Deciphering Depression: Epigenetic Mechanisms and Treatment Strategies
Abstract
Simple Summary
Abstract
1. Introduction
Background and Significance
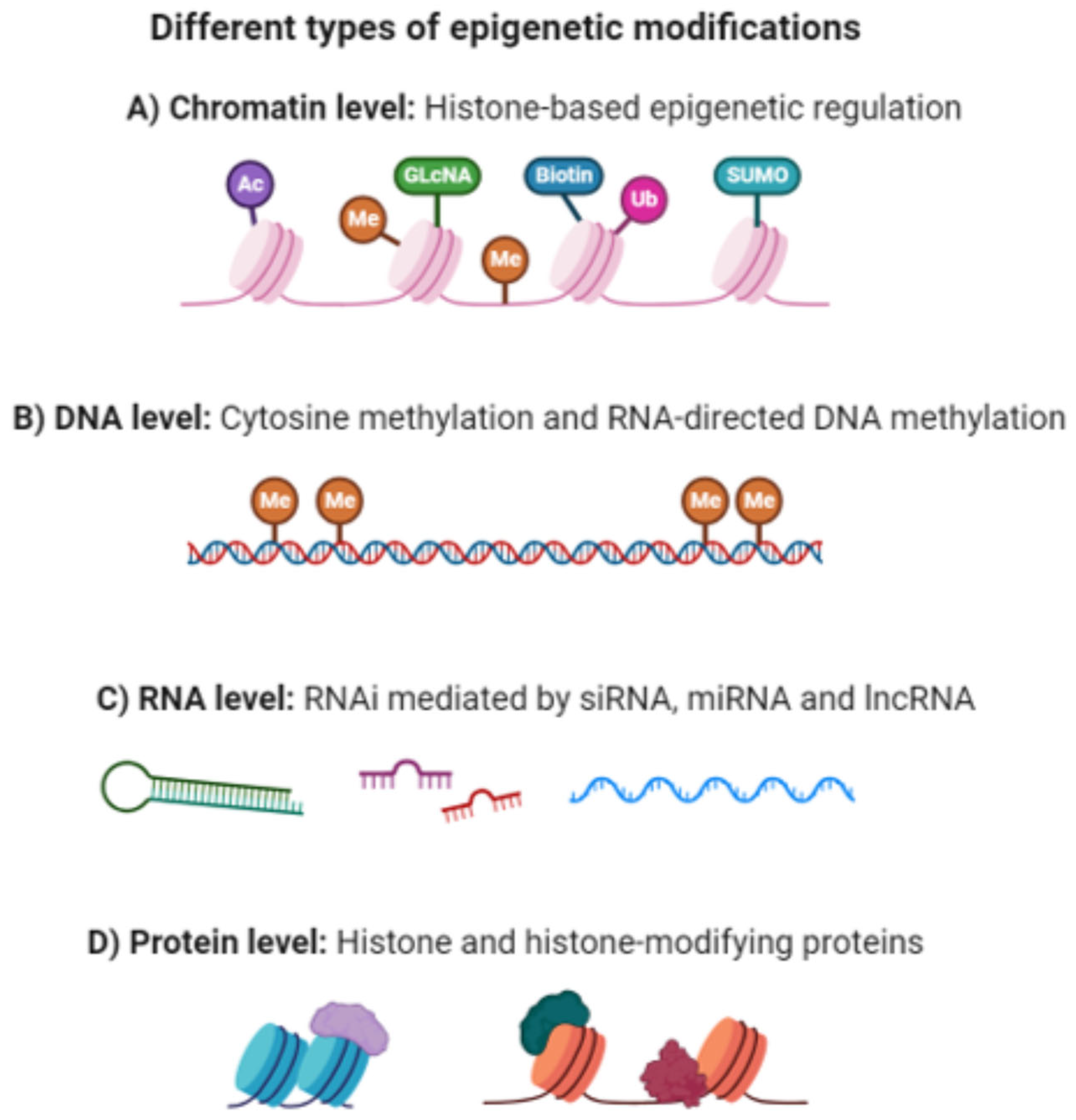
2. Epigenetic Mechanisms (EMs)
2.1. DNA Methylation
2.2. Histone Modifications
2.3. Non-Coding RNAs
3. Epigenetic Mechanisms in Depression
3.1. DNA Methylation and Depression
3.2. Histone Modifications and Depression
3.3. Non-Coding RNAs and Depression
3.4. Future Directions
4. Dysregulation of Non-Coding RNAs in Depression
4.1. Functional Consequences
4.2. Impact of Non-Coding RNAs on Gene Expression
5. Environmental Influences on Epigenetic Modifications
5.1. Prenatal Environment
5.2. Early Life Stress
5.3. Parental Care
5.4. Stress and Depression
6. Epigenetics and Treatment Approaches
Clinical Applications of Epigenetic Research
7. Recent Advances in Epigenetic Technologies
8. Summary and Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Peña, C.J.; Nestler, E.J. Progress in epigenetics of depression. Prog. Mol. Biol. Transl. Sci. 2018, 157, 41–66. [Google Scholar]
- Cui, L.; Li, S.; Wang, S.; Wu, X.; Liu, Y.; Yu, W.; Wang, Y.; Tang, Y.; Xia, M.; Li, B. Major depressive disorder: Hypothesis, mechanism, prevention and treatment. Signal Transduct. Target. Ther. 2024, 9, 30. [Google Scholar] [CrossRef] [PubMed]
- Marx, W.; Penninx, B.; Solmi, M.; Furukawa, T.A.; Firth, J.; Carvalho, A.F.; Berk, M. Major depressive disorder. Nat. Rev. Dis. Primers 2023, 9, 44. [Google Scholar] [CrossRef] [PubMed]
- Alshaya, D.S. Genetic and epigenetic factors associated with depression: An updated overview. Saudi J. Biol. Sci. 2022, 29, 103311. [Google Scholar] [CrossRef]
- Gibney, E.R.; Nolan, C.M. Epigenetics and gene expression. Heredity 2010, 105, 4–13. [Google Scholar] [CrossRef]
- Al Aboud, N.M.; Tupper, C.; Jialal, I. Genetics, epigenetic mechanism. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2018. [Google Scholar]
- Mohammadi, A.H.; Karimian, M.; Mirzaei, H.; Milajerdi, A. Epigenetic modifications and obsessive–compulsive disorder: What do we know? Brain Struct. Funct. 2023, 228, 1295–1305. [Google Scholar] [CrossRef]
- Jiang, S.; Postovit, L.; Cattaneo, A.; Binder, E.B.; Aitchison, K.J. Epigenetic Modifications in Stress Response Genes Associated with Childhood Trauma. Front. Psychiatry 2019, 10, 808. [Google Scholar] [CrossRef]
- Levy, M.J.F.; Boulle, F.; Steinbusch, H.W.; van den Hove, D.L.A.; Kenis, G.; Lanfumey, L. Neurotrophic factors and neuroplasticity pathways in the pathophysiology and treatment of depression. Psychopharmacology 2018, 235, 2195–2220. [Google Scholar] [CrossRef]
- Ortega, M.A.; Alvarez-Mon, M.A.; García-Montero, C.; Fraile-Martinez, O.; Lahera, G.; Monserrat, J.; Muñoz-Merida, L.; Mora, F.; Rodríguez-Jiménez, R.; Fernandez-Rojo, S.; et al. MicroRNAs as Critical Biomarkers of Major Depressive Disorder: A Comprehensive Perspective. Biomedicines 2021, 9, 1659. [Google Scholar] [CrossRef] [PubMed]
- Elhag, D.A.; Al Khodor, S. Exploring the potential of microRNA as a diagnostic tool for gestational diabetes. J. Transl. Med. 2023, 21, 392. [Google Scholar] [CrossRef]
- Martins de Carvalho, L.; Chen, W.Y.; Lasek, A.W. Epigenetic mechanisms underlying stress-induced depression. Int. Rev. Neurobiol. 2021, 156, 87–126. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.S.; Li, X.J.; Liu, C.Y.; Xu, Q.; Huang, J.Q.; Gu, S.; Chen, J.X. Effects of Histone Modification in Major Depressive Disorder. Curr. Neuropharmacol. 2022, 20, 1261–1277. [Google Scholar] [CrossRef] [PubMed]
- Penner-Goeke, S.; Binder, E.B. Epigenetics and depression. Dialogues Clin. Neurosci. 2019, 21, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Torres-Berrío, A.; Issler, O.; Parise, E.M.; Nestler, E.J. Unraveling the epigenetic landscape of depression: Focus on early life stress. Dialogues Clin. Neurosci. 2019, 21, 341–357. [Google Scholar] [CrossRef] [PubMed]
- Santaló, J.; Berdasco, M. Ethical Implications of Epigenetics in the Era of Personalized Medicine. Clin. Epigenetics 2022, 14, 44. [Google Scholar] [CrossRef]
- Roy, M.-C.; Dupras, C.; Ravitsky, V. The Epigenetic Effects of Assisted Reproductive Technologies: Ethical Considerations. J. Dev. Orig. Health Dis. 2017, 8, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Dyke, S.O.M.; Saulnier, K.M.; Dupras, C.; Webster, A.D.B.; Maschke, K.J.; Rothstein, M.A.; Siebert, R.; Walter, J.; Beck, S.; Pastinen, T.; et al. Points-to-Consider on the Return of Results in Epigenetic Research. Genome Med. 2019, 11, 31. [Google Scholar] [CrossRef] [PubMed]
- Dupras, C.; Knoppers, T.; Palmour, N.; Beauchamp, E.; Liosi, S.; Siebert, R.; Berner, A.; Beck, S.; Charest, I.; Joly, Y. Researcher Perspectives on Ethics Considerations in Epigenetics: An International Survey. Clin. Epigenetics 2022, 14, 110. [Google Scholar] [CrossRef] [PubMed]
- Taki, F.; de Melo-Martín, I. Conducting Epigenetics Research With Refugees and Asylum Seekers: Attending to the Ethical Challenges. Clin. Epigenetics 2021, 13, 105. [Google Scholar] [CrossRef] [PubMed]
- Notterman, D.A.; Mitchell, C. Epigenetics and Understanding the Impact of Social Determinants of Health. Pediatr. Clin. N. Am. 2015, 62, 1227–1240. [Google Scholar] [CrossRef] [PubMed]
- Meloni, M. The social brain meets the reactive genome: Neuroscience, epigenetics and the new social biology. Front. Hum. Neurosci. 2014, 8, 309. [Google Scholar] [CrossRef]
- Beckedorff, F.C.; Amaral, M.S.; Deocesano-Pereira, C.; Verjovski-Almeida, S. Long non-coding RNAs and their implications in cancer epigenetics. Biosci. Rep. 2013, 33, e00061. [Google Scholar] [CrossRef]
- Berra, L. Existential Depression: A Nonpathological and Philosophical-Existential Approach. J. Humanist. Psychol. 2019, 61, 757–765. [Google Scholar] [CrossRef]
- Sjösten, M.; Hörberg, U.; Fagerström, C.; Tuvesson, H. Supporting recovery in persons with stress-related disorders: A reflective lifeworld research study of health care professionals in primary health care in Sweden. Int. J. Qual. Stud. Health Well-Being 2023, 18, 2209967. [Google Scholar] [CrossRef] [PubMed]
- Restifo, S. Existential depression: A meaningful diagnostic entity? Australas. Psychiatry 2023, 31, 502–504. [Google Scholar] [CrossRef]
- Bang, Y.; Lim, J.; Choi, H.J. Recent advances in the pathology of prodromal non-motor symptoms olfactory deficit and depression in Parkinson’s disease: Clues to early diagnosis and effective treatment. Arch. Pharmacal Res. 2021, 44, 588–604. [Google Scholar] [CrossRef] [PubMed]
- Lian, T.-H.; Guo, P.; Zuo, L.-J.; Hu, Y.; Yu, S.-Y.; Liu, L.; Jin, Z.; Yu, Q.-J.; Wang, R.-D.; Li, L.-X. An investigation on the clinical features and neurochemical changes in Parkinson’s disease with depression. Front. Psychiatry 2019, 9, 723. [Google Scholar] [CrossRef]
- Hemmerle, A.M.; Herman, J.P.; Seroogy, K.B. Stress, depression and Parkinson’s disease. Exp. Neurol. 2012, 233, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Collaborators, G.M.D. Global, regional, and national burden of 12 mental disorders in 204 countries and territories, 1990–2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet Psychiatry 2022, 9, 137–150. [Google Scholar] [CrossRef]
- Toth, M. Epigenetic neuropharmacology: Drugs affecting the epigenome in the brain. Annu. Rev. Pharmacol. Toxicol. 2021, 61, 181–201. [Google Scholar] [CrossRef] [PubMed]
- Nestler, E.J.; Peña, C.J.; Kundakovic, M.; Mitchell, A.; Akbarian, S. Epigenetic basis of mental illness. Neuroscientist 2016, 22, 447–463. [Google Scholar] [CrossRef] [PubMed]
- Handy, D.E.; Castro, R.; Loscalzo, J. Epigenetic modifications: Basic mechanisms and role in cardiovascular disease. Circulation 2011, 123, 2145–2156. [Google Scholar] [CrossRef] [PubMed]
- Kanwal, R.; Gupta, S. Epigenetic modifications in cancer. Clin. Genet. 2012, 81, 303–311. [Google Scholar] [CrossRef]
- Bure, I.V.; Nemtsova, M.V.; Kuznetsova, E.B. Histone Modifications and Non-Coding RNAs: Mutual Epigenetic Regulation and Role in Pathogenesis. Int. J. Mol. Sci. 2022, 23, 5801. [Google Scholar] [CrossRef] [PubMed]
- Salinas, R.D.; Connolly, D.R.; Song, H. Invited Review: Epigenetics in neurodevelopment. Neuropathol. Appl. Neurobiol. 2020, 46, 6–27. [Google Scholar] [CrossRef] [PubMed]
- Giacoman-Lozano, M.; Meléndez-Ramírez, C.; Martinez-Ledesma, E.; Cuevas-Diaz Duran, R.; Velasco, I. Epigenetics of neural differentiation: Spotlight on enhancers. Front. Cell Dev. Biol. 2022, 10, 1001701. [Google Scholar] [CrossRef]
- Klengel, T.; Binder, E.B. Epigenetics of Stress-Related Psychiatric Disorders and Gene × Environment Interactions. Neuron 2015, 86, 1343–1357. [Google Scholar] [CrossRef]
- Lee, S.-A.; Huang, K.-C. Epigenetic profiling of human brain differential DNA methylation networks in schizophrenia. BMC Med. Genom. 2016, 9, 68. [Google Scholar] [CrossRef]
- Cecil, C.A.M.; Neumann, A.; Walton, E. Epigenetics applied to child and adolescent mental health: Progress, challenges and opportunities. JCPP Adv. 2023, 3, e12133. [Google Scholar] [CrossRef]
- Peedicayil, J. Genome–Environment Interactions and Psychiatric Disorders. Biomedicines 2023, 11, 1209. [Google Scholar] [CrossRef]
- Tirado-Magallanes, R.; Rebbani, K.; Lim, R.; Pradhan, S.; Benoukraf, T. Whole genome DNA methylation: Beyond genes silencing. Oncotarget 2017, 8, 5629. [Google Scholar] [CrossRef] [PubMed]
- Fries, G.R.; Li, Q.; McAlpin, B.; Rein, T.; Walss-Bass, C.; Soares, J.C.; Quevedo, J. The role of DNA methylation in the pathophysiology and treatment of bipolar disorder. Neurosci. Biobehav. Rev. 2016, 68, 474–488. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Rivero, M.; Gutiérrez-Fragoso, K.; Stoll, M.; Baune, B.T.; Berger, K. DNA methylation links between depression and immunity. J. Affect. Disord. Rep. 2023, 12, 100546. [Google Scholar] [CrossRef]
- Saavedra, K.; Molina-Márquez, A.M.; Saavedra, N.; Zambrano, T.; Salazar, L.A. Epigenetic Modifications of Major Depressive Disorder. Int. J. Mol. Sci. 2016, 17, 1279. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Sun, T.; Li, G.; Xu, G.; Cheng, J.; Gao, S. The role of BDNF exon I region methylation in the treatment of depression with sertraline and its clinical diagnostic value. J. Clin. Lab. Anal. 2021, 35, e23993. [Google Scholar] [CrossRef]
- Arosio, B.; Guerini, F.R.; Voshaar, R.C.O.; Aprahamian, I. Blood Brain-Derived Neurotrophic Factor (BDNF) and Major Depression: Do We Have a Translational Perspective? Front. Behav. Neurosci. 2021, 15, 626906. [Google Scholar] [CrossRef]
- Šalamon Arčan, I.; Kouter, K.; Videtič Paska, A. Depressive disorder and antidepressants from an epigenetic point of view. World J. Psychiatry 2022, 12, 1150–1168. [Google Scholar] [CrossRef] [PubMed]
- de Mendoza, A.; Nguyen, T.V.; Ford, E.; Poppe, D.; Buckberry, S.; Pflueger, J.; Grimmer, M.R.; Stolzenburg, S.; Bogdanovic, O.; Oshlack, A.; et al. Large-scale manipulation of promoter DNA methylation reveals context-specific transcriptional responses and stability. Genome Biol. 2022, 23, 163. [Google Scholar] [CrossRef]
- Czarny, P.; Białek, K.; Ziółkowska, S.; Strycharz, J.; Barszczewska, G.; Sliwinski, T. The Importance of Epigenetics in Diagnostics and Treatment of Major Depressive Disorder. J. Pers. Med. 2021, 11, 167. [Google Scholar] [CrossRef]
- Zhu, J.-H.; Bo, H.-H.; Liu, B.-P.; Jia, C.-X. The associations between DNA methylation and depression: A systematic review and meta-analysis. J. Affect. Disord. 2023, 327, 439–450. [Google Scholar] [CrossRef]
- Hüls, A.; Robins, C.; Conneely, K.N.; De Jager, P.L.; Bennett, D.A.; Epstein, M.P.; Wingo, T.S.; Wingo, A.P. Association between DNA methylation levels in brain tissue and late-life depression in community-based participants. Transl. Psychiatry 2020, 10, 262. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Meng, L.; Pei, F.; Zheng, Y.; Leng, J. A review of DNA methylation in depression. J. Clin. Neurosci. 2017, 43, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Goldberg, J.; Bremner, J.D.; Vaccarino, V. Association between promoter methylation of serotonin transporter gene and depressive symptoms: A monozygotic twin study. Psychosom. Med. 2013, 75, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Gao, L.; Song, J. Structural basis of DNMT1 and DNMT3A-mediated DNA methylation. Genes 2018, 9, 620. [Google Scholar] [CrossRef] [PubMed]
- Fuchikami, M.; Morinobu, S.; Segawa, M.; Okamoto, Y.; Yamawaki, S.; Ozaki, N.; Inoue, T.; Kusumi, I.; Koyama, T.; Tsuchiyama, K.; et al. DNA methylation profiles of the brain-derived neurotrophic factor (BDNF) gene as a potent diagnostic biomarker in major depression. PLoS ONE 2011, 6, e23881. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.; Tsai, S.J. Epigenetics and Depression: An Update. Psychiatry Investig. 2019, 16, 654. [Google Scholar] [CrossRef] [PubMed]
- Holloway, C.; González-Maeso, J. Epigenetic Mechanisms of Serotonin Signaling. ACS Chem. Neurosci. 2015, 6, 1099–1109. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Hu, Z.; Xiao, H.; Zhou, F.; Yang, B. The tale of histone modifications and its role in multiple sclerosis. Hum. Genom. 2018, 12, 31. [Google Scholar] [CrossRef]
- Lu, C.; Coradin, M.; Porter, E.G.; Garcia, B.A. Accelerating the Field of Epigenetic Histone Modification through Mass Spectrometry-Based Approaches. Mol. Cell. Proteom. 2021, 20, 100006. [Google Scholar] [CrossRef]
- Cavalieri, V. The Expanding Constellation of Histone Post-Translational Modifications in the Epigenetic Landscape. Genes 2021, 12, 1596. [Google Scholar] [CrossRef]
- Gammoh, O.; Aljabali, A.A.A.; Tambuwala, M.M. Plasma amino acids in major depressive disorder: Between pathology to pharmacology. EXCLI J. 2024, 23, 62–78. [Google Scholar] [CrossRef]
- Park, J.; Lee, K.; Kim, K.; Yi, S.-J. The role of histone modifications: From neurodevelopment to neurodiseases. Signal Transduct. Target. Ther. 2022, 7, 217. [Google Scholar] [CrossRef] [PubMed]
- Gujral, P.; Mahajan, V.; Lissaman, A.C.; Ponnampalam, A.P. Histone acetylation and the role of histone deacetylases in normal cyclic endometrium. Reprod. Biol. Endocrinol. 2020, 18, 84. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.L.; Grant, P.A. The role of DNA methylation and histone modifications in transcriptional regulation in humans. Subcell. Biochem. 2013, 61, 289–317. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Zhang, T.; Cooper, S.; Brockdorff, N. The interplay of histone modifications-writers that read. EMBO Rep. 2015, 16, 1467–1481. [Google Scholar] [CrossRef] [PubMed]
- Lutz, P.-E.; Chay, M.-A.; Pacis, A.; Chen, G.G.; Aouabed, Z.; Maffioletti, E.; Théroux, J.-F.; Grenier, J.-C.; Yang, J.; Aguirre, M.; et al. Non-CG methylation and multiple histone profiles associate child abuse with immune and small GTPase dysregulation. Nat. Commun. 2021, 12, 1132. [Google Scholar] [CrossRef] [PubMed]
- Sanbonmatsu, D.M.; Cooley, E.H.; Butner, J.E. The Impact of Complexity on Methods and Findings in Psychological Science. Front. Psychol. 2020, 11, 580111. [Google Scholar] [CrossRef]
- Dong, X.; Weng, Z. The correlation between histone modifications and gene expression. Epigenomics 2013, 5, 113–116. [Google Scholar] [CrossRef]
- Dent, S.Y.R. Grand challenge in chromatin epigenomics: Everything, everywhere, all at once. Front. Epigenet. Epigenom. 2023, 1, 1195690. [Google Scholar] [CrossRef]
- Galow, A.M.; Peleg, S. How to Slow down the Ticking Clock: Age-Associated Epigenetic Alterations and Related Interventions to Extend Life Span. Cells 2022, 11, 468. [Google Scholar] [CrossRef] [PubMed]
- Erfanian, N.; Heydari, A.A.; Feriz, A.M.; Iañez, P.; Derakhshani, A.; Ghasemigol, M.; Farahpour, M.; Razavi, S.M.; Nasseri, S.; Safarpour, H.; et al. Deep learning applications in single-cell genomics and transcriptomics data analysis. Biomed. Pharmacother. 2023, 165, 115077. [Google Scholar] [CrossRef]
- Ratti, M.; Lampis, A.; Ghidini, M.; Salati, M.; Mirchev, M.B.; Valeri, N.; Hahne, J.C. MicroRNAs (miRNAs) and Long Non-Coding RNAs (lncRNAs) as New Tools for Cancer Therapy: First Steps from Bench to Bedside. Target. Oncol. 2020, 15, 261–278. [Google Scholar] [CrossRef] [PubMed]
- Sati, S.; Ghosh, S.; Jain, V.; Scaria, V.; Sengupta, S. Genome-wide analysis reveals distinct patterns of epigenetic features in long non-coding RNA loci. Nucleic Acids Res. 2012, 40, 10018–10031. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. (Lausanne) 2018, 9, 402. [Google Scholar] [CrossRef]
- Annese, T.; Tamma, R.; De Giorgis, M.; Ribatti, D. microRNAs Biogenesis, Functions and Role in Tumor Angiogenesis. Front. Oncol. 2020, 10, 581007. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, W.; Wang, M.; Zhou, X. The role of long noncoding RNAs in gene expression regulation. In Gene Expression Profiling in Cancer; IntechOpen Limited: London, UK, 2019; pp. 1–17. [Google Scholar]
- Hanly, D.J.; Esteller, M.; Berdasco, M. Interplay between long non-coding RNAs and epigenetic machinery: Emerging targets in cancer? Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20170074. [Google Scholar] [CrossRef] [PubMed]
- Aliperti, V.; Skonieczna, J.; Cerase, A. Long Non-Coding RNA (lncRNA) Roles in Cell Biology, Neurodevelopment and Neurological Disorders. Non-Coding RNA 2021, 7, 36. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Lan, Y.; Xie, A.; Shi, J.; Zhao, H.; Xu, L.; Zhu, S.; Luo, T.; Zhao, T.; Xiao, Y. Comprehensive analysis of long noncoding RNA (lncRNA)-chromatin interactions reveals lncRNA functions dependent on binding diverse regulatory elements. J. Biol. Chem. 2019, 294, 15613–15622. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Liu, X.; Yu, L.; Lin, S.; Zhang, C.; Xu, H.; Leng, Z.; Huang, W.; Lei, J.; Li, T. Comprehensive landscape of epigenetic-dysregulated lncRNAs reveals a profound role of enhancers in carcinogenesis in BC subtypes. Mol. Ther.-Nucleic Acids 2021, 23, 667–681. [Google Scholar] [CrossRef]
- Dahrendorff, J.; Currier, G.; Uddin, M. Leveraging DNA methylation to predict treatment response in major depressive disorder: A critical review. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2024, e32985. [Google Scholar] [CrossRef]
- Kowiański, P.; Lietzau, G.; Czuba, E.; Waśkow, M.; Steliga, A.; Moryś, J. BDNF: A Key Factor with Multipotent Impact on Brain Signaling and Synaptic Plasticity. Cell. Mol. Neurobiol. 2018, 38, 579–593. [Google Scholar] [CrossRef] [PubMed]
- Notaras, M.; van den Buuse, M. Neurobiology of BDNF in fear memory, sensitivity to stress, and stress-related disorders. Mol. Psychiatry 2020, 25, 2251–2274. [Google Scholar] [CrossRef] [PubMed]
- Patnaik, E.; Madu, C.; Lu, Y. Epigenetic modulators as therapeutic agents in cancer. Int. J. Mol. Sci. 2023, 24, 14964. [Google Scholar] [CrossRef]
- Maze, I.; Covington, H.E., 3rd; Dietz, D.M.; LaPlant, Q.; Renthal, W.; Russo, S.J.; Mechanic, M.; Mouzon, E.; Neve, R.L.; Haggarty, S.J.; et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science 2010, 327, 213–216. [Google Scholar] [CrossRef]
- Castillo-Aguilera, O.; Depreux, P.; Halby, L.; Arimondo, P.B.; Goossens, L. DNA Methylation Targeting: The DNMT/HMT Crosstalk Challenge. Biomolecules 2017, 7, 3. [Google Scholar] [CrossRef]
- Uddin, M.G.; Fandy, T.E. Chapter Five-DNA methylation inhibitors: Retrospective and perspective view. In Advances in Cancer Research; Tew, K.D., Fisher, P.B., Eds.; Academic Press: Cambridge, MA, USA, 2021; Volume 152, pp. 205–223. [Google Scholar]
- Haggerty, C.; Kretzmer, H.; Riemenschneider, C.; Kumar, A.S.; Mattei, A.L.; Bailly, N.; Gottfreund, J.; Giesselmann, P.; Weigert, R.; Brändl, B.; et al. Dnmt1 has de novo activity targeted to transposable elements. Nat. Struct. Mol. Biol. 2021, 28, 594–603. [Google Scholar] [CrossRef]
- Wan, Q.; Gao, K.; Rong, H.; Wu, M.; Wang, H.; Wang, X.; Wang, G.; Liu, Z. Histone modifications of the Crhr1 gene in a rat model of depression following chronic stress. Behav. Brain Res. 2014, 271, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Tsankova, N.M.; Kumar, A.; Nestler, E.J. Histone modifications at gene promoter regions in rat hippocampus after acute and chronic electroconvulsive seizures. J. Neurosci. 2004, 24, 5603–5610. [Google Scholar] [CrossRef] [PubMed]
- Cruceanu, C.; Alda, M.; Nagy, C.; Freemantle, E.; Rouleau, G.A.; Turecki, G. H3K4 tri-methylation in synapsin genes leads to different expression patterns in bipolar disorder and major depression. Int. J. Neuropsychopharmacol. 2013, 16, 289–299. [Google Scholar] [CrossRef]
- Rinn, J.L.; Chang, H.Y. Long Noncoding RNAs: Molecular Modalities to Organismal Functions. Annu. Rev. Biochem. 2020, 89, 283–308. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, H.; Niu, M.; Wang, Y.; Xu, R.; Guo, Y.; Zhang, C. Roles of long noncoding RNAs in human inflammatory diseases. Cell Death Discov. 2024, 10, 235. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Pan, J.; Chen, W.; Jiang, J.; Huang, J. Chronic stress-induced immune dysregulation in cancer: Implications for initiation, progression, metastasis, and treatment. Am. J. Cancer Res. 2020, 10, 1294–1307. [Google Scholar] [PubMed]
- Hakami, M.A.; Hazazi, A.; Abdulaziz, O.; Almasoudi, H.H.; Alhazmi, A.Y.M.; Alkhalil, S.S.; Alharthi, N.S.; Alhuthali, H.M.; Almalki, W.H.; Gupta, G.; et al. HOTAIR: A key regulator of the Wnt/β-catenin signaling cascade in cancer progression and treatment. Pathol. Res. Pract. 2024, 253, 154957. [Google Scholar] [CrossRef] [PubMed]
- Anilkumar, A.K.; Vij, P.; Lopez, S.; Leslie, S.M.; Doxtater, K.; Khan, M.M.; Yallapu, M.M.; Chauhan, S.C.; Maestre, G.E.; Tripathi, M.K. Long Non-Coding RNAs: New Insights in Neurodegenerative Diseases. Int. J. Mol. Sci. 2024, 25, 2268. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Sebastian-delaCruz, M.; Gonzalez-Moro, I.; Olazagoitia-Garmendia, A.; Castellanos-Rubio, A.; Santin, I. The Role of lncRNAs in Gene Expression Regulation through mRNA Stabilization. Non-Coding RNA 2021, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Long, F.; Li, X.; Pan, J.; Ye, H.; Di, C.; Huang, Y.; Li, J.; Zhou, X.; Yi, H.; Huang, Q.; et al. The role of lncRNA NEAT1 in human cancer chemoresistance. Cancer Cell Int. 2024, 24, 236. [Google Scholar] [CrossRef] [PubMed]
- Yrondi, A.; Fiori, L.M.; Nogovitsyn, N.; Hassel, S.; Théroux, J.F.; Aouabed, Z.; Frey, B.N.; Lam, R.W.; Milev, R.; Müller, D.J.; et al. Association between the expression of lncRNA BASP-AS1 and volume of right hippocampal tail moderated by episode duration in major depressive disorder: A CAN-BIND 1 report. Transl. Psychiatry 2021, 11, 469. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.C.R.; Acuña, S.M.; Aoki, J.I.; Floeter-Winter, L.M.; Muxel, S.M. Long Non-Coding RNAs in the Regulation of Gene Expression: Physiology and Disease. Non-Coding RNA 2019, 5, 17. [Google Scholar] [CrossRef]
- Lin, R.; Turecki, G. Noncoding RNAs in Depression. In Neuroepigenomics in Aging and Disease; Delgado-Morales, R., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 197–210. [Google Scholar]
- Spadaro, P.A.; Bredy, T.W. Emerging role of non-coding RNA in neural plasticity, cognitive function, and neuropsychiatric disorders. Front. Genet. 2012, 3, 132. [Google Scholar] [CrossRef]
- Gomes, A.Q.; Nolasco, S.; Soares, H. Non-coding RNAs: Multi-tasking molecules in the cell. Int. J. Mol. Sci. 2013, 14, 16010–16039. [Google Scholar] [CrossRef]
- Li, Q.; Su, Z.; Xu, X.; Liu, G.; Song, X.; Wang, R.; Sui, X.; Liu, T.; Chang, X.; Huang, D. AS1DHRS4, a head-to-head natural antisense transcript, silences the DHRS4 gene cluster in cis and trans. Proc. Natl. Acad. Sci. USA 2012, 109, 14110–14115. [Google Scholar] [CrossRef]
- Løkhammer, S.; Stavrum, A.-K.; Polushina, T.; Aas, M.; Ottesen, A.A.; Andreassen, O.A.; Melle, I.; Le Hellard, S. An epigenetic association analysis of childhood trauma in psychosis reveals possible overlap with methylation changes associated with PTSD. Transl. Psychiatry 2022, 12, 177. [Google Scholar] [CrossRef] [PubMed]
- Booij, L.; Szyf, M.; Carballedo, A.; Frey, E.-M.; Morris, D.; Dymov, S.; Vaisheva, F.; Ly, V.; Fahey, C.; Meaney, J. DNA methylation of the serotonin transporter gene in peripheral cells and stress-related changes in hippocampal volume: A study in depressed patients and healthy controls. PLoS ONE 2015, 10, e0119061. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, A.; Labad, J.; Salvat-Pujol, N.; Barrachina, M.; Costas, J.; Urretavizcaya, M.; de Arriba-Arnau, A.; Crespo, J.M.; Soriano-Mas, C.; Carracedo, Á.; et al. BDNF genetic variants and methylation: Effects on cognition in major depressive disorder. Transl. Psychiatry 2019, 9, 265. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, W.; Wu, Y.; Tian, X.; Duan, H.; Li, S.; Tan, Q.; Zhang, D. Genome-wide DNA methylation and gene expression analyses in monozygotic twins identify potential biomarkers of depression. Transl. Psychiatry 2021, 11, 416. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Strachan, E.; Fowler, E.; Bacus, T.; Roy-Byrne, P.; Zhao, J. Genome-wide profiling of DNA methylome and transcriptome in peripheral blood monocytes for major depression: A Monozygotic Discordant Twin Study. Transl. Psychiatry 2019, 9, 215. [Google Scholar] [CrossRef]
- Sato, S.; Bunney, B.; Mendoza-Viveros, L.; Bunney, W.; Borrelli, E.; Sassone-Corsi, P.; Orozco-Solis, R. Rapid-acting antidepressants and the circadian clock. Neuropsychopharmacology 2022, 47, 805–816. [Google Scholar] [CrossRef]
- Dwivedi, Y. MicroRNAs in depression and suicide: Recent insights and future perspectives. J. Affect. Disord. 2018, 240, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Roy, B.; Ochi, S.; Dwivedi, Y. Potential of Circulating miRNAs as Molecular Markers in Mood Disorders and Associated Suicidal Behavior. Int. J. Mol. Sci. 2023, 24, 4664. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Lutz, P.E.; Wang, Y.C.; Ragoussis, J.; Turecki, G. Global long non-coding RNA expression in the rostral anterior cingulate cortex of depressed suicides. Transl. Psychiatry 2018, 8, 224. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, Q.; Song, R.; Kong, Y.; Zhang, Z. Non-coding RNAs in depression: Promising diagnostic and therapeutic biomarkers. EBioMedicine 2021, 71, 103569. [Google Scholar] [CrossRef] [PubMed]
- Uchida, S.; Yamagata, H.; Seki, T.; Watanabe, Y. Epigenetic mechanisms of major depression: Targeting neuronal plasticity. Psychiatry Clin. Neurosci. 2018, 72, 212–227. [Google Scholar] [CrossRef]
- Thapa, R.; Sparks, F.T.; Hanif, W.; Gulbrandsen, T.; Sutherland, R.J. Recent memory for socially transmitted food preferences in rats does not depend on the hippocampus. Neurobiol. Learn. Mem. 2014, 114, 113–116. [Google Scholar] [CrossRef]
- Egervari, G. Chromatin accessibility in neuropsychiatric disorders. Neurobiol. Learn. Mem. 2021, 181, 107438. [Google Scholar] [CrossRef]
- Huisman, S.G.; van der Veen, R.C.A.; Sun, C.; Lohse, D. Multiple states in highly turbulent Taylor–Couette flow. Nat. Commun. 2014, 5, 3820. [Google Scholar] [CrossRef] [PubMed]
- Aberg, K.A.; Dean, B.; Shabalin, A.A.; Chan, R.F.; Han, L.K.M.; Zhao, M.; van Grootheest, G.; Xie, L.Y.; Milaneschi, Y.; Clark, S.L.; et al. Methylome-wide association findings for major depressive disorder overlap in blood and brain and replicate in independent brain samples. Mol. Psychiatry 2020, 25, 1344–1354. [Google Scholar] [CrossRef]
- Yamagata, H.; Ogihara, H.; Matsuo, K.; Uchida, S.; Kobayashi, A.; Seki, T.; Kobayashi, M.; Harada, K.; Chen, C.; Miyata, S.; et al. Distinct epigenetic signatures between adult-onset and late-onset depression. Sci. Rep. 2021, 11, 2296. [Google Scholar] [CrossRef]
- Starnawska, A.; Demontis, D. Role of DNA Methylation in Mediating Genetic Risk of Psychiatric Disorders. Front. Psychiatry 2021, 12, 596821. [Google Scholar] [CrossRef]
- Barbon, A.; Magri, C. RNA Editing and Modifications in Mood Disorders. Genes 2020, 11, 872. [Google Scholar] [CrossRef] [PubMed]
- Salvetat, N.; Checa-Robles, F.J.; Patel, V.; Cayzac, C.; Dubuc, B.; Chimienti, F.; Abraham, J.D.; Dupré, P.; Vetter, D.; Méreuze, S.; et al. A game changer for bipolar disorder diagnosis using RNA editing-based biomarkers. Transl. Psychiatry 2022, 12, 182. [Google Scholar] [CrossRef] [PubMed]
- Hu, G. Evaluation of 3D Chromatin Interactions Using Hi-C. Methods Mol. Biol. 2020, 2117, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, V.; Nestler, E.J. Animal models of depression: Molecular perspectives. Curr. Top. Behav. Neurosci. 2011, 7, 121–147. [Google Scholar] [CrossRef] [PubMed]
- Shadrina, M.; Bondarenko, E.A.; Slominsky, P.A. Genetics Factors in Major Depression Disease. Front. Psychiatry 2018, 9, 334. [Google Scholar] [CrossRef] [PubMed]
- Dion, A.; Muñoz, P.T.; Franklin, T.B. Epigenetic mechanisms impacted by chronic stress across the rodent lifespan. Neurobiol. Stress 2022, 17, 100434. [Google Scholar] [CrossRef]
- Li, M.; Fu, X.; Xie, W.; Guo, W.; Li, B.; Cui, R.; Yang, W. Effect of Early Life Stress on the Epigenetic Profiles in Depression. Front. Cell Dev. Biol. 2020, 8, 867. [Google Scholar] [CrossRef]
- Ochi, S.; Dwivedi, Y. Dissecting early life stress-induced adolescent depression through epigenomic approach. Mol. Psychiatry 2023, 28, 141–153. [Google Scholar] [CrossRef]
- Nestler, E.J. Epigenetic mechanisms of depression. JAMA Psychiatry 2014, 71, 454–456. [Google Scholar] [CrossRef]
- Schell, G.; Roy, B.; Prall, K.; Dwivedi, Y. miR-218: A Stress-Responsive Epigenetic Modifier. Non-Coding RNA 2022, 8, 55. [Google Scholar] [CrossRef]
- Zhu, Z.; Huang, X.; Du, M.; Wu, C.; Fu, J.; Tan, W.; Wu, B.; Zhang, J.; Liao, Z.B. Recent advances in the role of miRNAs in post-traumatic stress disorder and traumatic brain injury. Mol. Psychiatry 2023, 28, 2630–2644. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wang, L.; Zhang, L.; Zhao, H. Phytochemicals targeting lncRNAs: A novel direction for neuroprotection in neurological disorders. Biomed. Pharmacother. 2023, 162, 114692. [Google Scholar] [CrossRef]
- Zhang, S.-F.; Gao, J.; Liu, C.-M. The role of non-coding RNAs in neurodevelopmental disorders. Front. Genet. 2019, 10, 1033. [Google Scholar] [CrossRef] [PubMed]
- Rahmani, S.; Kadkhoda, S.; Ghafouri-Fard, S. Synaptic plasticity and depression: The role of miRNAs dysregulation. Mol. Biol. Rep. 2022, 49, 9759–9765. [Google Scholar] [CrossRef]
- Ding, R.; Su, D.; Zhao, Q.; Wang, Y.; Wang, J.-Y.; Lv, S.; Ji, X. The role of microRNAs in depression. Front. Pharmacol. 2023, 14, 1129186. [Google Scholar] [CrossRef]
- Mongelli, A.; Martelli, F.; Farsetti, A.; Gaetano, C. The Dark That Matters: Long Non-Coding RNAs as Master Regulators of Cellular Metabolism in Non-Communicable Diseases. Front. Physiol. 2019, 10, 369. [Google Scholar] [CrossRef]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics-challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef]
- Khorkova, O.; Stahl, J.; Joji, A.; Volmar, C.-H.; Wahlestedt, C. Amplifying gene expression with RNA-targeted therapeutics. Nat. Rev. Drug Discov. 2023, 22, 539–561. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Gao, L.; Cheng, L.; Lv, G.; Sun, B.; Wang, G.; Tang, Q. The roles of N6-methyladenosine and its target regulatory noncoding RNAs in tumors: Classification, mechanisms, and potential therapeutic implications. Exp. Mol. Med. 2023, 55, 487–501. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, D.; Liu, T.; Chen, J.; Yu, J.; Yi, P. N6-methyladenosine-mediated gene regulation and therapeutic implications. Trends Mol. Med. 2023, 29, 454–467. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Roy, B.; Dwivedi, Y.K. Co-Expression Network Modeling Identifies Key Long Non-Coding RNA and mRNA Modules in Altering Molecular Phenotype to Develop Stress-Induced Depression in Rats. Transl. Psychiatry 2019, 9, 125. [Google Scholar] [CrossRef] [PubMed]
- Glăvan, D.; Gheorman, V.; Gresita, A.; Hermann, D.M.; Udriștoiu, I.; Popa-Wagner, A. Identification of Transcriptome Alterations in the Prefrontal Cortex, Hippocampus, Amygdala and Hippocampus of Suicide Victims. Sci. Rep. 2021, 11, 18853. [Google Scholar] [CrossRef] [PubMed]
- García-Fonseca, Á.; Martin-Jimenez, C.; Barreto, G.E.; Pachón, A.F.A.; González, J. The Emerging Role of Long Non-Coding RNAs and MicroRNAs in Neurodegenerative Diseases: A Perspective of Machine Learning. Biomolecules 2021, 11, 1132. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.R.; Olive, M.F. Early-life stress interactions with the epigenome: Potential mechanisms driving vulnerability toward psychiatric illness. Behav. Pharmacol. 2014, 25, 341–351. [Google Scholar] [CrossRef]
- Cheng, Z.; Su, J.; Zhang, K.; Jiang, H.-B.; Li, B. Epigenetic Mechanism of Early Life Stress-Induced Depression: Focus on the Neurotransmitter Systems. Front. Cell Dev. Biol. 2022, 10, 929732. [Google Scholar] [CrossRef] [PubMed]
- Uchida, S.; Hara, K.; Kobayashi, A.; Otsuki, K.; Yamagata, H.; Hobara, T.; Suzuki, T.; Miyata, N.; Watanabe, Y. Epigenetic Status of GDNF in the Ventral Striatum Determines Susceptibility and Adaptation to Daily Stressful Events. Neuron 2011, 69, 359–372. [Google Scholar] [CrossRef] [PubMed]
- Kundakovic, M.; Jaric, I. The Epigenetic Link between Prenatal Adverse Environments and Neurodevelopmental Disorders. Genes 2017, 8, 104. [Google Scholar] [CrossRef] [PubMed]
- Tiffon, C. The Impact of Nutrition and Environmental Epigenetics on Human Health and Disease. Int. J. Mol. Sci. 2018, 19, 3425. [Google Scholar] [CrossRef]
- Kertes, D.A.; Kamin, H.S.; Hughes, D.A.; Rodney, N.C.; Bhatt, S.; Mulligan, C.J. Prenatal Maternal Stress Predicts Methylation of Genes Regulating the Hypothalamic-Pituitary-Adrenocortical System in Mothers and Newborns in the Democratic Republic of Congo. Child Dev. 2016, 87, 61–72. [Google Scholar] [CrossRef]
- Perera, F.; Herbstman, J. Prenatal environmental exposures, epigenetics, and disease. Reprod. Toxicol. 2011, 31, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Kundakovic, M.; Champagne, F.A. Early-life experience, epigenetics, and the developing brain. Neuropsychopharmacology 2015, 40, 141–153. [Google Scholar] [CrossRef]
- Parade, S.H.; Huffhines, L.; Daniels, T.E.; Stroud, L.R.; Nugent, N.R.; Tyrka, A.R. A systematic review of childhood maltreatment and DNA methylation: Candidate gene and epigenome-wide approaches. Transl. Psychiatry 2021, 11, 134. [Google Scholar] [CrossRef] [PubMed]
- Quidé, Y.; Tozzi, L.; Corcoran, M.; Cannon, D.M.; Dauvermann, M.R. The Impact of Childhood Trauma on Developing Bipolar Disorder: Current Understanding and Ensuring Continued Progress. Neuropsychiatr. Dis. Treat. 2020, 16, 3095–3115. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.; Wen, L.; Song, J.; Wang, N.; Huang, L.; Gao, L.; Qu, M. Emerging trends in epigenetic and childhood trauma: Bibliometrics and visual analysis. Front. Psychiatry 2022, 13, 925273. [Google Scholar] [CrossRef]
- Thumfart, K.M.; Jawaid, A.; Bright, K.; Flachsmann, M.; Mansuy, I.M. Epigenetics of childhood trauma: Long term sequelae and potential for treatment. Neurosci. Biobehav. Rev. 2022, 132, 1049–1066. [Google Scholar] [CrossRef] [PubMed]
- Murgatroyd, C.; Spengler, D. Epigenetics of early child development. Front. Psychiatry 2011, 2, 16. [Google Scholar] [CrossRef]
- Soga, T.; Teo, C.H.; Parhar, I. Genetic and Epigenetic Consequence of Early-Life Social Stress on Depression: Role of Serotonin-Associated Genes. Front. Genet. 2021, 11, 601868. [Google Scholar] [CrossRef]
- Park, C.; Rosenblat, J.D.; Brietzke, E.; Pan, Z.; Lee, Y.; Cao, B.; Zuckerman, H.; Kalantarova, A.; McIntyre, R.S. Stress, epigenetics and depression: A systematic review. Neurosci. Biobehav. Rev. 2019, 102, 139–152. [Google Scholar] [CrossRef]
- Poon, C.H.; Heng, B.C.; Lim, L.W. New insights on brain-derived neurotrophic factor epigenetics: From depression to memory extinction. Ann. N. Y. Acad. Sci. 2021, 1484, 9–31. [Google Scholar] [CrossRef]
- Schroeder, M.; Krebs, M.O.; Bleich, S.; Frieling, H. Epigenetics and depression: Current challenges and new therapeutic options. Curr. Opin. Psychiatry 2010, 23, 588–592. [Google Scholar] [CrossRef] [PubMed]
- Menke, A.; Binder, E.B. Epigenetic alterations in depression and antidepressant treatment. Dialogues Clin. Neurosci. 2014, 16, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Jaini, P.A.; Papa, F. An Epigenetic Perspective on Lifestyle Medicine for Depression: Implications for Primary Care Practice. Am. J. Lifestyle Med. 2022, 16, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Cai, H.; Wang, M.; Ma, X.; Mo, F.; Yang, S.; Han, J.; Wei, X. Targeting Epigenetic Regulators for Cancer Therapy: Mechanisms and Advances in Clinical Trials. Signal Transduct. Target. Ther. 2019, 4, 62. [Google Scholar] [CrossRef]
- Cramer, S.A.; Adjei, I.M.; Labhasetwar, V. Advancements in the Delivery of Epigenetic Drugs. Expert Opin. Drug Deliv. 2015, 12, 1501–1512. [Google Scholar] [CrossRef] [PubMed]
- Cacabelos, R.; Torrellas, C. Epigenetics of Aging and Alzheimer’s Disease: Implications for Pharmacogenomics and Drug Response. Int. J. Mol. Sci. 2015, 16, 30483–30543. [Google Scholar] [CrossRef] [PubMed]
- Peedicayil, J. Role of Epigenetics in Pharmacotherapy, Psychotherapy and Nutritional Management of Mental Disorders. J. Clin. Pharm. Ther. 2012, 37, 499–501. [Google Scholar] [CrossRef]
- Kelly, T.K.; Carvalho, D.D.D.; Jones, P.A. Epigenetic Modifications as Therapeutic Targets. Nat. Biotechnol. 2010, 28, 1069–1078. [Google Scholar] [CrossRef]
- Puri, B.K. Calcium Signaling and Gene Expression. In Calcium Signaling; Islam, M.S., Ed.; Springer International Publishing: Cham, Switzerland, 2020; pp. 537–545. [Google Scholar]
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in Cancer. Carcinogenesis 2009, 31, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Gladkova, M.G.; Leidmaa, E.; Anderzhanova, E.A. Epidrugs in the therapy of central nervous system disorders: A way to drive on? Cells 2023, 12, 1464. [Google Scholar] [CrossRef]
- Krishnan, V.; Nestler, E.J. Linking molecules to mood: New insight into the biology of depression. Am. J. Psychiatry 2010, 167, 1305–1320. [Google Scholar] [CrossRef]
- Radosavljevic, M.; Svob Strac, D.; Jancic, J.; Samardzic, J. The role of pharmacogenetics in personalizing the antidepressant and anxiolytic therapy. Genes 2023, 14, 1095. [Google Scholar] [CrossRef]
- Huang, Z.; Jiang, H.; Liu, X.; Chen, Y.; Wong, J.; Wang, Q.; Huang, W.; Shi, T.; Zhang, J. HEMD: An integrated tool of human epigenetic enzymes and chemical modulators for therapeutics. PLoS ONE 2012, 7, e39917. [Google Scholar] [CrossRef] [PubMed]
- Sales, A.J.; Biojone, C.; Terceti, M.S.; Guimarães, F.S.; Gomes, M.V.M.; Joca, S.R.L. Antidepressant-like effect induced by systemic and intra-hippocampal administration of DNA methylation inhibitors. Br. J. Pharmacol. 2011, 164, 1711–1721. [Google Scholar] [CrossRef]
- Luo, H.; Zhao, Q.; Wei, W.; Zheng, L.; Yi, S.; Li, G.; Wang, W.; Sheng, H.; Pu, H.; Mo, H.; et al. Circulating tumor DNA methylation profiles enable early diagnosis, prognosis prediction, and screening for colorectal cancer. Sci. Transl. Med. 2020, 12, eaax7533. [Google Scholar] [CrossRef] [PubMed]
- Nohesara, S.; Abdolmaleky, H.M.; Zhou, J.-R.; Thiagalingam, S. Microbiota-Induced Epigenetic Alterations in Depressive Disorders Are Targets for Nutritional and Probiotic Therapies. Genes 2023, 14, 2217. [Google Scholar] [CrossRef] [PubMed]
- Nassiri, F.; Chakravarthy, A.; Feng, S.; Shen, S.Y.; Nejad, R.; Zuccato, J.A.; Voisin, M.R.; Patil, V.; Horbinski, C.; Aldape, K.; et al. Detection and discrimination of intracranial tumors using plasma cell-free DNA methylomes. Nat. Med. 2020, 26, 1044–1047. [Google Scholar] [CrossRef]
- Yuan, M.; Yang, B.; Rothschild, G.; Mann, J.J.; Sanford, L.D.; Tang, X.; Huang, C.; Wang, C.; Zhang, W. Epigenetic regulation in major depression and other stress-related disorders: Molecular mechanisms, clinical relevance and therapeutic potential. Signal Transduct. Target. Ther. 2023, 8, 309. [Google Scholar] [CrossRef]
- Li, M.; D’Arcy, C.; Li, X.; Zhang, T.; Joober, R.; Meng, X. What do DNA methylation studies tell us about depression? A systematic review. Transl. Psychiatry 2019, 9, 68. [Google Scholar] [CrossRef]
- Duan, Z.; Lu, J. DNA Methyltransferases in Depression: An Update. Front. Psychiatry 2020, 11, 538683. [Google Scholar] [CrossRef]
- Figueroa, M.E.; Abdel-Wahab, O.; Lu, C.; Ward, P.S.; Patel, J.; Shih, A.; Li, Y.; Bhagwat, N.; Vasanthakumar, A.; Fernandez, H.F.; et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 2010, 18, 553–567. [Google Scholar] [CrossRef]
- Berdasco, M.; Esteller, M. Clinical epigenetics: Seizing opportunities for translation. Nat. Rev. Genet. 2019, 20, 109–127. [Google Scholar] [CrossRef] [PubMed]
- Scangos, K.W.; State, M.W.; Miller, A.H.; Baker, J.T.; Williams, L.M. New and emerging approaches to treat psychiatric disorders. Nat. Med. 2023, 29, 317–333. [Google Scholar] [CrossRef]
- Jones, P.A.; Issa, J.-P.J.; Baylin, S. Targeting the cancer epigenome for therapy. Nat. Rev. Genet. 2016, 17, 630–641. [Google Scholar] [CrossRef] [PubMed]
- Morel, D.; Jeffery, D.; Aspeslagh, S.; Almouzni, G.; Postel-Vinay, S. Combining epigenetic drugs with other therapies for solid tumours—Past lessons and future promise. Nat. Rev. Clin. Oncol. 2020, 17, 91–107. [Google Scholar] [CrossRef]
- Topper, M.J.; Vaz, M.; Chiappinelli, K.B.; Shields, C.E.D.; Niknafs, N.; Yen, R.-W.C.; Wenzel, A.; Hicks, J.; Ballew, M.; Stone, M. Epigenetic therapy ties MYC depletion to reversing immune evasion and treating lung cancer. Cell 2017, 171, 1284–1300. [Google Scholar] [CrossRef] [PubMed]
- Rothstein, M.A.; Cai, Y.; Marchant, G.E. The ghost in our genes: Legal and ethical implications of epigenetics. Health Matrix (Cleveland, Ohio: 1991) 2009, 19, 1. [Google Scholar]
- The BLUEPRINT consortium. Quantitative comparison of DNA methylation assays for biomarker development and clinical applications. Nat. Biotechnol. 2016, 34, 726–737. [Google Scholar] [CrossRef]
- Yu, X.; Zhao, H.; Wang, R.; Chen, Y.; Ouyang, X.; Li, W.; Sun, Y.; Peng, A. Cancer epigenetics: From laboratory studies and clinical trials to precision medicine. Cell Death Discov. 2024, 10, 28. [Google Scholar] [CrossRef]
- Majchrzak-Celińska, A.; Warych, A.; Szoszkiewicz, M. Novel Approaches to Epigenetic Therapies: From Drug Combinations to Epigenetic Editing. Genes 2021, 12, 208. [Google Scholar] [CrossRef]
- Mabe, N.W.; Perry, J.A.; Malone, C.F.; Stegmaier, K. Pharmacological targeting of the cancer epigenome. Nat. Cancer 2024, 5, 844–865. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Gao, Y.; Dominguez, A.A.; Qi, L.S. CRISPR technologies for precise epigenome editing. Nat. Cell Biol. 2021, 23, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Aljabali, A.A.A.; El-Tanani, M.; Tambuwala, M.M. Principles of CRISPR-Cas9 technology: Advancements in genome editing and emerging trends in drug delivery. J. Drug Deliv. Sci. Technol. 2024, 92, 105338. [Google Scholar] [CrossRef]
- Hong, T.H.; Park, W.-Y. Single-cell genomics technology: Perspectives. Exp. Mol. Med. 2020, 52, 1407–1408. [Google Scholar] [CrossRef] [PubMed]
- Adil, A.; Kumar, V.; Jan, A.T.; Asger, M. Single-Cell Transcriptomics: Current Methods and Challenges in Data Acquisition and Analysis. Front. Neurosci. 2021, 15, 591122. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Ma, N.; Li, X.; Gou, Y.; Duan, Y.; Liu, B.; Xia, J.; Zhao, X.; Wang, X.; Li, Q.; et al. Advances in single-cell RNA sequencing and its applications in cancer research. J. Hematol. Oncol. 2023, 16, 98. [Google Scholar] [CrossRef] [PubMed]
- Gammoh, O.; Qnais, E.Y.; Athamneh, R.Y.; Al-Jaidi, B.; Al-Tawalbeh, D.; Altaber, S.; Alqudah, A.; Aljabali, A.A.A.; Tambuwala, M.M. Unraveling the Potential of Isorhamnetin as an Adjuvant in Depression Treatment with Escitalopram. Curr. Issues Mol. Biol. 2023, 45, 7668–7679. [Google Scholar] [CrossRef]
- Vadivalagan, C.; Krishnan, A.; Chen, S.-J.; Hseu, Y.-C.; Muthu, S.; Dhar, R.; Aljabali, A.A.A.; Tambuwala, M.M. The Warburg effect in osteoporosis: Cellular signaling and epigenetic regulation of energy metabolic events to targeting the osteocalcin for phenotypic alteration. Cell. Signal. 2022, 100, 110488. [Google Scholar] [CrossRef]
- Chen, X.; Xu, H.; Shu, X.; Song, C.-X. Mapping epigenetic modifications by sequencing technologies. Cell Death Differ. 2023. [Google Scholar] [CrossRef]
- Füllgrabe, J.; Gosal, W.S.; Creed, P.; Liu, S.; Lumby, C.K.; Morley, D.J.; Ost, T.W.B.; Vilella, A.J.; Yu, S.; Bignell, H.; et al. Simultaneous sequencing of genetic and epigenetic bases in DNA. Nat. Biotechnol. 2023, 41, 1457–1464. [Google Scholar] [CrossRef]
- Zhang, Q.; Ma, S.; Liu, Z.; Zhu, B.; Zhou, Z.; Li, G.; Meana, J.J.; González-Maeso, J.; Lu, C. Droplet-based bisulfite sequencing for high-throughput profiling of single-cell DNA methylomes. Nat. Commun. 2023, 14, 4672. [Google Scholar] [CrossRef] [PubMed]
- Vaisvila, R.; Ponnaluri, V.K.C.; Sun, Z.; Langhorst, B.W.; Saleh, L.; Guan, S.; Dai, N.; Campbell, M.A.; Sexton, B.S.; Marks, K.; et al. Enzymatic methyl sequencing detects DNA methylation at single-base resolution from picograms of DNA. Genome Res. 2021, 31, 1280–1289. [Google Scholar] [CrossRef]
- Wu, Y.-L.; Lin, Z.-J.; Li, C.-C.; Lin, X.; Shan, S.-K.; Guo, B.; Zheng, M.-H.; Li, F.; Yuan, L.-Q.; Li, Z.-h. Epigenetic regulation in metabolic diseases: Mechanisms and advances in clinical study. Signal Transduct. Target. Ther. 2023, 8, 98. [Google Scholar] [CrossRef] [PubMed]
- Farsetti, A.; Illi, B.; Gaetano, C. How epigenetics impacts on human diseases. Eur. J. Intern. Med. 2023, 114, 15–22. [Google Scholar] [CrossRef] [PubMed]




| Epigenetic Mechanisms (EM) | Relationship to Depression | Relevant Studies |
|---|---|---|
| DNA Methylation | Altered DNA methylation patterns in genes associated with neurodevelopment and neurotransmission have been linked to depression susceptibility and treatment response. | - Investigation of DNA methylation in the serotonin transporter gene (SLC6A4) and its association with depression risk and MDD and antidepressant response. The focus appears to be on understanding the relationships between MDD, childhood trauma, and biological factors such as DNA methylation and hippocampal volume [110]. - Study on DNA methylation changes in brain-derived neurotrophic factor (BDNF) and their role in depression pathophysiology [111]. - Genome-wide DNA methylation profiling in MDD, identifying epigenetic biomarkers and biological pathways involved in the disorder. Methylation levels of 14 DMRs in genes predisposing toward major depressive disorder were positively associated with depression scores, giving a possible mechanism for how methylation affects depression. Genes of interest were the following: BMP2, PRDM7, KCNIP1, GRIK2, TPRN2, GATA2, HELZ2, and ZNF624 [112,113]. |
| Histone Modifications | Dysregulation of histone-modifying enzymes has been observed in depression, affecting genes involved in synaptic plasticity and stress response. | - The role of circadian genes and histone modifications in MDD such as those affecting sleep, temperature, hormonal secretions, and mood are associated with MDD and antidepressant treatment [114]. - Investigation of chromatin modifications and their impact on cognitive behaviors in depression [12]. - Epigenetic regulation of genes implicated in synaptic function and neuronal plasticity in MDD [45]. |
| Non-coding RNAs (miRNAs) | Dysregulation of miRNAs has been associated with neuroinflammation, synaptic plasticity, and stress response pathways, making them potential diagnostic biomarkers and therapeutic targets for depression. | - MicroRNAs in MDD, depression, and suicide disorder [115]. - Altered miRNA expression in peripheral blood mononuclear cells in depression patients, revealing potential circulating biomarkers [116]. |
| Long Non-coding RNAs (lncRNAs) | Aberrant expression of lncRNAs has been implicated in depression, modulating gene expression and affecting key pathways in the brain. | - Comprehensive profiling of lncRNA expression in postmortem brain tissues of depression patients, revealing potential therapeutic targets [117,118]. - Meta-analysis of lncRNA expression patterns in blood samples of patients with MDD [103]. |
| Chromatin Remodeling | Altered chromatin remodeling complexes have been associated with depression, influencing the accessibility of genes involved in neuroplasticity and stress response. | - The roles of histone acetylation, DNA methylation, and non-coding RNA. Behavioral response to stress, depressive behaviors, and response to antidepressants [119]. - Chromatin medication and their cognitive behaviors in depression [120]. - Identification of altered chromatin accessibility in MDD patient-derived neurons, providing insights into the transcriptional dysregulation by studying open and closed chromatin states in the brain, with emphasis on neuropsychiatric disorders [121]. - Altered chromatin accessibility in MDD patient-derived neurons [122] |
| DNA Hydroxy methylation | Changes in DNA hydroxy methylation have been observed in depression, potentially influencing gene expression and neurodevelopmental processes. | - Investigation of DNA hydroxy methylation dynamics in depression using postmortem brain tissues [123]. - Altered DNA hydroxy methylation patterns in peripheral blood samples of depression patients [124]. - The role of DNA hydroxy methylation in depression susceptibility and treatment response [125]. |
| RNA Editing | Dysregulation of RNA editing enzymes has been implicated in depression, leading to altered transcriptomes and neurobiological processes. | - RNA editing landscape in MDD patient brains, highlighting potential editing sites in genes relevant to depression [126]. - Study on the impact of RNA editing adenosine to inosine (A-to-I) RNA editing and m6A methylations on MDD [126]. - Altered RNA editing profiles in peripheral blood samples of patients with treatment-resistant depression and bipolar disorder. The association of RNA editing variant modifications with depression subtypes [127]. |
| Chromosomal Conformation | Changes in chromosomal conformation have been associated with depression, affecting gene interactions and regulatory networks in the brain. | - Hi-C analysis of chromosomal interactions in MDD, dysthymic disorder, and depression in bipolar disorder patient brain tissues, identifying altered 3D chromatin organization [128]. - Investigation of chromosomal interactions and their role in gene regulation in a mouse model of depression [129]. - Altered chromosomal conformation associated with genes involved in synaptic function and depressive behaviors in MDD [130]. |
| Transgenerational Epigenetic Inheritance | Epigenetic changes in parental germ cells have been implicated in the risk of depression in offspring, suggesting transgenerational epigenetic inheritance. | - Investigation of transgenerational effects of stress-induced DNA methylation changes on chronic stress susceptibility in rat offspring [131]. - Human study exploring the potential impact of parental early life stress on DNA methylation patterns in MDD, dysthymic disorder, and depression in bipolar disorder patients and their children [132,133]. - Evidence for transgenerational epigenetic inheritance in depression and its underlying depression mechanisms [134]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aljabali, A.A.A.; Alkaraki, A.K.; Gammoh, O.; Tambuwala, M.M.; Mishra, V.; Mishra, Y.; Hassan, S.S.; El-Tanani, M. Deciphering Depression: Epigenetic Mechanisms and Treatment Strategies. Biology 2024, 13, 638. https://doi.org/10.3390/biology13080638
Aljabali AAA, Alkaraki AK, Gammoh O, Tambuwala MM, Mishra V, Mishra Y, Hassan SS, El-Tanani M. Deciphering Depression: Epigenetic Mechanisms and Treatment Strategies. Biology. 2024; 13(8):638. https://doi.org/10.3390/biology13080638
Chicago/Turabian StyleAljabali, Alaa A. A., Almuthanna K. Alkaraki, Omar Gammoh, Murtaza M. Tambuwala, Vijay Mishra, Yachana Mishra, Sk. Sarif Hassan, and Mohamed El-Tanani. 2024. "Deciphering Depression: Epigenetic Mechanisms and Treatment Strategies" Biology 13, no. 8: 638. https://doi.org/10.3390/biology13080638
APA StyleAljabali, A. A. A., Alkaraki, A. K., Gammoh, O., Tambuwala, M. M., Mishra, V., Mishra, Y., Hassan, S. S., & El-Tanani, M. (2024). Deciphering Depression: Epigenetic Mechanisms and Treatment Strategies. Biology, 13(8), 638. https://doi.org/10.3390/biology13080638






