol. Macromol. 2020, 161, 155–167. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, C.; Li, Z.; Sun, J.; Wang, D.; Xu, L.; Li, X.; Guo, Y. Identification and analysis of bZIP family genes in potato and their potential roles in stress responses. Front. Plant Sci. 2021, 12, 637343. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zhang, L.; Sun, Y.; Xue, J.; Gao, C.; Yuan, L.; Wang, J.; Jia, X.; Li, R. Genome-wide characterization of bZIP transcription factors in foxtail millet and their expression profiles in response to drought and salt stresses. Chin. Bull. Bot. 2016, 51, 473. [Google Scholar]
- Qu, D.; Wu, F.; Zhao, X.; Zhu, D.; Gu, L.; Yang, L.; Zhao, W.; Sun, Y.; Yang, J.; Tian, W. A bZIP transcription factor VabZIP12 from blueberry induced by dark septate endocyte improving the salt tolerance of transgenic Arabidopsis. Plant Sci. 2022, 315, 111135. [Google Scholar] [CrossRef]
- Jiang, M.; Wang, Z.; Ren, W.; Yan, S.; Xing, N.; Zhang, Z.; Li, H.; Ma, W. Identification of the bZIP gene family and regulation of metabolites under salt stress in Isatis indigotica. Front. Plant Sci. 2022, 13, 1011616. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Zhai, H.; He, S.; Zhao, N.; Liu, Q. A novel sweetpotato bZIP transcription factor gene, IbbZIP1, is involved in salt and drought tolerance in transgenic Arabidopsis. Plant Cell Rep. 2019, 38, 1373–1382. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Bao, X.; Zhi, Y.; Wu, Q.; Guo, Y.; Yin, X.; Zeng, L.; Li, J.; Zhang, J.; He, W. Overexpression of a MYB family gene, OsMYB6, increases drought and salinity stress tolerance in transgenic rice. Front. Plant Sci. 2019, 10, 168. [Google Scholar] [CrossRef]
- Xiong, H.; Li, J.; Liu, P.; Duan, J.; Zhao, Y.; Guo, X.; Li, Y.; Zhang, H.; Ali, J.; Li, Z. Overexpression of OsMYB48-1, a novel MYB-related transcription factor, enhances drought and salinity tolerance in rice. PLoS ONE 2014, 9, e92913. [Google Scholar] [CrossRef]
- Yoo, J.H.; Park, C.Y.; Kim, J.C.; Do Heo, W.; Cheong, M.S.; Park, H.C.; Kim, M.C.; Moon, B.C.; Choi, M.S.; Kang, Y.H. Direct interaction of a divergent CaM isoform and the transcription factor, MYB2, enhances salt tolerance in Arabidopsis. J. Biol. Chem. 2005, 280, 3697–3706. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, G.; Zhao, G.; Xia, C.; Jia, J.; Liu, X.; Kong, X. Characterization of a wheat R2R3-MYB transcription factor gene, TaMYB19, involved in enhanced abiotic stresses in Arabidopsis. Plant Cell Physiol. 2014, 55, 1802–1812. [Google Scholar] [CrossRef]
- Liao, Y.; Zou, H.-F.; Wang, H.-W.; Zhang, W.-K.; Ma, B.; Zhang, J.-S.; Chen, S.-Y. Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants. Cell Res. 2008, 18, 1047–1060. [Google Scholar] [CrossRef]
- Zhao, K.; Cheng, Z.; Guo, Q.; Yao, W.; Liu, H.; Zhou, B.; Jiang, T. Characterization of the poplar R2R3-MYB gene family and over-expression of PsnMYB108 confers salt tolerance in transgenic tobacco. Front. Plant Sci. 2020, 11, 571881. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, L.; Shi, Q.; Ren, Z. SlMYB102, an R2R3-type MYB gene, confers salt tolerance in transgenic tomato. Plant Sci. 2020, 291, 110356. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Li, X.; Huang, X.; Ma, T.; Liang, Y.; Ma, X.; Peng, X.; Jia, J.; Chen, S.; Chen, Y. Overexpression of sheepgrass R1-MYB transcription factor LcMYB1 confers salt tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2013, 70, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Nulu, J.K.; AM, A.J.; Venkatesh, B.; Jayamma, N.; Reddy, B.M.; Pandurangaiah, M. R2R3 MYB Transcription Factor, AhMYB94 Plays a Crucial Role in Stress adaptation of a Salt Susceptible Groundnut Cultivar-K6. 2022; preprint. [Google Scholar] [CrossRef]
- Wu, J.; Jiang, Y.; Liang, Y.; Chen, L.; Chen, W.; Cheng, B. Expression of the maize MYB transcription factor ZmMYB3R enhances drought and salt stress tolerance in transgenic plants. Plant Physiol. Biochem. 2019, 137, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Liu, X.; Li, D.; Gao, T.; Song, Y. Transcriptional profiling reveals that a MYB transcription factor MsMYB4 contributes to the salinity stress response of alfalfa. PLoS ONE 2018, 13, e0204033. [Google Scholar] [CrossRef]
- Ren, C.; Li, Z.; Song, P.; Wang, Y.; Liu, W.; Zhang, L.; Li, X.; Li, W.; Han, D. Overexpression of a grape MYB transcription factor gene VhMYB2 increases salinity and drought tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2023, 24, 10743. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Fan, C.; Wei, Y.; Meng, J.; Li, Z.; Zhong, C. Genome-wide analysis of MYB transcription factors and their responses to salt stress in Casuarina equisetifolia. BMC Plant Biol. 2021, 21, 328. [Google Scholar] [CrossRef]
- Ullah, A.; Ul Qamar, M.T.; Nisar, M.; Hazrat, A.; Rahim, G.; Khan, A.H.; Hayat, K.; Ahmed, S.; Ali, W.; Khan, A. Characterization of a novel cotton MYB gene, GhMYB108-like responsive to abiotic stresses. Mol. Biol. Rep. 2020, 47, 1573–1581. [Google Scholar] [CrossRef]
- ZHANG, Y.-b.; Wei, T.; WANG, L.-h.; HU, Y.-w.; LIU, X.-w.; LIU, Y.-s. Kiwifruit (Actinidia chinensis) R1R2R3-MYB transcription factor AcMYB3R enhances drought and salinity tolerance in Arabidopsis thaliana. J. Integr. Agric. 2019, 18, 417–427. [Google Scholar] [CrossRef]
- Shan, H.; Chen, S.; Jiang, J.; Chen, F.; Chen, Y.; Gu, C.; Li, P.; Song, A.; Zhu, X.; Gao, H. Heterologous expression of the chrysanthemum R2R3-MYB transcription factor CmMYB2 enhances drought and salinity tolerance, increases hypersensitivity to ABA and delays flowering in Arabidopsis thaliana. Mol. Biotechnol. 2012, 51, 160–173. [Google Scholar] [CrossRef]
- Shen, X.; Guo, X.; Guo, X.; Zhao, D.; Zhao, W.; Chen, J.; Li, T. PacMYBA, a sweet cherry R2R3-MYB transcription factor, is a positive regulator of salt stress tolerance and pathogen resistance. Plant Physiol. Biochem. 2017, 112, 302–311. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.-H.; Zhang, S.-Z.; Wang, R.-K.; Zhang, R.-F.; Hao, Y.-J. Genome wide analysis of the apple MYB transcription factor family allows the identification of MdoMYB121 gene confering abiotic stress tolerance in plants. PLoS ONE 2013, 8, e69955. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, K.; Sheng, S.; Wang, M.; Hua, P.; Wang, Y.; Chen, P.; Wang, K.; Zhao, M.; Wang, Y. Transcriptome analysis of MYB transcription factors family and PgMYB genes involved in salt stress resistance in Panax ginseng. BMC Plant Biol. 2022, 22, 479. [Google Scholar] [CrossRef] [PubMed]
- Yong, Y.; Zhang, Y.; Lyu, Y. A MYB-related transcription factor from Lilium lancifolium L.(LlMYB3) is involved in anthocyanin biosynthesis pathway and enhances multiple abiotic stress tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2019, 20, 3195. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Cui, Z.; Li, Y.; Kang, Y.; Song, X.; Wang, J.; Zhou, Y. Genome-wide identification and expression analysis of MYB transcription factor superfamily in Dendrobium catenatum. Front. Genet. 2021, 12, 714696. [Google Scholar] [CrossRef]
- Su, Y.; Liu, Y.; Xiao, S.; Wang, Y.; Deng, Y.; Zhao, L.; Wang, Y.; Zhao, D.; Dai, X.; Zhou, Z. Isolation, characterization, and functional verification of salt stress response genes of NAC transcription factors in Ipomoea pes-caprae. Front. Plant Sci. 2023, 14, 1119282. [Google Scholar] [CrossRef]
- Liao, Y.-D.; Lin, K.-H.; Chen, C.-C.; Chiang, C.-M. Oryza sativa protein phosphatase 1a (OsPP1a) involved in salt stress tolerance in transgenic rice. Mol. Breed. 2016, 36, 22. [Google Scholar] [CrossRef]
- Li, M.; Wu, Z.; Gu, H.; Cheng, D.; Guo, X.; Li, L.; Shi, C.; Xu, G.; Gu, S.; Abid, M. AvNAC030, a NAC domain transcription factor, enhances salt stress tolerance in Kiwifruit. Int. J. Mol. Sci. 2021, 22, 11897. [Google Scholar] [CrossRef]
- Liu, X.; Baird, W.V. Differential expression of genes regulated in response to drought or salinity stress in sunflower. Crop Sci. 2003, 43, 678–687. [Google Scholar] [CrossRef]
- Peng, H.; Yu, X.; Cheng, H.; Shi, Q.; Zhang, H.; Li, J.; Ma, H. Cloning and characterization of a novel NAC family gene CarNAC1 from chickpea (Cicer arietinum L.). Mol. Biotechnol. 2010, 44, 30–40. [Google Scholar] [CrossRef]
- Zhong, H.; Guo, Q.-Q.; Chen, L.; Ren, F.; Wang, Q.-Q.; Zheng, Y.; Li, X.-B. Two Brassica napus genes encoding NAC transcription factors are involved in response to high-salinity stress. Plant Cell Rep. 2012, 31, 1991–2003. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; He, M.; Dong, F.; Cai, Y.; Gao, W.; Zhou, Y.; Huang, H.; Dai, S. The Chrysanthemum lavandulifolium ClNAC9 gene positively regulates saline, alkaline, and drought stress in transgenic Chrysanthemum grandiflora. J. Am. Soc. Hortic. Sci. 2019, 144, 280–288. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, S.; Guan, C.; Kong, X.; Wang, Y.; Cui, Y.; Liu, B.; Zhou, Y.; Zhang, Y. Overexpressing the NAC transcription factor LpNAC13 from Lilium pumilum in tobacco negatively regulates the drought response and positively regulates the salt response. Plant Physiol. Biochem. 2020, 149, 96–110. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Ying, S.; Zhang, D.-F.; Shi, Y.-S.; Song, Y.-C.; Wang, T.-Y.; Li, Y. A maize stress-responsive NAC transcription factor, ZmSNAC1, confers enhanced tolerance to dehydration in transgenic Arabidopsis. Plant Cell Rep. 2012, 31, 1701–1711. [Google Scholar] [CrossRef]
- Dudhate, A.; Shinde, H.; Yu, P.; Tsugama, D.; Gupta, S.K.; Liu, S.; Takano, T. Comprehensive analysis of NAC transcription factor family uncovers drought and salinity stress response in pearl millet (Pennisetum glaucum). BMC Genom. 2021, 22, 70. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Huang, J.; Wang, X.; Dang, H.; Jiang, T.; Han, Y. Expression analysis of the NAC transcription factor family of populus in response to salt stress. Forests 2019, 10, 688. [Google Scholar] [CrossRef]
- Li, M.; Chen, R.; Jiang, Q.; Sun, X.; Zhang, H.; Hu, Z. GmNAC06, a NAC domain transcription factor enhances salt stress tolerance in soybean. Plant Mol. Biol. 2021, 105, 333–345. [Google Scholar] [CrossRef]
- Wei, S.; Gao, L.; Zhang, Y.; Zhang, F.; Yang, X.; Huang, D. Genome-wide investigation of the NAC transcription factor family in melon (Cucumis melo L.) and their expression analysis under salt stress. Plant Cell Rep. 2016, 35, 1827–1839. [Google Scholar] [CrossRef]
- Tak, H.; Negi, S.; Ganapathi, T. Banana NAC transcription factor MusaNAC042 is positively associated with drought and salinity tolerance. Protoplasma 2017, 254, 803–816. [Google Scholar] [CrossRef]
- Yu, X.; Liu, Y.; Wang, S.; Tao, Y.; Wang, Z.; Shu, Y.; Peng, H.; Mijiti, A.; Wang, Z.; Zhang, H. CarNAC4, a NAC-type chickpea transcription factor conferring enhanced drought and salt stress tolerances in Arabidopsis. Plant Cell Rep. 2016, 35, 613–627. [Google Scholar] [CrossRef]
- An, J.P.; Yao, J.F.; Xu, R.R.; You, C.X.; Wang, X.F.; Hao, Y.J. An apple NAC transcription factor enhances salt stress tolerance by modulating the ethylene response. Physiol. Plant. 2018, 164, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Punia, H.; Tokas, J.; Malik, A.; Sangwan, S.; Rani, A.; Yashveer, S.; Alansi, S.; Hashim, M.J.; El-Sheikh, M.A. Genome-wide transcriptome profiling, characterization, and functional identification of NAC transcription factors in sorghum under salt stress. Antioxidants 2021, 10, 1605. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Du, M.; Zhou, Z.; Wang, S.; Li, T.; Han, J.; Xu, T.; Yang, G. Overexpression of a Malus baccata NAC transcription factor gene MbNAC25 increases cold and salinity tolerance in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 1198. [Google Scholar] [CrossRef]
- Wang, L.; Li, Z.; Lu, M.; Wang, Y. ThNAC13, a NAC transcription factor from Tamarix hispida, confers salt and osmotic stress tolerance to transgenic Tamarix and Arabidopsis. Front. Plant Sci. 2017, 8, 635. [Google Scholar] [CrossRef]
- Min, X.; Jin, X.; Zhang, Z.; Wei, X.; Ndayambaza, B.; Wang, Y.; Liu, W. Genome-Wide Identification of NAC Transcription Factor Family and Functional Analysis of the Abiotic Stress-Responsive Genes in Medicago sativa L. J. Plant Growth Regul. 2020, 39, 324–337. [Google Scholar] [CrossRef]
- Bokolia, M.; Singh, B.; Kumar, A.; Goyal, N.; Singh, K.; Chhabra, R. Genome-wide identification of NAC transcription factors in Avena sativa under salinity stress. Plant Stress 2023, 10, 100276. [Google Scholar] [CrossRef]
- Jia, H.; Wang, C.; Wang, F.; Liu, S.; Li, G.; Guo, X. GhWRKY68 reduces resistance to salt and drought in transgenic Nicotiana benthamiana. PLoS ONE 2015, 10, e0120646. [Google Scholar] [CrossRef]
- Yokotani, N.; Ichikawa, T.; Kondou, Y.; Matsui, M.; Hirochika, H.; Iwabuchi, M.; Oda, K. Tolerance to various environmental stresses conferred by the salt-responsive rice gene ONAC063 in transgenic Arabidopsis. Planta 2009, 229, 1065–1075. [Google Scholar] [CrossRef]
- Nakashima, K.; Ito, Y.; Yamaguchi-Shinozaki, K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009, 149, 88–95. [Google Scholar] [CrossRef]
- Zou, M.; Guan, Y.; Ren, H.; Zhang, F.; Chen, F. Characterization of alternative splicing products of bZIP transcription factors OsABI5. Biochem. Biophys. Res. Commun. 2007, 360, 307–313. [Google Scholar] [CrossRef]
- Chaudhry, U.K.; Gökçe, Z.N.Ö.; Gökçe, A.F. The influence of salinity stress on plants and their molecular mechanisms. Biol. Life Sci. Forum 2021, 11, 31. [Google Scholar]
- Guan, Y.; Hwarari, D.; Korboe, H.M.; Ahmad, B.; Cao, Y.; Movahedi, A.; Yang, L. Low temperature stress-induced perception and molecular signaling pathways in plants. Environ. Exp. Bot. 2023, 207, 105190. [Google Scholar] [CrossRef]
- Long, L.; Gu, L.; Wang, S.; Cai, H.; Wu, J.; Wang, J.; Yang, M. Progress in the understanding of WRKY transcription factors in woody plants. Int. J. Biol. Macromol. 2023, 242, 124379. [Google Scholar] [CrossRef]
- Kamali, S.; Singh, A. Genomic and transcriptomic approaches to developing abiotic stress-resilient crops. Agronomy 2023, 13, 2903. [Google Scholar] [CrossRef]
- Samarina, L.; Wang, S.; Malyukova, L.; Bobrovskikh, A.; Doroshkov, A.; Koninskaya, N.; Shkhalakhova, R.; Matskiv, A.; Fedorina, J.; Fizikova, A. Long-term cold, freezing and drought: Overlapping and specific regulatory mechanisms and signal transduction in tea plant (Camellia sinensis (L.) Kuntze). Front. Plant Sci. 2023, 14, 1145793. [Google Scholar] [CrossRef] [PubMed]
- Ha, C.V.; Nasr Esfahani, M.; Watanabe, Y.; Tran, U.T.; Sulieman, S.; Mochida, K.; Van Nguyen, D.; Tran, L.-S.P. Genome-wide identification and expression analysis of the CaNAC family members in chickpea during development, dehydration and ABA treatments. PLoS ONE 2014, 9, e114107. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, E.G.G.; Leite, J.P.; Marin, S.R.R.; Marinho, J.P.; de Fátima Corrêa Carvalho, J.; Fuganti-Pagliarini, R.; Farias, J.R.B.; Neumaier, N.; Marcelino-Guimarães, F.C.; de Oliveira, M.C.N. Overexpression of the ABA-dependent AREB1 transcription factor from Arabidopsis thaliana improves soybean tolerance to water deficit. Plant Mol. Biol. Report. 2013, 31, 719–730. [Google Scholar] [CrossRef]
- Kim, J.-S.; Mizoi, J.; Yoshida, T.; Fujita, Y.; Nakajima, J.; Ohori, T.; Todaka, D.; Nakashima, K.; Hirayama, T.; Shinozaki, K. An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A gene, which encodes a transcription factor regulating drought-inducible genes in Arabidopsis. Plant Cell Physiol. 2011, 52, 2136–2146. [Google Scholar] [CrossRef]
- Narusaka, Y.; Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Furihata, T.; Abe, H.; Narusaka, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003, 34, 137–148. [Google Scholar] [CrossRef]
- Xu, Z.-Y.; Kim, S.Y.; Hyeon, D.Y.; Kim, D.H.; Dong, T.; Park, Y.; Jin, J.B.; Joo, S.-H.; Kim, S.-K.; Hong, J.C. The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell 2013, 25, 4708–4724. [Google Scholar] [CrossRef]
- Oh, S.-J.; Song, S.I.; Kim, Y.S.; Jang, H.-J.; Kim, S.Y.; Kim, M.; Kim, Y.-K.; Nahm, B.H.; Kim, J.-K. Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol. 2005, 138, 341–351. [Google Scholar] [CrossRef]
- Wu, Y.; Deng, Z.; Lai, J.; Zhang, Y.; Yang, C.; Yin, B.; Zhao, Q.; Zhang, L.; Li, Y.; Yang, C. Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 2009, 19, 1279–1290. [Google Scholar] [CrossRef] [PubMed]
- Vannini, C.; Campa, M.; Iriti, M.; Genga, A.; Faoro, F.; Carravieri, S.; Rotino, G.L.; Rossoni, M.; Spinardi, A.; Bracale, M. Evaluation of transgenic tomato plants ectopically expressing the rice Osmyb4 gene. Plant Sci. 2007, 173, 231–239. [Google Scholar] [CrossRef]
- Qiu, Y.; Yu, D. Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environ. Exp. Bot. 2009, 65, 35–47. [Google Scholar] [CrossRef]
- Xiao, F.; Zhou, H. Plant salt response: Perception, signaling, and tolerance. Front. Plant Sci. 2023, 13, 1053699. [Google Scholar] [CrossRef]
- Waadt, R.; Seller, C.A.; Hsu, P.-K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef]
- Fahad, S.; Hussain, S.; Matloob, A.; Khan, F.A.; Khaliq, A.; Saud, S.; Hassan, S.; Shan, D.; Khan, F.; Ullah, N. Phytohormones and plant responses to salinity stress: A review. Plant Growth Regul. 2015, 75, 391–404. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 2000, 3, 217–223. [Google Scholar] [CrossRef]
- Swamy, P.; Smith, B.N. Role of abscisic acid in plant stress tolerance. Curr. Sci. 1999, 1220–1227. [Google Scholar]
- Zhao, Y.; Zhang, Z.; Gao, J.; Wang, P.; Hu, T.; Wang, Z.; Hou, Y.J.; Wan, Y.; Liu, W.; Xie, S.; et al. Arabidopsis Duodecuple Mutant of PYL ABA Receptors Reveals PYL Repression of ABA-Independent SnRK2 Activity. Cell Rep. 2018, 23, 3340–3351.e3345. [Google Scholar] [CrossRef]
- Aleman, F.; Yazaki, J.; Lee, M.; Takahashi, Y.; Kim, A.Y.; Li, Z.; Kinoshita, T.; Ecker, J.R.; Schroeder, J.I. An ABA-increased interaction of the PYL6 ABA receptor with MYC2 transcription factor: A putative link of ABA and JA signaling. Sci. Rep. 2016, 6, 28941. [Google Scholar] [CrossRef]
- Qin, F.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol. 2011, 52, 1569–1582. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, R.; Watanabe, Y.; Leyva-Gonzalez, M.A.; Van Ha, C.; Fujita, Y.; Tanaka, M.; Seki, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Herrera-Estrella, L. Arabidopsis AHP2, AHP3, and AHP5 histidine phosphotransfer proteins function as redundant negative regulators of drought stress response. Proc. Natl. Acad. Sci. USA 2013, 110, 4840–4845. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, S.; Kudoyarova, G.R.; Veselov, D.S.; Arkhipova, T.N.; Davies, W.J. Plant hormone interactions: Innovative targets for crop breeding and management. J. Exp. Bot. 2012, 63, 3499–3509. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.; Azhar, M.T.; Hinze, L.; Qayyum, A.; Li, H.; Peng, Z.; Qin, G.; Jia, Y.; Pan, Z.; He, S.; et al. Insight into abscisic acid perception and signaling to increase plant tolerance to abiotic stress. J. Plant Interact. 2021, 16, 222–237. [Google Scholar] [CrossRef]
- Ko, J.H.; Yang, S.H.; Han, K.H. Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J. 2006, 47, 343–355. [Google Scholar] [CrossRef]
- Achard, P.; Genschik, P. Releasing the brakes of plant growth: How GAs shutdown DELLA proteins. J. Exp. Bot. 2009, 60, 1085–1092. [Google Scholar] [CrossRef]
- Xi, W.; Liu, C.; Hou, X.; Yu, H. MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 2010, 22, 1733–1748. [Google Scholar] [CrossRef]
- Jusovic, M.; Velitchkova, M.Y.; Misheva, S.P.; Börner, A.; Apostolova, E.L.; Dobrikova, A.G. Photosynthetic responses of a wheat mutant (Rht-B1c) with altered DELLA proteins to salt stress. J. Plant Growth Regul. 2018, 37, 645–656. [Google Scholar] [CrossRef]
- Zhao, C.; Jiang, W.; Zayed, O.; Liu, X.; Tang, K.; Nie, W.; Li, Y.; Xie, S.; Li, Y.; Long, T. The LRXs-RALFs-FER module controls plant growth and salt stress responses by modulating multiple plant hormones. Natl. Sci. Rev. 2021, 8, nwaa149. [Google Scholar] [CrossRef]
- Tao, J.-J.; Chen, H.-W.; Ma, B.; Zhang, W.-K.; Chen, S.-Y.; Zhang, J.-S. The role of ethylene in plants under salinity stress. Front. Plant Sci. 2015, 6, 1059. [Google Scholar] [CrossRef]
- Gieniec, M.; Miszalski, Z.; Rozpądek, P.; Jędrzejczyk, R.J.; Czernicka, M.; Nosek, M. How the Ethylene Biosynthesis Pathway of Semi-Halophytes Is Modified with Prolonged Salinity Stress Occurrence? Int. J. Mol. Sci. 2024, 25, 4777. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; He, S.; Gong, W.; Sun, J.; Pan, Z.; Xu, F.; Lu, Y.; Du, X. Comprehensive analysis of differentially expressed genes and transcriptional regulation induced by salt stress in two contrasting cotton genotypes. BMC Genom. 2014, 15, 760. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zou, X.; Chen, G.; Meng, Y.; Ma, Q.; Chen, Q.; Wang, Z.; Li, F. Potential Roles of 1-Aminocyclopropane-1-carboxylic Acid Synthase Genes in the Response of Gossypium Species to Abiotic Stress by Genome-Wide Identification and Expression Analysis. Plants 2022, 11, 1524. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zhang, J.; Chen, L.; Shi, X.; Lui, Z.; Li, C. Heterologous expression of ACC deaminase from Trichoderma asperellum improves the growth performance of Arabidopsis thaliana under normal and salt stress conditions. Plant Physiol. Biochem. 2015, 94, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.H.; Liu, J.; Zhou, Q.Y.; Cao, Y.R.; Zheng, S.F.; Du, B.X.; Zhang, J.S.; Chen, S.Y. Expression of tobacco ethylene receptor NTHK1 alters plant responses to salt stress. Plant Cell Environ. 2006, 29, 1210–1219. [Google Scholar] [CrossRef]
- Shen, X.; Wang, Z.; Song, X.; Xu, J.; Jiang, C.; Zhao, Y.; Ma, C.; Zhang, H. Transcriptomic profiling revealed an important role of cell wall remodeling and ethylene signaling pathway during salt acclimation in Arabidopsis. Plant Mol. Biol. 2014, 86, 303–317. [Google Scholar] [CrossRef]
- Ryu, H.; Cho, Y.-G. Plant hormones in salt stress tolerance. J. Plant Biol. 2015, 58, 147–155. [Google Scholar] [CrossRef]
- Saeng-ngam, S.; Takpirom, W.; Buaboocha, T.; Chadchawan, S. The role of the OsCam1-1 salt stress sensor in ABA accumulation and salt tolerance in rice. J. Plant Biol. 2012, 55, 198–208. [Google Scholar] [CrossRef]
- Moons, A.; Bauw, G.; Prinsen, E.; Van Montagu, M.; Van Der Straeten, D. Molecular and physiological responses to abscisic acid and salts in roots of salt-sensitive and salt-tolerant Indica rice varieties. Plant Physiol. 1995, 107, 177–186. [Google Scholar] [CrossRef]
- He, T.; Cramer, G.R. Abscisic acid concentrations are correlated with leaf area reductions in two salt-stressed rapid-cycling Brassica species. Plant Soil 1996, 179, 25–33. [Google Scholar] [CrossRef]
- Cramer, G.R.; Quarrie, S.A. Abscisic acid is correlated with the leaf growth inhibition of four genotypes of maize differing in their response to salinity. Funct. Plant Biol. 2002, 29, 111–115. [Google Scholar] [CrossRef]
- Rao, K.P.; Richa, T.; Kumar, K.; Raghuram, B.; Sinha, A.K. In silico analysis reveals 75 members of mitogen-activated protein kinase kinase kinase gene family in rice. DNA Res. 2010, 17, 139–153. [Google Scholar] [CrossRef]
- Jung, J.-H.; Park, C.-M. Auxin modulation of salt stress signaling in Arabidopsis seed germination. Plant Signal. Behav. 2011, 6, 1198–1200. [Google Scholar] [CrossRef] [PubMed]
- Marzi, D.; Brunetti, P.; Saini, S.S.; Yadav, G.; Puglia, G.D.; Dello Ioio, R. Role of transcriptional regulation in auxin-mediated response to abiotic stresses. Front. Genet. 2024, 15, 1394091. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K. Auxin and the integration of environmental signals into plant root development. Ann. Bot. 2013, 112, 1655–1665. [Google Scholar] [CrossRef] [PubMed]
- Talaat, N.B. Abiotic stresses-induced physiological alteration in wheat. Wheat Prod. Chang. Environ. Responses Adapt. Toler. 2019, 1–30. [Google Scholar]
- Nishiyama, R.; Watanabe, Y.; Fujita, Y.; Le, D.T.; Kojima, M.; Werner, T.; Vankova, R.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Kakimoto, T. Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell 2011, 23, 2169–2183. [Google Scholar] [CrossRef]
- Weidemüller, P.; Kholmatov, M.; Petsalaki, E.; Zaugg, J.B. Transcription factors: Bridge between cell signaling and gene regulation. Proteomics 2021, 21, 2000034. [Google Scholar] [CrossRef]
- Khadem, A.; Moshtaghi, N.; Bagheri, A. Regulatory networks of hormone-involved transcription factors and their downstream pathways during somatic embryogenesis of Arabidopsis thaliana. 3 Biotech 2023, 13, 132. [Google Scholar] [CrossRef]
- Altmann, M.; Altmann, S.; Rodriguez, P.A.; Weller, B.; Elorduy Vergara, L.; Palme, J.; Marín-de la Rosa, N.; Sauer, M.; Wenig, M.; Villaécija-Aguilar, J.A. Extensive signal integration by the phytohormone protein network. Nature 2020, 583, 271–276. [Google Scholar] [CrossRef]
- Cluis, C.P.; Mouchel, C.F.; Hardtke, C.S. The Arabidopsis transcription factor HY5 integrates light and hormone signaling pathways. Plant J. 2004, 38, 332–347. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef] [PubMed]
- Depuydt, S.; Hardtke, C.S. Hormone signalling crosstalk in plant growth regulation. Curr. Biol. 2011, 21, R365–R373. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Bano, A.; Ali, S.; Babar, M.A. Crosstalk amongst phytohormones from planta and PGPR under biotic and abiotic stresses. Plant Growth Regul. 2020, 90, 189–203. [Google Scholar] [CrossRef]
- Figueroa-Macías, J.P.; García, Y.C.; Núñez, M.; Díaz, K.; Olea, A.F.; Espinoza, L. Plant growth-defense trade-offs: Molecular processes leading to physiological changes. Int. J. Mol. Sci. 2021, 22, 693. [Google Scholar] [CrossRef]
- Yao, J.; Waters, M.T. Perception of karrikins by plants: A continuing enigma. J. Exp. Bot. 2020, 71, 1774–1781. [Google Scholar] [CrossRef] [PubMed]
- Jaillais, Y.; Chory, J. Unraveling the paradoxes of plant hormone signaling integration. Nat. Struct. Mol. Biol. 2010, 17, 642–645. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.-L.; Yao, J.; Mei, C.-S.; Tong, X.-H.; Zeng, L.-J.; Li, Q.; Xiao, L.-T.; Sun, T.-p.; Li, J.; Deng, X.-W. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 2012, 109, E1192–E1200. [Google Scholar] [CrossRef]
- Nemhauser, J.L.; Hong, F.; Chory, J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 2006, 126, 467–475. [Google Scholar] [CrossRef]
- Zander, M.; Lewsey, M.G.; Clark, N.M.; Yin, L.; Bartlett, A.; Saldierna Guzmán, J.P.; Hann, E.; Langford, A.E.; Jow, B.; Wise, A. Integrated multi-omics framework of the plant response to jasmonic acid. Nat. Plants 2020, 6, 290–302. [Google Scholar] [CrossRef]
- Clark, N.M.; Nolan, T.M.; Wang, P.; Song, G.; Montes, C.; Valentine, C.T.; Guo, H.; Sozzani, R.; Yin, Y.; Walley, J.W. Integrated omics networks reveal the temporal signaling events of brassinosteroid response in Arabidopsis. Nat. Commun. 2021, 12, 5858. [Google Scholar] [CrossRef]
- Moreno, J.E.; Shyu, C.; Campos, M.L.; Patel, L.C.; Chung, H.S.; Yao, J.; He, S.Y.; Howe, G.A. Negative feedback control of jasmonate signaling by an alternative splice variant of JAZ10. Plant Physiol. 2013, 162, 1006–1017. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Liu, T. Use model-based analysis of ChIP-Seq (MACS) to analyze short reads generated by sequencing protein–DNA interactions in embryonic stem cells. Stem Cell Transcr. Netw. Methods Protoc. 2014, 81–95. [Google Scholar]
- Strader, L.; Weijers, D.; Wagner, D. Plant transcription factors—being in the right place with the right company. Curr. Opin. Plant Biol. 2022, 65, 102136. [Google Scholar] [CrossRef]
- Singh, K.B.; Foley, R.C.; Oñate-Sánchez, L. Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 2002, 5, 430–436. [Google Scholar] [CrossRef]
- He, C.; Yan, J.; Shen, G.; Fu, L.; Holaday, A.S.; Auld, D.; Blumwald, E.; Zhang, H. Expression of an Arabidopsis vacuolar sodium/proton antiporter gene in cotton improves photosynthetic performance under salt conditions and increases fiber yield in the field. Plant Cell Physiol. 2005, 46, 1848–1854. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.-S.P.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 2004, 16, 2481–2498. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Tran, L.S.P.; Van Nguyen, D.; Fujita, M.; Maruyama, K.; Todaka, D.; Ito, Y.; Hayashi, N.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007, 51, 617–630. [Google Scholar] [CrossRef]
- Hu, H.; You, J.; Fang, Y.; Zhu, X.; Qi, Z.; Xiong, L. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol. Biol. 2008, 67, 169–181. [Google Scholar] [CrossRef]
- Peng, H.; Cheng, H.-Y.; Yu, X.-W.; Shi, Q.-H.; Zhang, H.; Li, J.-G.; Ma, H. Characterization of a chickpea (Cicer arietinum L.) NAC family gene, CarNAC5, which is both developmentally-and stress-regulated. Plant Physiol. Biochem. 2009, 47, 1037–1045. [Google Scholar] [CrossRef]
- Meng, C.; Cai, C.; Zhang, T.; Guo, W. Characterization of six novel NAC genes and their responses to abiotic stresses in Gossypium hirsutum L. Plant Sci. 2009, 176, 352–359. [Google Scholar] [CrossRef]
- Xia, N.; Zhang, G.; Liu, X.-Y.; Deng, L.; Cai, G.-L.; Zhang, Y.; Wang, X.-J.; Zhao, J.; Huang, L.-L.; Kang, Z.-S. Characterization of a novel wheat NAC transcription factor gene involved in defense response against stripe rust pathogen infection and abiotic stresses. Mol. Biol. Rep. 2010, 37, 3703–3712. [Google Scholar] [CrossRef] [PubMed]
- Puranik, S.; Bahadur, R.P.; Srivastava, P.S.; Prasad, M. Molecular cloning and characterization of a membrane associated NAC family gene, SiNAC from foxtail millet [Setaria italica (L.) P. Beauv.]. Mol. Biotechnol. 2011, 49, 138–150. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Li, S.; An, X.; Liu, X.; Qin, H.; Wang, D. Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J. Genet. Genom. 2009, 36, 17–29. [Google Scholar] [CrossRef]
- Archana, K.; Rama, N.; Mamrutha, H.; Nataraja, K.N. Down-Regulation of an Abiotic Stress Related Nicotiana Benthamiana WRKY Transcription Factor Induces Physiological Abnormalities; CSIR: Paris, France, 2009. [Google Scholar]
- Zhou, Q.Y.; Tian, A.G.; Zou, H.F.; Xie, Z.M.; Lei, G.; Huang, J.; Wang, C.M.; Wang, H.W.; Zhang, J.S.; Chen, S.Y. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 2008, 6, 486–503. [Google Scholar] [CrossRef]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought-and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef]
- Lee, S.-j.; Kang, J.-y.; Park, H.-J.; Kim, M.D.; Bae, M.S.; Choi, H.-i.; Kim, S.Y. DREB2C interacts with ABF2, a bZIP protein regulating abscisic acid-responsive gene expression, and its overexpression affects abscisic acid sensitivity. Plant Physiol. 2010, 153, 716–727. [Google Scholar] [CrossRef]
- Dubouzet, J.G.; Sakuma, Y.; Ito, Y.; Kasuga, M.; Dubouzet, E.G.; Miura, S.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt-and cold-responsive gene expression. Plant J. 2003, 33, 751–763. [Google Scholar] [CrossRef]
- Wang, Q.; Guan, Y.; Wu, Y.; Chen, H.; Chen, F.; Chu, C. Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Mol. Biol. 2008, 67, 589–602. [Google Scholar] [CrossRef]
- Xue, G.P.; Loveridge, C.W. HvDRF1 is involved in abscisic acid-mediated gene regulation in barley and produces two forms of AP2 transcriptional activators, interacting preferably with a CT-rich element. Plant J. 2004, 37, 326–339. [Google Scholar] [CrossRef]
- Xu, Z.-S.; Ni, Z.-Y.; Li, Z.-Y.; Li, L.-C.; Chen, M.; Gao, D.-Y.; Yu, X.-D.; Liu, P.; Ma, Y.-Z. Isolation and functional characterization of HvDREB1—a gene encoding a dehydration-responsive element binding protein in Hordeum vulgare. J. Plant Res. 2009, 122, 121–130. [Google Scholar] [CrossRef]
- Qin, F.; Kakimoto, M.; Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Tran, L.S.P.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J. 2007, 50, 54–69. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.; Agarwal, P.K.; Nair, S.; Sopory, S.; Reddy, M. Stress-inducible DREB2A transcription factor from Pennisetum glaucum is a phosphoprotein and its phosphorylation negatively regulates its DNA-binding activity. Mol. Genet. Genom. 2007, 277, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Lata, C.; Bhutty, S.; Bahadur, R.P.; Majee, M.; Prasad, M. Association of an SNP in a novel DREB2-like gene SiDREB2 with stress tolerance in foxtail millet [Setaria italica (L.)]. J. Exp. Bot. 2011, 62, 3387–3401. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.-P.; Kim, W.T. Isolation and functional characterization of the Ca-DREBLP1 gene encoding a dehydration-responsive element binding-factor-like protein 1 in hot pepper (Capsicum annuum L. cv. Pukang). Planta 2005, 220, 875–888. [Google Scholar] [CrossRef]
- Shen, Y.-G.; Zhang, W.-K.; Yan, D.-Q.; Du, B.-X.; Zhang, J.-S.; Liu, Q.; Chen, S.-Y. Characterization of a DRE-binding transcription factor from a halophyte Atriplex hortensis. Theor. Appl. Genet. 2003, 107, 155–161. [Google Scholar] [CrossRef]
- Li, X.-P.; Tian, A.-G.; Luo, G.-Z.; Gong, Z.-Z.; Zhang, J.-S.; Chen, S.-Y. Soybean DRE-binding transcription factors that are responsive to abiotic stresses. Theor. Appl. Genet. 2005, 110, 1355–1362. [Google Scholar] [CrossRef]
- Chen, M.; Wang, Q.-Y.; Cheng, X.-G.; Xu, Z.-S.; Li, L.-C.; Ye, X.-G.; Xia, L.-Q.; Ma, Y.-Z. GmDREB2, a soybean DRE-binding transcription factor, conferred drought and high-salt tolerance in transgenic plants. Biochem. Biophys. Res. Commun. 2007, 353, 299–305. [Google Scholar] [CrossRef]
- Shukla, R.K.; Raha, S.; Tripathi, V.; Chattopadhyay, D. Expression of CAP2, an APETALA2-family transcription factor from chickpea, enhances growth and tolerance to dehydration and salt stress in transgenic tobacco. Plant Physiol. 2006, 142, 113–123. [Google Scholar] [CrossRef]
- Gupta, K.; Agarwal, P.K.; Reddy, M.; Jha, B. SbDREB2A, an A-2 type DREB transcription factor from extreme halophyte Salicornia brachiata confers abiotic stress tolerance in Escherichia coli. Plant Cell Rep. 2010, 29, 1131–1137. [Google Scholar] [CrossRef]
- Choi, H.-I.; Hong, J.-H.; Ha, J.-O.; Kang, J.-Y.; Kim, S.Y. ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 2000, 275, 1723–1730. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Zou, H.-F.; Wei, W.; Hao, Y.-J.; Tian, A.-G.; Huang, J.; Liu, Y.-F.; Zhang, J.-S.; Chen, S.-Y. Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 2008, 228, 225–240. [Google Scholar] [CrossRef]
- Kobayashi, F.; Maeta, E.; Terashima, A.; Kawaura, K.; Ogihara, Y.; Takumi, S. Development of abiotic stress tolerance via bZIP-type transcription factor LIP19 in common wheat. J. Exp. Bot. 2008, 59, 891–905. [Google Scholar] [CrossRef]
- Zou, M.; Guan, Y.; Ren, H.; Zhang, F.; Chen, F. A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol. Biol. 2008, 66, 675–683. [Google Scholar] [CrossRef]
- Jia, Z.; Lian, Y.; Zhu, Y.; He, J.; Cao, Z.; Wang, G. Cloning and characterization of a putative transcription factor induced by abiotic stress in Zea mays. Afr. J. Biotechnol. 2009, 8. [Google Scholar]
- Hsieh, T.-H.; Li, C.-W.; Su, R.-C.; Cheng, C.-P.; Sanjaya; Tsai, Y.-C.; Chan, M.-T. A tomato bZIP transcription factor, SlAREB, is involved in water deficit and salt stress response. Planta 2010, 231, 1459–1473. [Google Scholar] [CrossRef]
- Nongpiur, R.C.; Singla-Pareek, S.L.; Pareek, A. The quest for osmosensors in plants. J. Exp. Bot. 2020, 71, 595–607. [Google Scholar] [CrossRef] [PubMed]
- Shah, W.H.; Rasool, A.; Saleem, S.; Mushtaq, N.U.; Tahir, I.; Hakeem, K.R.; Rehman, R.U. Understanding the integrated pathways and mechanisms of transporters, protein kinases, and transcription factors in plants under salt stress. Int. J. Genom. 2021, 2021. [Google Scholar] [CrossRef]
- Ci, D.; Song, Y.; Tian, M.; Zhang, D. Methylation of miRNA genes in the response to temperature stress in Populus simonii. Front. Plant Sci. 2015, 6, 921. [Google Scholar] [CrossRef]
- Kim, J.-M.; Sasaki, T.; Ueda, M.; Sako, K.; Seki, M. Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 2015, 6, 114. [Google Scholar] [CrossRef]
- Baulcombe, D.C.; Dean, C. Epigenetic regulation in plant responses to the environment. Cold Spring Harb. Perspect. Biol. 2014, 6, a019471. [Google Scholar] [CrossRef]
- Liu, J.; He, Z. Small DNA methylation, big player in plant abiotic stress responses and memory. Front. Plant Sci. 2020, 11, 595603. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Ji, D.; Li, S.; Wang, P.; Li, Q.; Xiang, F. The dynamic changes of DNA methylation and histone modifications of salt responsive transcription factor genes in soybean. PLoS ONE 2012, 7, e41274. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.J.; Azevedo, V.; Maroco, J.; Oliveira, M.M.; Santos, A.P. Salt tolerant and sensitive rice varieties display differential methylome flexibility under salt stress. PLoS ONE 2015, 10, e0124060. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Beena, A.S.; Awana, M.; Singh, A. Salt-induced tissue-specific cytosine methylation downregulates expression of HKT genes in contrasting wheat (Triticum aestivum L.) genotypes. DNA Cell Biol. 2017, 36, 283–294. [Google Scholar] [CrossRef]
- Kaldis, A.; Tsementzi, D.; Tanriverdi, O.; Vlachonasios, K.E. Arabidopsis thaliana transcriptional co-activators ADA2b and SGF29a are implicated in salt stress responses. Planta 2011, 233, 749–762. [Google Scholar] [CrossRef]
- Pandey, G.; Sharma, N.; Pankaj Sahu, P.; Prasad, M. Chromatin-based epigenetic regulation of plant abiotic stress response. Curr. Genom. 2016, 17, 490–498. [Google Scholar] [CrossRef]
- Banerjee, A.; Roychoudhury, A. Epigenetic regulation during salinity and drought stress in plants: Histone modifications and DNA methylation. Plant Gene 2017, 11, 199–204. [Google Scholar] [CrossRef]
- Bilichak, A.; Ilnystkyy, Y.; Hollunder, J.; Kovalchuk, I. The progeny of Arabidopsis thaliana plants exposed to salt exhibit changes in DNA methylation, histone modifications and gene expression. PLoS ONE 2012, 7, e30515. [Google Scholar] [CrossRef]
- Tanveer, M.; Shabala, S. Targeting redox regulatory mechanisms for salinity stress tolerance in crops. Salin. Responses Toler. Plants Vol. 1 Target. Sens. Transp. Signal. Mech. 2018, 213–234. [Google Scholar]
- Costa, S.F.; Martins, D.; Agacka-Mołdoch, M.; Czubacka, A.; de Sousa Araújo, S. Strategies to alleviate salinity stress in plants. Salin. Responses Toler. Plants Vol. 1 Target. Sens. Transp. Signal. Mech. 2018, 307–337. [Google Scholar]
- Morton, M.J.; Awlia, M.; Al-Tamimi, N.; Saade, S.; Pailles, Y.; Negrão, S.; Tester, M. Salt stress under the scalpel–dissecting the genetics of salt tolerance. Plant J. 2019, 97, 148–163. [Google Scholar] [CrossRef] [PubMed]
- Bansal, J.; Gupta, K.; Rajkumar, M.S.; Garg, R.; Jain, M. Draft genome and transcriptome analyses of halophyte rice Oryza coarctata provide resources for salinity and submergence stress response factors. Physiol. Plant. 2021, 173, 1309–1322. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Roychoudhury, A. Transcript profiling of stress-responsive genes and metabolic changes during salinity in indica and japonica rice exhibit distinct varietal difference. Physiol. Plant. 2021, 173, 1434–1447. [Google Scholar] [CrossRef] [PubMed]
- Elnaggar, A.; Mosa, K.A.; Ramamoorthy, K.; El-Keblawy, A.; Navarro, T.; Soliman, S.S. De novo transcriptome sequencing, assembly, and gene expression profiling of a salt-stressed halophyte (Salsola drummondii) from a saline habitat. Physiol. Plant. 2021, 173, 1695–1714. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, M.S.; Jain, M.; Garg, R. Discovery of DNA polymorphisms via whole genome resequencing and their functional relevance in salinity stress response in chickpea. Physiol. Plant. 2021, 173, 1573–1586. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).