Dinoflagellate–Bacteria Interactions: Physiology, Ecology, and Evolution
Simple Summary
Abstract
1. Introduction
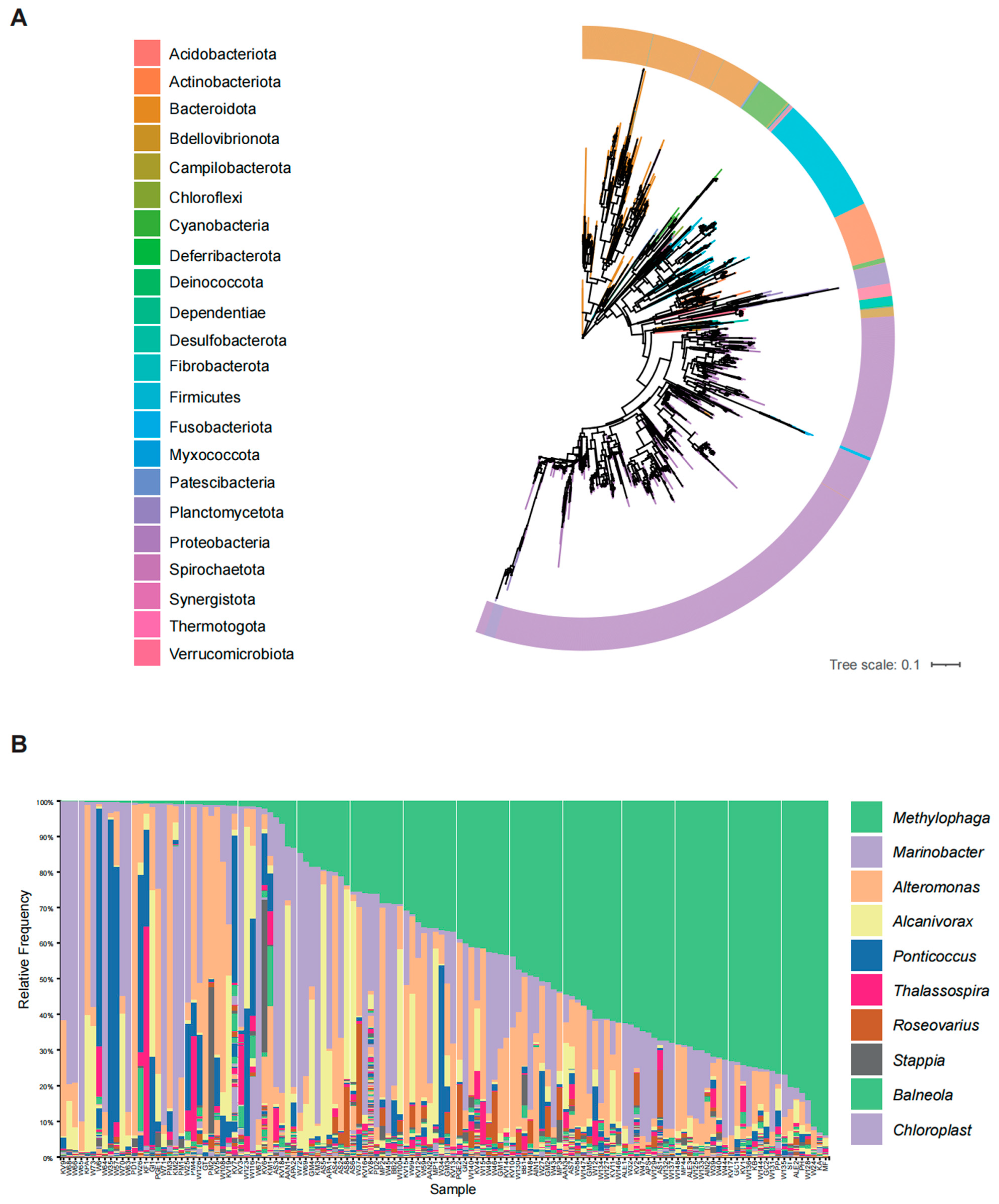
2. Dinoflagellates Harbor Distinct Bacterial Communities
3. Dinoflagellates Recruit Bacteria through Dissolved Organic Matter (DOM)
4. Growth-Promoting Metabolic Substances Generated by Bacteria for the Growth of Dinoflagellates
5. Synergistic Utilization of Nitrogen (N) and Phosphorus (P) Nutrients by Dinoflagellates and Bacteria
6. Algicidal Bacteria Inhibit the Growth of Dinoflagellates
| Bacterial Genus | Bacterial Class | Bacterial Phylum | Target Dinoflagellate | Reference |
|---|---|---|---|---|
| Microbacterium (1), Brevibacterium (4), Bacillus (3), Halobacillus (1), Virgobacillus (1), Mangrovimonas (1), Sulfitobacter (2), Pelagibaca (3), Citreicella (1), Mameliella (1), Halomonas (4), Pseudomonas (4), Vibrio (7), Alteromonas (3), Pseudoalteromonas (7) | Actinomycetes, Baccilli, Flavobacteriia, Alphaproteobacteria, Gammaproteobacteria | Actinobacteria, Firmicutes, Bacteroidetes, Proteobacteria | Pyrodinium bahamense L.Plate, 1906 | [83] |
| Muricauda (2) | Flavobacteriia | Bacteroidota | Ak. sanguinea | [84] |
| Pseudoalteromonas | Gammaproteobacteria | Proteobacteria | Alexandrium (A) tamarense (Lebour) Balech, 1995 | [85] |
| Acetinobacter | Gammaproteobacteria | Proteobacteria | A. tamarense | [86] |
| Bacillus | Bacilli | Bacillota | A. minutum | [87] |
| Bacillus | Bacilli | Bacillota | Scrippsiella trochoidea (F.Stein) A.R.Loeblich III, 1976, Prorocentrum micans Ehrenberg, 1834, Peridinium umbonatum Karsten, 1907 | [88] |
| Sulfitobacter | Alphaproteobacteria | Proteobacteria | P. donghaiense | [74] |
| Vibrio (2) | Gammaproteobacteria | Proteobacteria | Ak. sanguinea | [89] |
| Bacillus | Bacilli | Bacillota | Gy. catenatum H.W. Graham | [90] |
| Bacillus | Bacilli | Bacillota | C. polykrikoides | [76] |
| Cochlodiniinecator | Alphaproteobacteria | Pseudomonadota | C. polykrikoides | [91] |
| Paracoccus | Alphaproteobacteria | Proteobacteria | Ka. mikimotoi | [92] |
| Pseudoruegeria | Alphaproteobacteria | Proteobacteria | A. catenella | [93] |
| Stenotrophomonas | Gammaproteobacteria | Proteobacteria | A. tamarense | [94] |
| Sulfitobacter | Alphaproteobacteria | Proteobacteria | A. tamarense | [95] |
| Vibrio | Gammaproteobacteria | Proteobacteria | Ak. sanguinea | [96] |
| Vibrio | Gammaproteobacteria | Proteobacteria | Prorocentrum | [97] |
| Alteromonas | Gammaproteobacteria | Pseudomonadota | P. donghaiense | [98] |
| Marinobacter, Pseudomonas | Alphaproteobacteria, Gammaproteobacteria | Pseudomonadota | Ka. mikimotoi | [99] |
| Pseudoalteromonas | Gammaproteobacteria | Pseudomonadota | Ka. mikimotoi | [100] |
| Pseudoalteromonas | Gammaproteobacteria | Pseudomonadota | Noctiluca scintillans (Macartney) Kofoid & Swezy, 1921 | [101] |
| Pseudoruegeria | Alphaproteobacteria | Proteobacteria | A. catenella | [102] |
| Shewanella | Gammaproteobacteria | Proteobacteria | Alexandrium pacificum R.W.Litaker, 2014 | [81] |
| Shewanella | Gammaproteobacteria | Proteobacteria | Karlodinium veneficum (D.Ballantine) J.Larsen, 2000 | [70] |
| Vibrio | Gammaproteobacteria | Proteobacteria | Ak. sanguinea | [103] |
| Alteromonas | Gammaproteobacteria | Pseudomonadota | Symbiodinium | [104] |
| Marinobacter | Alphaproteobacteria | Pseudomonadota | Ka. mikimotoi | [105] |
| Microbulbifer | Gammaproteobacteria | Pseudomonadota | Amphidinium carterae D-044, P. minimum D-127 | [106] |
| Pseudoalteromonas | Gammaproteobacteria | Pseudomonadota | Ka. mikimotoi, A. tamarense | [107] |
| Pseudomonas | Gammaproteobacteria | Pseudomonadota | Gy. catenatum, Ka. mikimotoi | [108] |
| Qipengyuania | Alphaproteobacteria | Pseudomonadota | Margalefidinium polykrikoides (Margalef) F.Gómez, Richlen & D.M.Anderson, 2017 | [109] |
| Shewanella | Gammaproteobacteria | Proteobacteria | Prorocentrum triestinum J.Schiller, 1918 | [110] |
| Tenacibaculum | Flavobacteriia | Bacteroidota | Ka. mikimotoi | [111] |
| Arenibacter | Flavobacteriia | Bacteroidota | Ak. sanguinea | [112] |
| Maribacter | Flavobacteriia | Bacteroidota | Ka. mikimotoi | [113] |
7. The Defense of Dinoflagellate with Bacteria Challenging
| Antibacteria Compound | Producer | Type | Bacterial Targets | Reference |
|---|---|---|---|---|
| 1-(2,6,6-trimethy-4-hydroxycyclohexenyl)-1,3-butanedione | P. minimum | β-diketone | Vibrio sp., Flavobacter sp., Chromobacterium sp. | [118] |
| Luteophanol D | Amphidinium sp. strain Y-52 | Polyketide | Micrococcus luteus Cohn 1872 | [119] |
| Amphidinolide Q | Amphidinium sp. 2012-7-4A strain | Macrolide | S. aureus, B. subtilis, E. coli | [115] |
| Amphidinin A | Amphidinium sp. | Polyketide | B. subtilis | [120] |
| Amphidinin E | Amphidinium sp. (2012-7-4A strain) | Polyketide | S. aureus, B. subtilis | [115] |
| Amphidinin C | Amphidinium sp. (2012-7-4A strain) | Polyketide | S. aureus | [115] |
| F5 | H. circularisquama | Porphyrin | S. aureus | [121] |
| Amphidinol dehydroAM-A | A. carterae strain LACW11 | Polyketide | S. aureus, En. faecalis | [116] |
| Amphidinol AM-A | A. carterae strain LACW11 | Polyketide | S. aureus, En. faecalis | [116] |
8. Bacteria Involved in Dinoflagellate Toxin Production
9. Other Interactions
10. Bacteria in Dinoflagellate Genome Evolution
11. Biofilms
12. Conclusions
13. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chénard, C.; Wijaya, W.; Vaulot, D.; Lopes dos Santos, A.; Martin, P.; Kaur, A.; Lauro, F.M. Temporal and Spatial Dynamics of Bacteria, Archaea and Protists in Equatorial Coastal Waters. Sci. Rep. 2019, 9, 16390. [Google Scholar] [CrossRef]
- Steele, J.A.; Countway, P.D.; Xia, L.; Vigil, P.D.; Beman, J.M.; Kim, D.Y.; Chow, C.-E.T.; Sachdeva, R.; Jones, A.C.; Schwalbach, M.S.; et al. Marine Bacterial, Archaeal and Protistan Association Networks Reveal Ecological Linkages. ISME J. 2011, 5, 1414–1425. [Google Scholar] [CrossRef] [PubMed]
- Krzmarzick, M.J.; Taylor, D.K.; Fu, X.; McCutchan, A.L. Diversity and Niche of Archaea in Bioremediation. Archaea 2018, 2018, 3194108. [Google Scholar] [CrossRef] [PubMed]
- Fallahi, A.; Rezvani, F.; Asgharnejad, H.; Khorshidi Nazloo, E.; Hajinajaf, N.; Higgins, B. Interactions of Microalgae-Bacteria Consortia for Nutrient Removal from Wastewater: A Review. Chemosphere 2021, 272, 129878. [Google Scholar] [CrossRef]
- Pauna, V.H.; Buonocore, E.; Renzi, M.; Russo, G.F.; Franzese, P.P. The Issue of Microplastics in Marine Ecosystems: A Bibliometric Network Analysis. Mar. Pollut. Bull. 2019, 149, 110612. [Google Scholar] [CrossRef]
- Fuentes, J.; Garbayo, I.; Cuaresma, M.; Montero, Z.; González-del-Valle, M.; Vílchez, C. Impact of Microalgae-Bacteria Interactions on the Production of Algal Biomass and Associated Compounds. Mar. Drugs 2016, 14, 100. [Google Scholar] [CrossRef]
- Deng, Y.; Wang, K.; Hu, Z.; Tang, Y.-Z. Abundant Species Diversity and Essential Functions of Bacterial Communities Associated with Dinoflagellates as Revealed from Metabarcoding Sequencing for Laboratory-Raised Clonal Cultures. Int. J. Environ. Res. Public Health 2022, 19, 4446. [Google Scholar] [CrossRef]
- Guiry, M.D. How Many Species of Algae Are There? J. Phycol. 2012, 48, 1057–1063. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Wang, K.; Hu, Z.; Hu, Q.; Tang, Y.Z. Toxic and Non-Toxic Dinoflagellates Host Distinct Bacterial Communities in Their Phycospheres. Commun. Earth Environ. 2023, 4, 263. [Google Scholar] [CrossRef]
- Jung, S.W.; Kang, J.; Park, J.S.; Joo, H.M.; Suh, S.-S.; Kang, D.; Lee, T.-K.; Kim, H.-J. Dynamic Bacterial Community Response to Akashiwo sanguinea (Dinophyceae) Bloom in Indoor Marine Microcosms. Sci. Rep. 2021, 11, 6983. [Google Scholar] [CrossRef]
- Tortell, P.D. Evolutionary and Ecological Perspectives on Carbon Acquisition in Phytoplankton. Limnol. Oceanogr. 2000, 45, 744–750. [Google Scholar] [CrossRef]
- Zohary, T.; Güde, H.; Pollingher, U.; Kaplan, B.; Pinkas, R.; Hadas, O. The Role of Nutrients in Decomposition of a Thecate Dinoflagellate. Limnol. Oceanogr. 2000, 45, 123–130. [Google Scholar] [CrossRef]
- Cruz-López, R.; Maske, H. The Vitamin B1 and B12 Required by the Marine Dinoflagellate Lingulodinium Polyedrum Can Be Provided by Its Associated Bacterial Community in Culture. Front. Microbiol. 2016, 7, 560. [Google Scholar] [CrossRef] [PubMed]
- Morden, C.W.; Sherwood, A.R. Continued Evolutionary Surprises among Dinoflagellates. Proc. Natl. Acad. Sci. USA 2002, 99, 11558–11560. [Google Scholar] [CrossRef] [PubMed]
- Wisecaver, J.H.; Brosnahan, M.L.; Hackett, J.D. Horizontal Gene Transfer Is a Significant Driver of Gene Innovation in Dinoflagellates. Genome Biol. Evol. 2013, 5, 2368–2381. [Google Scholar] [CrossRef] [PubMed]
- Alavi, M.; Miller, T.; Erlandson, K.; Schneider, R.; Belas, R. Bacterial Community Associated with Pfiesteria-like Dinoflagellate Cultures. Environ. Microbiol. 2001, 3, 380–396. [Google Scholar] [CrossRef] [PubMed]
- Gold, K.; Pollingher, U. Occurrence of Endosymbiotic Bacteria in Marine Dinoflagellates. J. Phycol. 1971, 7, 264–265. [Google Scholar] [CrossRef]
- Sakami, T.; Nakahara, H.; Chinain, M.; Ishida, Y. Effects of Epiphytic Bacteria on the Growth of the Toxic Dinoflagellate Gambierdiscus toxicus (Dinophyceae). J. Exp. Mar. Biol. Ecol. 1999, 233, 231–246. [Google Scholar] [CrossRef]
- Bolch, C.J.S.; Subramanian, T.A.; Green, D.H. The Toxic Dinoflagellate Gymnodinium catenatum (Dinophyceae) Requires Marine Bacteria For Growth. J. Phycol. 2011, 47, 1009–1022. [Google Scholar] [CrossRef]
- Lawson, C.A.; Raina, J.; Kahlke, T.; Seymour, J.R.; Suggett, D.J. Defining the Core Microbiome of the Symbiotic Dinoflagellate, Symbiodinium. Environ. Microbiol. Rep. 2018, 10, 7–11. [Google Scholar] [CrossRef]
- Wu, Z.; Lee, W.H.; Liu, Z.; Lin, S.; Lam, P.K.S. Microbiome Associated with Gambierdiscus balechii Cultures under Different Toxicity Conditions. Front. Mar. Sci. 2022, 9, 760553. [Google Scholar] [CrossRef]
- Sanchez-Garcia, S.; Wang, H.; Wagner-Döbler, I. The Microbiome of the Dinoflagellate Prorocentrum cordatum in Laboratory Culture and Its Changes at Higher Temperatures. Front. Microbiol. 2022, 13, 952238. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Fandino, L.; Riemann, L.; Steward, G.; Long, R.; Azam, F. Variations in Bacterial Community Structure during a Dinoflagellate Bloom Analyzed by DGGE and 16S RDNA Sequencing. Aquat. Microb. Ecol. 2001, 23, 119–130. [Google Scholar] [CrossRef]
- Zhou, J.; Richlen, M.L.; Sehein, T.R.; Kulis, D.M.; Anderson, D.M.; Cai, Z. Microbial Community Structure and Associations During a Marine Dinoflagellate Bloom. Front. Microbiol. 2018, 9, 1201. [Google Scholar] [CrossRef]
- Camarena-Gómez, M.T.; Ruiz-González, C.; Piiparinen, J.; Lipsewers, T.; Sobrino, C.; Logares, R.; Spilling, K. Bacterioplankton Dynamics Driven by Interannual and Spatial Variation in Diatom and Dinoflagellate Spring Bloom Communities in the Baltic Sea. Limnol. Oceanogr. 2021, 66, 255–271. [Google Scholar] [CrossRef]
- Van Dolah, F.M.; Lidie, K.B.; Monroe, E.A.; Bhattacharya, D.; Campbell, L.; Doucette, G.J.; Kamykowski, D. The Florida Red Tide Dinoflagellate Karenia Brevis: New Insights into Cellular and Molecular Processes Underlying Bloom Dynamics. Harmful Algae 2009, 8, 562–572. [Google Scholar] [CrossRef]
- Yu, L.; Li, T.; Li, H.; Ma, M.; Li, L.; Lin, S. In Situ Molecular Ecological Analyses Illuminate Distinct Factors Regulating Formation and Demise of a Harmful Dinoflagellate Bloom. Microbiol. Spectr. 2023, 11, e05157-22. [Google Scholar] [CrossRef]
- Buchan, A.; LeCleir, G.R.; Gulvik, C.A.; González, J.M. Master Recyclers: Features and Functions of Bacteria Associated with Phytoplankton Blooms. Nat. Rev. Microbiol. 2014, 12, 686–698. [Google Scholar] [CrossRef]
- Raina, J.-B.; Giardina, M.; Brumley, D.R.; Clode, P.L.; Pernice, M.; Guagliardo, P.; Bougoure, J.; Mendis, H.; Smriga, S.; Sonnenschein, E.C.; et al. Chemotaxis Increases Metabolic Exchanges between Marine Picophytoplankton and Heterotrophic Bacteria. Nat. Microbiol. 2023, 8, 510–521. [Google Scholar] [CrossRef] [PubMed]
- Purina, I.; Balode, M.; Béchemin, C.; Põder, T.; Vérité, C.; Maestrini, S. Influence of Dissolved Organic Matter from Terrestrial Origin on the Changes of Dinoflagellate Species Composition in the Gulf of Riga, Baltic Sea. Hydrobiologia 2004, 514, 127–137. [Google Scholar] [CrossRef]
- Thornton, D.C.O. Dissolved Organic Matter (DOM) Release by Phytoplankton in the Contemporary and Future Ocean. Eur. J. Phycol. 2014, 49, 20–46. [Google Scholar] [CrossRef]
- Lin, Q.; Shang, L.; Wang, X.; Hu, Z.; Du, H.; Wang, H. Different Dimethylsulphoniopropionate-Producing Ability of Dinoflagellates Could Affect the Structure of Their Associated Bacterial Community. Algal Res. 2021, 57, 102359. [Google Scholar] [CrossRef]
- Miller, T.R.; Hnilicka, K.; Dziedzic, A.; Desplats, P.; Belas, R. Chemotaxis of Silicibacter sp. Strain TM1040 toward Dinoflagellate Products. Appl. Environ. Microbiol. 2004, 70, 4692–4701. [Google Scholar] [CrossRef]
- Han, Y.; Jiao, N.; Zhang, Y.; Zhang, F.; He, C.; Liang, X.; Cai, R.; Shi, Q.; Tang, K. Opportunistic Bacteria with Reduced Genomes Are Effective Competitors for Organic Nitrogen Compounds in Coastal Dinoflagellate Blooms. Microbiome 2021, 9, 71. [Google Scholar] [CrossRef] [PubMed]
- Osbeck, C.M.G.; Lundin, D.; Karlsson, C.; Teikari, J.E.; Moran, M.A.; Pinhassi, J. Divergent Gene Expression Responses in Two Baltic Sea Heterotrophic Model Bacteria to Dinoflagellate Dissolved Organic Matter. PLoS ONE 2022, 17, e0243406. [Google Scholar] [CrossRef]
- Shultis, D.D.; Purdy, M.D.; Banchs, C.N.; Wiener, M.C. Outer Membrane Active Transport: Structure of the BtuB:TonB Complex. Science (1979) 2006, 312, 1396–1399. [Google Scholar] [CrossRef]
- Sweeney, B.M. Gymnodinium splendens, a Marine Dinoflagellate Requiring Vitamin B 12. Am. J. Bot. 1954, 41, 821. [Google Scholar] [CrossRef]
- Gobler, C.; Norman, C.; Panzeca, C.; Taylor, G.; Sañudo-Wilhelmy, S. Effect of B-Vitamins (B1, B12) and Inorganic Nutrients on Algal Bloom Dynamics in a Coastal Ecosystem. Aquat. Microb. Ecol. 2007, 49, 181–194. [Google Scholar] [CrossRef]
- Tang, Y.Z.; Koch, F.; Gobler, C.J. Most Harmful Algal Bloom Species Are Vitamin B1 and B12 Auxotrophs. Proc. Natl. Acad. Sci. USA 2010, 107, 20756–20761. [Google Scholar] [CrossRef] [PubMed]
- Sañudo-Wilhelmy, S.A.; Gómez-Consarnau, L.; Suffridge, C.; Webb, E.A. The Role of B Vitamins in Marine Biogeochemistry. Ann. Rev. Mar. Sci. 2014, 6, 339–367. [Google Scholar] [CrossRef] [PubMed]
- Croft, M.T.; Lawrence, A.D.; Raux-Deery, E.; Warren, M.J.; Smith, A.G. Algae Acquire Vitamin B12 through a Symbiotic Relationship with Bacteria. Nature 2005, 438, 90–93. [Google Scholar] [CrossRef]
- Lin, S.; Hu, Z.; Song, X.; Gobler, C.J.; Tang, Y.Z. Vitamin B12-Auxotrophy in Dinoflagellates Caused by Incomplete or Absent Cobalamin-Independent Methionine Synthase Genes (MetE). Fundam. Res. 2022, 2, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Lin, S. Genomic Understanding of Dinoflagellates. Res. Microbiol. 2011, 162, 551–569. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.J.; Yoo, Y.D.; Kang, N.S.; Lim, A.S.; Seong, K.A.; Lee, S.Y.; Lee, M.J.; Lee, K.H.; Kim, H.S.; Shin, W.; et al. Heterotrophic Feeding as a Newly Identified Survival Strategy of the Dinoflagellate Symbiodinium. Proc. Natl. Acad. Sci. USA 2012, 109, 12604–12609. [Google Scholar] [CrossRef] [PubMed]
- Glibert, P.; Burkholder, J.; Kana, T.; Alexander, J.; Skelton, H.; Shilling, C. Grazing by Karenia Brevis on Synechococcus Enhances Its Growth Rate and May Help to Sustain Blooms. Aquat. Microb. Ecol. 2009, 55, 17–30. [Google Scholar] [CrossRef]
- Seong, K.; Jeong, H.; Kim, S.; Kim, G.; Kang, J. Bacterivory by Co-Occurring Red-Tide Algae, Heterotrophic Nanoflagellates, and Ciliates. Mar. Ecol. Prog. Ser. 2006, 322, 85–97. [Google Scholar] [CrossRef]
- Jeong, H.; Park, J.; Nho, J.; Park, M.; Ha, J.; Seong, K.; Jeng, C.; Seong, C.; Lee, K.; Yih, W. Feeding by Red-Tide Dinoflagellates on the Cyanobacterium Synechococcus. Aquat. Microb. Ecol. 2005, 41, 131–143. [Google Scholar] [CrossRef]
- Tarangkoon, W.; Hansen, G.; Hansen, P. Spatial Distribution of Symbiont-Bearing Dinoflagellates in the Indian Ocean in Relation to Oceanographic Regimes. Aquat. Microb. Ecol. 2010, 58, 197–213. [Google Scholar] [CrossRef]
- HANSEN, P.J. The Role of Photosynthesis and Food Uptake for the Growth of Marine Mixotrophic Dinoflagellates1. J. Eukaryot. Microbiol. 2011, 58, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, T.; Nomura, M.; Takano, Y.; Tanifuji, G.; Shiba, K.; Inaba, K.; Inagaki, Y.; Kawata, M. Single-Cell Genomics Unveiled a Cryptic Cyanobacterial Lineage with a Worldwide Distribution Hidden by a Dinoflagellate Host. Proc. Natl. Acad. Sci. USA 2019, 116, 15973–15978. [Google Scholar] [CrossRef] [PubMed]
- Husnik, F.; Tashyreva, D.; Boscaro, V.; George, E.E.; Lukeš, J.; Keeling, P.J. Bacterial and Archaeal Symbioses with Protists. Curr. Biol. 2021, 31, R862–R877. [Google Scholar] [CrossRef] [PubMed]
- Moynihan, M.A.; Goodkin, N.F.; Morgan, K.M.; Kho, P.Y.Y.; Lopes dos Santos, A.; Lauro, F.M.; Baker, D.M.; Martin, P. Coral-Associated Nitrogen Fixation Rates and Diazotrophic Diversity on a Nutrient-Replete Equatorial Reef. ISME J. 2022, 16, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Lema, K.A.; Willis, B.L.; Bourne, D.G. Corals Form Characteristic Associations with Symbiotic Nitrogen-Fixing Bacteria. Appl. Environ. Microbiol. 2012, 78, 3136–3144. [Google Scholar] [CrossRef] [PubMed]
- Lesser, M.; Falcón, L.; Rodríguez-Román, A.; Enríquez, S.; Hoegh-Guldberg, O.; Iglesias-Prieto, R. Nitrogen Fixation by Symbiotic Cyanobacteria Provides a Source of Nitrogen for the Scleractinian Coral Montastraea cavernosa. Mar. Ecol. Prog. Ser. 2007, 346, 143–152. [Google Scholar] [CrossRef]
- Olson, N.D.; Ainsworth, T.D.; Gates, R.D.; Takabayashi, M. Diazotrophic Bacteria Associated with Hawaiian Montipora Corals: Diversity and Abundance in Correlation with Symbiotic Dinoflagellates. J. Exp. Mar. Biol. Ecol. 2009, 371, 140–146. [Google Scholar] [CrossRef]
- Biebl, H.; Tindall, B.J.; Pukall, R.; Lünsdorf, H.; Allgaier, M.; Wagner-Döbler, I. Hoeflea phototrophica sp. Nov., a Novel Marine Aerobic Alphaproteobacterium That Forms Bacteriochlorophyll a. Int. J. Syst. Evol. Microbiol. 2006, 56, 821–826. [Google Scholar] [CrossRef]
- Biebl, H.; Pukall, R.; Lünsdorf, H.; Schulz, S.; Allgaier, M.; Tindall, B.J.; Wagner-Döbler, I. Description of Labrenzia alexandrii Gen. Nov., sp. Nov., a Novel Alphaproteobacterium Containing Bacteriochlorophyll a, and a Proposal for Reclassification of Stappia aggregata as Labrenzia aggregata Comb. Nov., of Stappia marina as Labrenzia marina Comb. Nov. and of Stappia alba as Labrenzia alba Comb. Nov., and Emended Descriptions of the Genera Pannonibacter, Stappia and Roseibium, and of the Species Roseibium denhamense and Roseibium hamelinense. Int. J. Syst. Evol. Microbiol. 2007, 57, 1095–1107. [Google Scholar] [CrossRef] [PubMed]
- Palacios, L.; Arahal, D.R.; Reguera, B.; Marín, I. Hoeflea alexandrii sp. Nov., Isolated from the Toxic Dinoflagellate Alexandrium Minutum AL1V. Int. J. Syst. Evol. Microbiol. 2006, 56, 1991–1995. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wu, Z.; Yang, X.; Lin, S.; Lee, W.H.; Lam, P.K.S. A Rhizobium Bacterium and Its Population Dynamics under Different Culture Conditions of Its Associated Toxic Dinoflagellate Gambierdiscus balechii. Mar. Life Sci. Technol. 2021, 3, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; McCabe, K.; Fawcett, S.E.; Forrer, H.J.; Hashihama, F.; Jeandel, C.; Marconi, D.; Planquette, H.; Saito, M.A.; Sohm, J.A.; et al. A Global Ocean Dissolved Organic Phosphorus Concentration Database (DOPv2021). Sci. Data 2022, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Paytan, A.; McLaughlin, K. The Oceanic Phosphorus Cycle. Chem. Rev. 2007, 107, 563–576. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Shi, J.; Jia, Y.; Bai, F.; Yang, S.; Mi, W.; He, S.; Wu, Z. Unveiling the Impact of Glycerol Phosphate (DOP) in the Dinoflagellate Peridinium Bipes by Physiological and Transcriptomic Analysis. Environ. Sci. Eur. 2020, 32, 38. [Google Scholar] [CrossRef]
- Karl, D.M. Microbially Mediated Transformations of Phosphorus in the Sea: New Views of an Old Cycle. Ann. Rev. Mar. Sci. 2014, 6, 279–337. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Lin, X.; Li, L.; Lin, L.; Lin, S. Glyphosate Shapes a Dinoflagellate-Associated Bacterial Community While Supporting Algal Growth as Sole Phosphorus Source. Front. Microbiol. 2017, 8, 2530. [Google Scholar] [CrossRef]
- Hattenrath-Lehmann, T.K.; Marcoval, M.A.; Mittlesdorf, H.; Goleski, J.A.; Wang, Z.; Haynes, B.; Morton, S.L.; Gobler, C.J. Nitrogenous Nutrients Promote the Growth and Toxicity of Dinophysis Acuminata during Estuarine Bloom Events. PLoS ONE 2015, 10, e0124148. [Google Scholar] [CrossRef]
- Wang, Y.; Coyne, K.J. Metabolomic Insights of the Effects of Bacterial Algicide IRI-160AA on Dinoflagellate Karlodinium veneficum. Metabolites 2022, 12, 317. [Google Scholar] [CrossRef]
- Coyne, K.J.; Wang, Y.; Johnson, G. Algicidal Bacteria: A Review of Current Knowledge and Applications to Control Harmful Algal Blooms. Front. Microbiol. 2022, 13, 871177. [Google Scholar] [CrossRef] [PubMed]
- Meyer, N.; Bigalke, A.; Kaulfuß, A.; Pohnert, G. Strategies and Ecological Roles of Algicidal Bacteria. FEMS Microbiol. Rev. 2017, 41, 880–899. [Google Scholar] [CrossRef] [PubMed]
- Xue, G.; Wang, X.; Xu, C.; Song, B.; Chen, H. Removal of Harmful Algae by Shigella sp. H3 and Alcaligenes sp. H5: Algicidal Pathways and Characteristics. Environ. Technol. 2022, 43, 4341–4353. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Fan, Y.; Zhang, D.; Chen, S.; Bai, X.; Ma, X.; Xie, Z.; Xu, H. Effect and Mechanism of the Algicidal Bacterium Sulfitobacter Porphyrae ZFX1 on the Mitigation of Harmful Algal Blooms Caused by Prorocentrum donghaiense. Environ. Pollut. 2020, 263, 114475. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Ye, Q.; Chen, Q.; Yang, K.; Zhang, D.; Chen, Z.; Lu, S.; Shao, X.; Fan, Y.; Yao, L.; et al. Algicidal Activity of Novel Marine Bacterium Paracoccus sp. Strain Y42 against a Harmful Algal-Bloom-Causing Dinoflagellate, Prorocentrum donghaiense. Appl. Environ. Microbiol. 2018, 84, e01015-18. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.-Y.; Son, H.-J. Effects of Mycosubtilin Homolog Algicides from a Marine Bacterium, Bacillus sp. SY-1, against the Harmful Algal Bloom Species Cochlodinium polykrikoides. J. Microbiol. 2021, 59, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Zhou, B.; Xu, L.; Liu, L.; Wang, G.; Liu, X.; Tang, X. A Marine Algicidal Thalassospira and Its Active Substance against the Harmful Algal Bloom Species Karenia mikimotoi. Appl. Microbiol. Biotechnol. 2016, 100, 5131–5139. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Liu, L.; Li, Y.; Xiao, Y.; Ding, G.; Lin, S.; Chen, J. Isolation of an Algicidal Bacterium and Its Effects against the Harmful-Algal- Bloom Dinoflagellate Prorocentrum donghaiense (Dinophyceae). Harmful Algae 2018, 80, 72–79. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, H.; Zhang, H.; Chen, Z.; Tian, Y.; Xu, H.; Zheng, T.; Zheng, W. Toxicity of Algicidal Extracts from Mangrovimonas yunxiaonensis Strain LY01 on a HAB Causing Alexandrium tamarense. J. Hazard. Mater. 2014, 278, 372–381. [Google Scholar] [CrossRef]
- Imai, I.; Ishida, Y.; Hata, Y. Killing of Marine Phytoplankton by a Gliding Bacterium Cytophaga sp., Isolated from the Coastal Sea of Japan. Mar. Biol. 1993, 116, 527–532. [Google Scholar] [CrossRef]
- Furusawa, G.; Yoshikawa, T.; Yasuda, A.; Sakata, T. Algicidal Activity and Gliding Motility of Saprospira sp. SS98-5. Can. J. Microbiol. 2003, 49, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, D.; Wang, Y.; Sun, P.; Ma, S.; Chen, T. Algicidal Effects of a High-Efficiency Algicidal Bacterium Shewanella Y1 on the Toxic Bloom-Causing Dinoflagellate Alexandrium pacificum. Mar. Drugs 2022, 20, 239. [Google Scholar] [CrossRef] [PubMed]
- Dungca-Santos, J.C.R.; Caspe, F.J.O.; Tablizo, F.A.; Purganan, D.J.E.; Azanza, R.V.; Onda, D.F.L. Algicidal Potential of Cultivable Bacteria from Pelagic Waters against the Toxic Dinoflagellate Pyrodinium bahamense (Dinophyceae). J. Appl. Phycol. 2019, 31, 3721–3735. [Google Scholar] [CrossRef]
- Li, S.; Chen, M.; Chen, Y.; Tong, J.; Wang, L.; Xu, Y.; Hu, Z.; Chen, H. Epibiotic Bacterial Community Composition in Red-Tide Dinoflagellate Akashiwo Sanguinea Culture under Various Growth Conditions. FEMS Microbiol. Ecol. 2019, 95, fiz057. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Y.; Zhou, Y.; Li, Y.; Zhou, J.; Xu, Y. Optimized Culturing Conditions for an Algicidal Bacterium Pseudoalteromonas sp. SP 48 on Harmful Algal Blooms Caused by Alexandrium tamarense. Microbiologyopen 2019, 8, e00803. [Google Scholar] [CrossRef]
- Zhang, C.; Li, Y.; Meng, C.-X.; Yang, M.-J.; Wang, Y.-G.; Cai, Z.-H.; Zuo, P.; Zhou, J. Complete Genome Sequence of Acinetobacter Baumanni J1, a Quorum Sensing-Producing Algicidal Bacterium, Isolated from Eastern Pacific Ocean. Mar. Genom. 2020, 52, 100719. [Google Scholar] [CrossRef]
- Zhang, S.-J.; Du, X.-P.; Zhu, J.-M.; Meng, C.-X.; Zhou, J.; Zuo, P. The Complete Genome Sequence of the Algicidal Bacterium Bacillus Subtilis Strain JA and the Use of Quorum Sensing to Evaluate Its Antialgal Ability. Biotechnol. Rep. 2020, 25, e00421. [Google Scholar] [CrossRef]
- Hu, X.; Su, H.; Xu, Y.; Xu, W.; Li, S.; Huang, X.; Cao, Y.; Wen, G. Algicidal Properties of Fermentation Products from Bacillus Cereus Strain JZBC1 Dissolving Dominant Dinoflagellate Species Scrippsiella trochoidea, Prorocentrum micans, and Peridinium umbonatum. Biologia 2020, 75, 2015–2024. [Google Scholar] [CrossRef]
- Wang, Y.; Li, S.; Liu, G.; Li, X.; Yang, Q.; Xu, Y.; Hu, Z.; Chen, C.-Y.; Chang, J.-S. Continuous Production of Algicidal Compounds against Akashiwo sanguinea via a Vibrio sp. Co-Culture. Bioresour. Technol. 2020, 295, 122246. [Google Scholar] [CrossRef]
- Balaji Prasath, B.; Wang, Y.; Su, Y.; Zheng, W.; Lin, H.; Yang, H. Coagulant Plus Bacillus Nitratireducens Fermentation Broth Technique Provides a Rapid Algicidal Effect of Toxic Red Tide Dinoflagellate. J. Mar. Sci. Eng. 2021, 9, 395. [Google Scholar] [CrossRef]
- Van Le, V.; Ko, S.-R.; Lee, S.-A.; Jin, L.; Blom, J.; Ahn, C.-Y.; Oh, H.-M. Cochlodiniinecator piscidefendens Gen. Nov., sp. Nov., an Algicidal Bacterium against the Ichthyotoxic Dinoflagellate Cochlodinium polykrikoides. Int. J. Syst. Evol. Microbiol. 2021, 71, 005124. [Google Scholar] [CrossRef]
- Ding, N.; Du, W.; Feng, Y.; Song, Y.; Wang, C.; Li, C.; Zheng, N.; Gao, P.; Wang, R. Algicidal Activity of a Novel Indigenous Bacterial Strain of Paracoccus homiensis against the Harmful Algal Bloom Species, Karenia mikimotoi. Arch. Microbiol. 2021, 203, 4821–4828. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Cho, S.-H.; Ko, S.-R.; Jeong, Y.; Lee, E.; Jin, S.; Jeong, B.-S.; Oh, B.-H.; Oh, H.-M.; Ahn, C.-Y.; et al. Elucidation of the Algicidal Mechanism of the Marine Bacterium Pseudoruegeria sp. M32A2M against the Harmful Alga Alexandrium catenella Based on Time-Course Transcriptome Analysis. Front. Mar. Sci. 2021, 8, 728890. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, Y.; Zhou, J. Complete Genome Sequence of Stenotrophomonas rhizophila KC1, a Quorum Sensing–Producing Algicidal Bacterium Isolated from Mangrove Kandelia candel. Mol. Plant-Microbe Interact.® 2021, 34, 857–861. [Google Scholar] [CrossRef]
- Hu, T.; Wang, S.; Shan, Y.; Zhang, Y.; Zhu, Y.; Zheng, L. Complete Genome of Marine Microalgae Associated Algicidal Bacterium Sulfitobacter pseudonitzschiae H46 with Quorum Sensing System. Curr. Microbiol. 2021, 78, 3741–3750. [Google Scholar] [CrossRef]
- Ouyang, L.; Liu, Y.; Chen, H.; Zaynab, M.; Yang, X.; Wang, S.; Li, S. Encapsulation and Algicidal Properties of Fermentation Products from Vibrio brasiliensis H115. Front. Mar. Sci. 2021, 8, 676913. [Google Scholar] [CrossRef]
- Kim, D.D.; Wan, L.; Cao, X.; Klisarova, D.; Gerdzhikov, D.; Zhou, Y.; Song, C.; Yoon, S. Metagenomic Insights into Co-Proliferation of Vibrio spp. and Dinoflagellates prorocentrum during a Spring Algal Bloom in the Coastal East China Sea. Water Res. 2021, 204, 117625. [Google Scholar] [CrossRef]
- Wang, Q.; Shi, X.; Guo, Y.; Lv, P.; Zhong, Y.; Xie, H.; Chen, J. Optimization of Algicidal Activity for Alteromonas sp. FDHY-03 against Harmful Dinoflagellate Prorocentrum donghaiense. J. Mar. Sci. Eng. 2022, 10, 1274. [Google Scholar] [CrossRef]
- Ding, N.; Gao, P.; Xu, D.; Xing, E.; Li, Y.; Sun, L.; Wang, R.; Zhang, W. Characterization and Algicidal Activity of Bacteria from the Phycosphere of the Harmful Alga Karenia mikimotoi. Braz. J. Microbiol. 2022, 53, 891–901. [Google Scholar] [CrossRef]
- Shi, X.; Zou, Y.; Zheng, W.; Liu, L.; Xie, Y.; Ma, R.; Chen, J. A Novel Algicidal Bacterium and Its Effects against the Toxic Dinoflagellate Karenia mikimotoi (Dinophyceae). Microbiol. Spectr. 2022, 10, e00429-22. [Google Scholar] [CrossRef]
- Wang, J.; Yin, X.; Xu, M.; Chen, Y.; Ji, N.; Gu, H.; Cai, Y.; Shen, X. Isolation and Characterization of a High-Efficiency Algicidal Bacterium Pseudoalteromonas sp. LD-B6 against the Harmful Dinoflagellate Noctiluca scintillans. Front. Microbiol. 2022, 13, 1091561. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.-R.; Jeong, Y.; Cho, S.-H.; Lee, E.; Jeong, B.-S.; Baek, S.H.; Oh, B.-H.; Ahn, C.-Y.; Oh, H.-M.; Cho, B.-K.; et al. Functional Role of a Novel Algicidal Compound Produced by Pseudoruegeria sp. M32A2M on the Harmful Algae Alexandrium catenella. Chemosphere 2022, 300, 134535. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, S.; Xie, L.; Liu, Y.; Chen, H.; Feng, J.; Ouyang, L. Identification and Optimization of the Algicidal Activity of a Novel Marine Bacterium Against Akashiwo sanguinea. Front. Mar. Sci. 2022, 9, 798544. [Google Scholar] [CrossRef]
- Jia, Y.; Lu, J.; Wang, M.; Qin, W.; Chen, B.; Xu, H.; Ma, Z. Algicidal Bacteria in Phycosphere Regulate Free-Living Symbiodinium Fate via Triggering Oxidative Stress and Photosynthetic System Damage. Ecotoxicol. Environ. Saf. 2023, 263, 115369. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Ding, N.; Wang, R.; Gao, P.; Zhang, W.; Lin, F.; Sun, S.; Sun, Y.; Sun, X.; Zhang, Z.; et al. Characterization of Cell Death in Harmful Karenia mikimotoi under Algicidal Activity of Marinobacter sp. O-7. J. Sea Res. 2023, 191, 102326. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, D.; Xie, R.; Guo, R.; Nair, S.; Han, H.; Zhang, G.; Zhao, Q.; Zhang, L.; Jiao, N.; et al. Plastoquinone Synthesis Inhibition by Tetrabromo biphenyldiol as a Widespread Algicidal Mechanism of Marine Bacteria. ISME J. 2023, 17, 1979–1992. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, X.; Tang, W.; Zhou, K.; Wang, Z.; Shi, X. Decoding the Influence of Bacterial Community Structure and Algicidal Bacteria in a Karenia longicanalis Bloom Event. Front. Mar. Sci. 2023, 10, 1242319. [Google Scholar] [CrossRef]
- Zheng, L.; Lin, H.; Balaji-Prasath, B.; Su, Y.; Wang, Y.; Zheng, Y.; Yu, G. A Novel Algicidal Properties of Fermentation Products from Pseudomonas sp. Ps3 Strain on the Toxic Red Tide Dinoflagellate Species. Front. Microbiol. 2023, 14, 1146325. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.-R.; Van Le, V.; Srivastava, A.; Kang, M.; Oh, H.-M.; Ahn, C.-Y. Algicidal Activity of a Novel Bacterium, Qipengyuania sp. 3-20A1M, against Harmful Margalefidinium polykrikoides: Effects of Its Active Compound. Mar. Pollut. Bull. 2023, 186, 114397. [Google Scholar] [CrossRef]
- Cruz-Balladares, V.; Avalos, V.; Vera-Villalobos, H.; Cameron, H.; Gonzalez, L.; Leyton, Y.; Riquelme, C. Identification of a Shewanella halifaxensis Strain with Algicidal Effects on Red Tide Dinoflagellate Prorocentrum Triestinum in Culture. Mar. Drugs 2023, 21, 501. [Google Scholar] [CrossRef]
- Shi, J.; Wang, W.; Wang, F.; Lei, S.; Shao, S.; Wang, C.; Li, G.; An, T. Efficient Inactivation of Harmful Algae Karenia mikimotoi by a Novel Algicidal Bacterium via a Rare Direct Contact Pathway: Performances and Mechanisms. Sci. Total Environ. 2023, 892, 164401. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Tong, J.; Chen, J.; Chen, M.; Wang, L.; Li, S.; Hu, Z.; Chen, H. Characterization of a Novel Algicidal Bacteria Arenibacter sp. Strain 6A1 and Its Application to Eliminate Harmful Algal Blooms. Front. Mar. Sci. 2024, 10, 1287998. [Google Scholar] [CrossRef]
- Lee, T.C.; Lam, W.; Tam, N.F.; Xu, S.J.; Chung, W.L.; Lee, F.W. Revealing the Algicidal Characteristics of Maribacter dokdonensis: An Investigation into Bacterial Strain P4 Isolated from Karenia mikimotoi Bloom Water. J. Phycol. 2024, 60, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Orefice, I.; Balzano, S.; Romano, G.; Sardo, A. Amphidinium spp. as a Source of Antimicrobial, Antifungal, and Anticancer Compounds. Life 2023, 13, 2164. [Google Scholar] [CrossRef]
- Kubota, T.; Iwai, T.; Sakai, K.; Gonoi, T.; Kobayashi, J. Amphidinins C–F, Amphidinolide Q Analogues from Marine Dinoflagellate Amphidinium sp. Org. Lett. 2014, 16, 5624–5627. [Google Scholar] [CrossRef] [PubMed]
- Barone, M.E.; Murphy, E.; Parkes, R.; Fleming, G.T.A.; Campanile, F.; Thomas, O.P.; Touzet, N. Antibacterial Activity and Amphidinol Profiling of the Marine Dinoflagellate Amphidinium carterae (Subclade III). Int. J. Mol. Sci. 2021, 22, 12196. [Google Scholar] [CrossRef]
- Quijano-Scheggia, S.; Barajas-Gonzalez, M.; Lim, H.C.; Leaw, C.P.; Olivos-Ortiz, A.; Gaviño-Rodriguez, J.; Pérez, J.B.; Bates, S.S. The Inhibitory Effect of a Non-Yessotoxin-Producing Dinoflagellate, Lingulodinium polyedrum (Stein) Dodge, towards Vibrio vulnificus and Staphylococcus aureus. Rev. Biol. Trop. 2016, 64, 805–816. [Google Scholar] [CrossRef]
- Trick, C.G.; Andersen, R.J.; Harrison, P.J. Environmental Factors Influencing the Production of an Antibacterial Metabolite from a Marine Dinoflagellate, Prorocentrum minimum. Can. J. Fish. Aquat. Sci. 1984, 41, 423–432. [Google Scholar] [CrossRef]
- Kubota, T.; Takahashi, A.; Tsuda, M.; Kobayashi, J. Luteophanol D, New Polyhydroxyl Metabolite from Marine Dinoflagellate Amphidinium sp. Mar. Drugs 2005, 3, 113–118. [Google Scholar] [CrossRef]
- Iwai, T.; Kubota, T.; Kobayashi, J. Absolute Configuration of Amphidinin A. J. Nat. Prod. 2014, 77, 1541–1544. [Google Scholar] [CrossRef]
- Wencheng, L.; Cho, K.; Yamasaki, Y.; Takeshita, S.; Hwang, K.; Kim, D.; Oda, T. Photo-Induced Antibacterial Activity of a Porphyrin Derivative Isolated from the Harmful Dinoflagellate Heterocapsa circularisquama. Aquat. Toxicol. 2018, 201, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.; Heo, J.; Han, J.; Hong, H.D.; Jeon, H.; Hwang, H.-J.; Hong, C.-Y.; Kim, D.; Han, J.W.; Baek, K. Industrial Applications of Dinoflagellate Phycotoxins Based on Their Modes of Action: A Review. Toxins 2020, 12, 805. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.-Z. Neurotoxins from Marine Dinoflagellates: A Brief Review. Mar. Drugs 2008, 6, 349–371. [Google Scholar] [CrossRef] [PubMed]
- Usami, M.; Satake, M.; Ishida, S.; Inoue, A.; Kan, Y.; Yasumoto, T. Palytoxin Analogs from the Dinoflagellate Ostreopsis siamensis. J. Am. Chem. Soc. 1995, 117, 5389–5390. [Google Scholar] [CrossRef]
- Pitcher, G.C.; Foord, C.J.; Macey, B.M.; Mansfield, L.; Mouton, A.; Smith, M.E.; Osmond, S.J.; van der Molen, L. Devastating Farmed Abalone Mortalities Attributed to Yessotoxin-Producing Dinoflagellates. Harmful Algae 2019, 81, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Ogino, H.; Kumagai, M.; Yasumoto, T. Toxicologic Evaluation of Yessotoxin. Nat. Toxins 1997, 5, 255–259. [Google Scholar] [CrossRef]
- Orr, R.; Stüken, A.; Murray, S.; Jakobsen, K. Evolution and Distribution of Saxitoxin Biosynthesis in Dinoflagellates. Mar. Drugs 2013, 11, 2814–2828. [Google Scholar] [CrossRef] [PubMed]
- Kodama, M.; Ogata, T.; Sato, S. Bacterial Production of Saxitoxin. Agric. Biol. Chem. 1988, 52, 1075–1077. [Google Scholar] [CrossRef]
- Doucette, G.; Trick, C. Characterization of Bacteria Associated with Different Isolates of Alexundrium Tumarense. In Harmful Algal Blooms Technique and Documentation; Lassus, P., Arzul, G., Erard, E., Gentien, P., Marcaillou, C., Eds.; Lavoisier: Paris, Frances, 1995; pp. 33–38. [Google Scholar]
- Green, D.H.; Llewellyn, L.E.; Negri, A.P.; Blackburn, S.I.; Bolch, C.J.S. Phylogenetic and Functional Diversity of the Cultivable Bacterial Community Associated with the Paralytic Shellfish Poisoning Dinoflagellate Gymnodinium catenatum. FEMS Microbiol. Ecol. 2004, 47, 345–357. [Google Scholar] [CrossRef]
- Martins, C.A.; Alvito, P.; Tavares, M.J.; Pereira, P.; Doucette, G.; Franca, S. Reevaluation of Production of Paralytic Shellfish Toxin by Bacteria Associated with Dinoflagellates of the Portuguese Coast. Appl. Environ. Microbiol. 2003, 69, 5693–5698. [Google Scholar] [CrossRef]
- Stüken, A.; Orr, R.J.S.; Kellmann, R.; Murray, S.A.; Neilan, B.A.; Jakobsen, K.S. Discovery of Nuclear-Encoded Genes for the Neurotoxin Saxitoxin in Dinoflagellates. PLoS ONE 2011, 6, e20096. [Google Scholar] [CrossRef] [PubMed]
- Baker, T.R.; Doucette, G.J.; Powell, C.L.; Boyer, G.L.; Plumley, F.G. GTX4 Imposters: Characterization of Fluorescent Compounds Synthesized by Pseudomonas Stutzeri SF/PS and Pseudomonas/Alteromonas PTB-1, Symbionts of Saxitoxin-Producing Alexandrium spp. Toxicon 2003, 41, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Uribe, P.; Espejo, R.T. Effect of Associated Bacteria on the Growth and Toxicity of Alexandrium catenella. Appl. Environ. Microbiol. 2003, 69, 659–662. [Google Scholar] [CrossRef]
- Wang, B.; Yao, M.; Zhou, J.; Tan, S.; Jin, H.; Zhang, F.; Mak, Y.; Wu, J.; Lai Chan, L.; Cai, Z. Growth and Toxin Production of Gambierdiscus spp. Can Be Regulated by Quorum-Sensing Bacteria. Toxins 2018, 10, 257. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Tomasch, J.; Michael, V.; Bhuju, S.; Jarek, M.; Petersen, J.; Wagner-Döbler, I. Identification of Genetic Modules Mediating the Jekyll and Hyde Interaction of Dinoroseobacter shibae with the Dinoflagellate Prorocentrum minimum. Front. Microbiol. 2015, 6, 1262. [Google Scholar] [CrossRef] [PubMed]
- Saad, O.S.; Lin, X.; Ng, T.Y.; Li, L.; Ang, P.; Lin, S. Genome Size, RDNA Copy, and QPCR Assays for Symbiodiniaceae. Front. Microbiol. 2020, 11, 847. [Google Scholar] [CrossRef] [PubMed]
- Veldhuis, M.J.W.; Cucci, T.L.; Sieracki, M.E. Cellular DNA Content of Marine Phytoplankton Using Two New Fluorochromes: Taxonomic and Ecological Implications. J. Phycol. 1997, 33, 527–541. [Google Scholar] [CrossRef]
- LaJeunesse, T.C.; Lambert, G.; Andersen, R.A.; Coffroth, M.A.; Galbraith, D.W. Symbiodinium (Pyrrhophyta) Genome Sizes (DNA Content) Are Smallest among Dinoflagellates. J. Phycol. 2005, 41, 880–886. [Google Scholar] [CrossRef]
- Irwin, N.A.T.; Martin, B.J.E.; Young, B.P.; Browne, M.J.G.; Flaus, A.; Loewen, C.J.R.; Keeling, P.J.; Howe, L.J. Viral Proteins as a Potential Driver of Histone Depletion in Dinoflagellates. Nat. Commun. 2018, 9, 1535. [Google Scholar] [CrossRef]
- Minge, M.A.; Shalchian-Tabrizi, K.; Tørresen, O.K.; Takishita, K.; Probert, I.; Inagaki, Y.; Klaveness, D.; Jakobsen, K.S. A Phylogenetic Mosaic Plastid Proteome and Unusual Plastid-Targeting Signals in the Green-Colored Dinoflagellate Lepidodinium chlorophorum. BMC Evol. Biol. 2010, 10, 191. [Google Scholar] [CrossRef]
- Waller, R.F.; Slamovits, C.H.; Keeling, P.J. Lateral Gene Transfer of a Multigene Region from Cyanobacteria to Dinoflagellates Resulting in a Novel Plastid-Targeted Fusion Protein. Mol. Biol. Evol. 2006, 23, 1437–1443. [Google Scholar] [CrossRef] [PubMed]
- Keeling, P.J.; Palmer, J.D. Horizontal Gene Transfer in Eukaryotic Evolution. Nat. Rev. Genet. 2008, 9, 605–618. [Google Scholar] [CrossRef] [PubMed]
- Castelli, M.; Sabaneyeva, E.; Lanzoni, O.; Lebedeva, N.; Floriano, A.M.; Gaiarsa, S.; Benken, K.; Modeo, L.; Bandi, C.; Potekhin, A.; et al. Deianiraea, an Extracellular Bacterium Associated with the Ciliate Paramecium, Suggests an Alternative Scenario for the Evolution of Rickettsiales. ISME J. 2019, 13, 2280–2294. [Google Scholar] [CrossRef]
- You, X.; Xu, N.; Yang, X.; Sun, W. Pollutants Affect Algae-Bacteria Interactions: A Critical Review. Environ. Pollut. 2021, 276, 116723. [Google Scholar] [CrossRef]
- Jia, Q.; Chen, X.; Köllner, T.G.; Rinkel, J.; Fu, J.; Labbé, J.; Xiong, W.; Dickschat, J.S.; Gershenzon, J.; Chen, F. Terpene Synthase Genes Originated from Bacteria through Horizontal Gene Transfer Contribute to Terpenoid Diversity in Fungi. Sci. Rep. 2019, 9, 9223. [Google Scholar] [CrossRef]
- Oberstaller, J.; Otto, T.D.; Rayner, J.C.; Adams, J.H. Essential Genes of the Parasitic Apicomplexa. Trends Parasitol. 2021, 37, 304–316. [Google Scholar] [CrossRef]
- Li, Z.; Wang, X.; Zhang, Y.; Yu, Z.; Zhang, T.; Dai, X.; Pan, X.; Jing, R.; Yan, Y.; Liu, Y.; et al. Genomic Insights into the Phylogeny and Biomass-Degrading Enzymes of Rumen Ciliates. ISME J. 2022, 16, 2775–2787. [Google Scholar] [CrossRef] [PubMed]
- Whitney, S.M.; Shaw, D.C.; Yellowlees, D. Evidence That Some Dinoflagellates Contain a Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase Related to That of the α-Proteobacteria. Proc. R. Soc. Lond. B Biol. Sci. 1995, 259, 271–275. [Google Scholar] [CrossRef]
- Tabita, F.R.; Satagopan, S.; Hanson, T.E.; Kreel, N.E.; Scott, S.S. Distinct Form I, II, III, and IV Rubisco Proteins from the Three Kingdoms of Life Provide Clues about Rubisco Evolution and Structure/Function Relationships. J. Exp. Bot. 2007, 59, 1515–1524. [Google Scholar] [CrossRef]
- Rowan, R.; Whitney, S.M.; Fowler, A.; Yellowlees, D. Rubisco in Marine Symbiotic Dinoflagellates: Form II Enzymes in Eukaryotic Oxygenic Phototrophs Encoded by a Nuclear Multigene Family. Plant Cell 1996, 8, 539–553. [Google Scholar] [CrossRef]
- Béjà, O.; Spudich, E.N.; Spudich, J.L.; Leclerc, M.; DeLong, E.F. Proteorhodopsin Phototrophy in the Ocean. Nature 2001, 411, 786–789. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Li, L.; Guo, C.; Lin, X.; Li, M.; Lin, S. Rhodopsin Gene Expression Regulated by the Light Dark Cycle, Light Spectrum and Light Intensity in the Dinoflagellate Prorocentrum. Front. Microbiol. 2015, 6, 555. [Google Scholar] [CrossRef] [PubMed]
- Cooney, E.C.; Holt, C.C.; Hehenberger, E.; Adams, J.A.; Leander, B.S.; Keeling, P.J. Investigation of Heterotrophs Reveals New Insights in Dinoflagellate Evolution. Mol. Phylogenet. Evol. 2024, 196, 108086. [Google Scholar] [CrossRef] [PubMed]
- Slamovits, C.H.; Okamoto, N.; Burri, L.; James, E.R.; Keeling, P.J. A Bacterial Proteorhodopsin Proton Pump in Marine Eukaryotes. Nat. Commun. 2011, 2, 183. [Google Scholar] [CrossRef] [PubMed]
- Bachvaroff, T.; Williams, E.; Jagus, R.; Place, A. A Non-Cryptic Non-Canonical Multi-Module NRPS/PKS Found in Dinoflagellates. In Proceedings of the 16th International Conference on Harmful Algae, Wellington, New Zealand, 26–31 October 2014; MacKenzie, A., Ed.; New Zealand and the International Society for the Study of Harmful Algae: Nelson, BC, Canada, 2014; pp. 101–104. [Google Scholar]
- Nosenko, T.; Bhattacharya, D. Horizontal Gene Transfer in Chromalveolates. BMC Evol. Biol. 2007, 7, 173. [Google Scholar] [CrossRef] [PubMed]
- Moszczyński, K.; Mackiewicz, P.; Bodył, A. Evidence for Horizontal Gene Transfer from Bacteroidetes Bacteria to Dinoflagellate Minicircles. Mol. Biol. Evol. 2012, 29, 887–892. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Park, B.S.; Lim, W.-A.; Ki, J.-S. CpMCA, a Novel Metacaspase Gene from the Harmful Dinoflagellate Cochlodinium polykrikoides and Its Expression during Cell Death. Gene 2018, 651, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Leggat, W.; Hoegh-Guldberg, O.; Dove, S.; Yellowlees, D. Analysis of an EST Library from the Dinoflagellate (Symbiodinium sp.) Symbiont of Reef-building Corals. J. Phycol. 2007, 43, 1010–1021. [Google Scholar] [CrossRef]
- George, E.E.; Tashyreva, D.; Kwong, W.K.; Okamoto, N.; Horák, A.; Husnik, F.; Lukeš, J.; Keeling, P.J. Gene Transfer Agents in Bacterial Endosymbionts of Microbial Eukaryotes. Genome Biol. Evol. 2022, 14, evac099. [Google Scholar] [CrossRef]
- Ponce-Toledo, R.I.; López-García, P.; Moreira, D. Horizontal and Endosymbiotic Gene Transfer in Early Plastid Evolution. New Phytol. 2019, 224, 618–624. [Google Scholar] [CrossRef]
- Archibald, J.M. Endosymbiosis and Eukaryotic Cell Evolution. Curr. Biol. 2015, 25, R911–R921. [Google Scholar] [CrossRef] [PubMed]
- Mackiewicz, P.; Bodył, A.; Moszczyński, K. The Case of Horizontal Gene Transfer from Bacteria to the Peculiar Dinoflagellate Plastid Genome. Mob. Genet. Elem. 2013, 3, e25845. [Google Scholar] [CrossRef] [PubMed]
- Ford Doolittle, W. You Are What You Eat: A Gene Transfer Ratchet Could Account for Bacterial Genes in Eukaryotic Nuclear Genomes. Trends Genet. 1998, 14, 307–311. [Google Scholar] [CrossRef] [PubMed]
- Vesteg, M.; Hadariová, L.; Horváth, A.; Estraño, C.E.; Schwartzbach, S.D.; Krajčovič, J. Comparative Molecular Cell Biology of Phototrophic Euglenids and Parasitic Trypanosomatids Sheds Light on the Ancestor of Euglenozoa. Biol. Rev. 2019, 94, 1701–1721. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, S.; Suzaki, T.; Weber, A.P.; Archibald, J.M.; Nozaki, H. Eukaryote-to-Eukaryote Gene Transfer Gives Rise to Genome Mosaicism in Euglenids. BMC Evol. Biol. 2011, 11, 105. [Google Scholar] [CrossRef] [PubMed]
- Eichinger, L.; Pachebat, J.A.; Glöckner, G.; Rajandream, M.-A.; Sucgang, R.; Berriman, M.; Song, J.; Olsen, R.; Szafranski, K.; Xu, Q.; et al. The Genome of the Social Amoeba Dictyostelium discoideum. Nature 2005, 435, 43–57. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P. Role and Significance of Biofilm-Forming Microbes in Phytoremediation—A Review. Environ. Technol. Innov. 2022, 25, 102182. [Google Scholar] [CrossRef]
- Tong, C.Y.; Derek, C.J.C. The Role of Substrates towards Marine Diatom Cylindrotheca fusiformis Adhesion and Biofilm Development. J. Appl. Phycol. 2021, 33, 2845–2862. [Google Scholar] [CrossRef]
- Spengler, C.; Maikranz, E.; Santen, L.; Jacobs, K. Modeling Bacterial Adhesion to Unconditioned Abiotic Surfaces. Front. Mech. Eng. 2021, 7, 661370. [Google Scholar] [CrossRef]
- Herrling, M.P.; Lackner, S.; Nirschl, H.; Horn, H.; Guthausen, G. Recent NMR/MRI Studies of Biofilm Structures and Dynamics. Annu. Rep. NMR Spectrosc. 2019, 97, 163–213. [Google Scholar]
- Vestby, L.K.; Grønseth, T.; Simm, R.; Nesse, L.L. Bacterial Biofilm and Its Role in the Pathogenesis of Disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef]
- Niazi, P.; Monib, A.W.; Ozturk, H.; Mansoor, M.; Azizi, A.; Hassand, M.H. Review on Surface Elements and Bacterial Biofilms in Plant-Bacterial Associations. J. Res. Appl. Sci. Biotechnol. 2023, 2, 204–214. [Google Scholar] [CrossRef]
- Oruganti, R.K.; Katam, K.; Show, P.L.; Gadhamshetty, V.; Upadhyayula, V.K.K.; Bhattacharyya, D. A Comprehensive Review on the Use of Algal-Bacterial Systems for Wastewater Treatment with Emphasis on Nutrient and Micropollutant Removal. Bioengineered 2022, 13, 10412–10453. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Haga, M.; Imai, I.; Sakai, R.; Fujita, M.J. Function of the Algicidal Bacterium Pseudomonas sp. Go58 Isolated from the Biofilm on a Water Plant, and Its Active Compounds, Pyoluteorins. Sci. Total Environ. 2023, 872, 162088. [Google Scholar] [CrossRef] [PubMed]
- Frommlet, J.C.; Sousa, M.L.; Alves, A.; Vieira, S.I.; Suggett, D.J.; Serôdio, J. Coral Symbiotic Algae Calcify Ex Hospite in Partnership with Bacteria. Proc. Natl. Acad. Sci. USA 2015, 112, 6158–6163. [Google Scholar] [CrossRef]
- Durán, P.; Ellis, T.J.; Thiergart, T.; Ågren, J.; Hacquard, S. Climate Drives Rhizosphere Microbiome Variation and Divergent Selection between Geographically Distant Arabidopsis Populations. New Phytol. 2022, 236, 608–621. [Google Scholar] [CrossRef]
| Gene | Enzyme | Dinoflagellate Source | Localization | Bacterial Origin | Bacteria Phylum | Reference |
|---|---|---|---|---|---|---|
| aceE | Pyruvate dehydrogenase | Dinoflagellates | Genome | - | Actinobacteria | [15] |
| aroB | 3-dehydroquinate synthase | Oxyrrhis | Genome | - | Cyanobacteria | [142] |
| OMT | O-methyltransferase | Oxyrrhis | Genome | - | Cyanobacteria | [142] |
| aslA | Arylsulfatase A | Karenia (Ka) brevis (C.C.Davis) Gert Hansen & Moestrup, 2000 | Genome | - | - | [157] |
| ATS1 | Alpha-tubulin suppressor | Ka. brevis | Genome | - | - | [158] |
| avtA | Valine:pyruvate aminotransferase | Dinoflagellates | Genome | - | Actinobacteria | [15] |
| CAS-like | Clavaminic acid synthetase-like protein | Ka. brevis | Genome | - | Cyanobacteria | [157] |
| citE | Citrate lyase beta subunit | Dinoflagellates | Genome | - | Proteobacteria | [15] |
| Epimerase | NAD dependent epimerase/dehydratase | Ka. brevis | Genome | - | - | [157] |
| Fe-ADH | Iron-containing alcohol dehydrogenase | Ka. brevis | Genome | - | - | [157] |
| Form II Rubisco | Form II ribulose-1,5-bisphosphate carboxylase-oxygenase | Peridinin-containing dinoflagellates | Genome | - | Proteobacteria | [149] |
| grpE | Protein GrpE | Dinoflagellates | Genome | - | - | [15] |
| HLP | Histone-like protein | A. tamarense | Genome | - | - | [149] |
| ligI | Metal-dependent hydrolase, TIM-barrel fold | Ka. brevis | Genome | - | - | [157] |
| MQO | Monomeric NADP(+)-dependent isocitrate dehydrogenase | Ka. brevis | Genome | - | - | [158] |
| MVIM | MVIM-sugar aminotransferase | Ka. brevis | Genome | - | Proteobacteria | [157] |
| pbpB | Substrate-bound, membrane-associated, periplasmic binding protein | Ka. brevis | Genome | - | - | [157] |
| pdxA | Pyridoxal phosphate biosynthetic protein | Ka. brevis | Genome | - | - | [157] |
| ptdss | Phosphatidylserine synthase | Dinoflagellates | Genome | - | Proteobacteria | [15] |
| putA | NAD-dependent aldehyde dehydrogenases | Ka. brevis | Genome | - | - | [157] |
| RHO | Rhodopsin synthesis | Oxyrrhis (O) marina Dujardin, 1841 | Genome | - | - | [155] |
| rlmF | SAM-dependent methyltransferase | Ka. brevis | Genome | - | - | [157] |
| rpl28 | 60S ribosomal protein L28 | Pyrocystis (Py) lunula (Schütt) Schütt, 1896 | Plastid genome | Cytophaga | Bacteroidetes | [158] |
| rpl33 | Large ribosomal subunit protein bL33c | Py. lunula | Plastid genome | Cytophaga | Bacteroidetes | [158] |
| SIR2 | Silent information regulator 2 | Ka. brevis | Genome | - | Proteobacteria | [157] |
| SRP54 N domain | The signal recognition particle 54-kDa subunit | Pyrocystis | Plastid genome | - | Bacteroidetes | [158] |
| sxtA | 8-amino-7-oxononanoate synthase | Alexandrium and Pyrodinium | Genome | - | - | [128] |
| sxtG | Glycine amidinotransferase | Alexandrium species and Gy. catenatum | Genome | - | [127] | |
| WECE | Pyridoxal phosphate dependent aminotransferase | Ka. brevis | Genome | - | Proteobacteria | [157] |
| yaaA | DNA-binding and peroxide stress resistance | Ka. brevis | Genome | - | - | [157] |
| ycf16 | Probable ATP-dependent transporter ycf16 | Ceratium (Ce) horridum (Cleve) Gran, 1902 | Plastid genome | Algoriphagus | Bacteroidetes | [157] |
| ycf24 | Iron-sulfur cluster assembly SufBD family protein ycf24 | Ce. horridum | Plastid genome | Algoriphagus | Bacteroidetes | [158] |
| MCA | Metacaspase | C. polykrikoides | Genome | - | - | [159] |
| PKS/NRPS | Non-ribosomal peptide synthases/polyketide synthases | O. marina and core dinoflagellates | Genome | Burkholderiales | Pseudomonadota | [156] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, X.; Liu, Z.; Zhang, Y.; Shi, X.; Wu, Z. Dinoflagellate–Bacteria Interactions: Physiology, Ecology, and Evolution. Biology 2024, 13, 579. https://doi.org/10.3390/biology13080579
Yang X, Liu Z, Zhang Y, Shi X, Wu Z. Dinoflagellate–Bacteria Interactions: Physiology, Ecology, and Evolution. Biology. 2024; 13(8):579. https://doi.org/10.3390/biology13080579
Chicago/Turabian StyleYang, Xiaohong, Zijian Liu, Yanwen Zhang, Xinguo Shi, and Zhen Wu. 2024. "Dinoflagellate–Bacteria Interactions: Physiology, Ecology, and Evolution" Biology 13, no. 8: 579. https://doi.org/10.3390/biology13080579
APA StyleYang, X., Liu, Z., Zhang, Y., Shi, X., & Wu, Z. (2024). Dinoflagellate–Bacteria Interactions: Physiology, Ecology, and Evolution. Biology, 13(8), 579. https://doi.org/10.3390/biology13080579







