ZmDST44 Gene Is a Positive Regulator in Plant Drought Stress Tolerance
Abstract
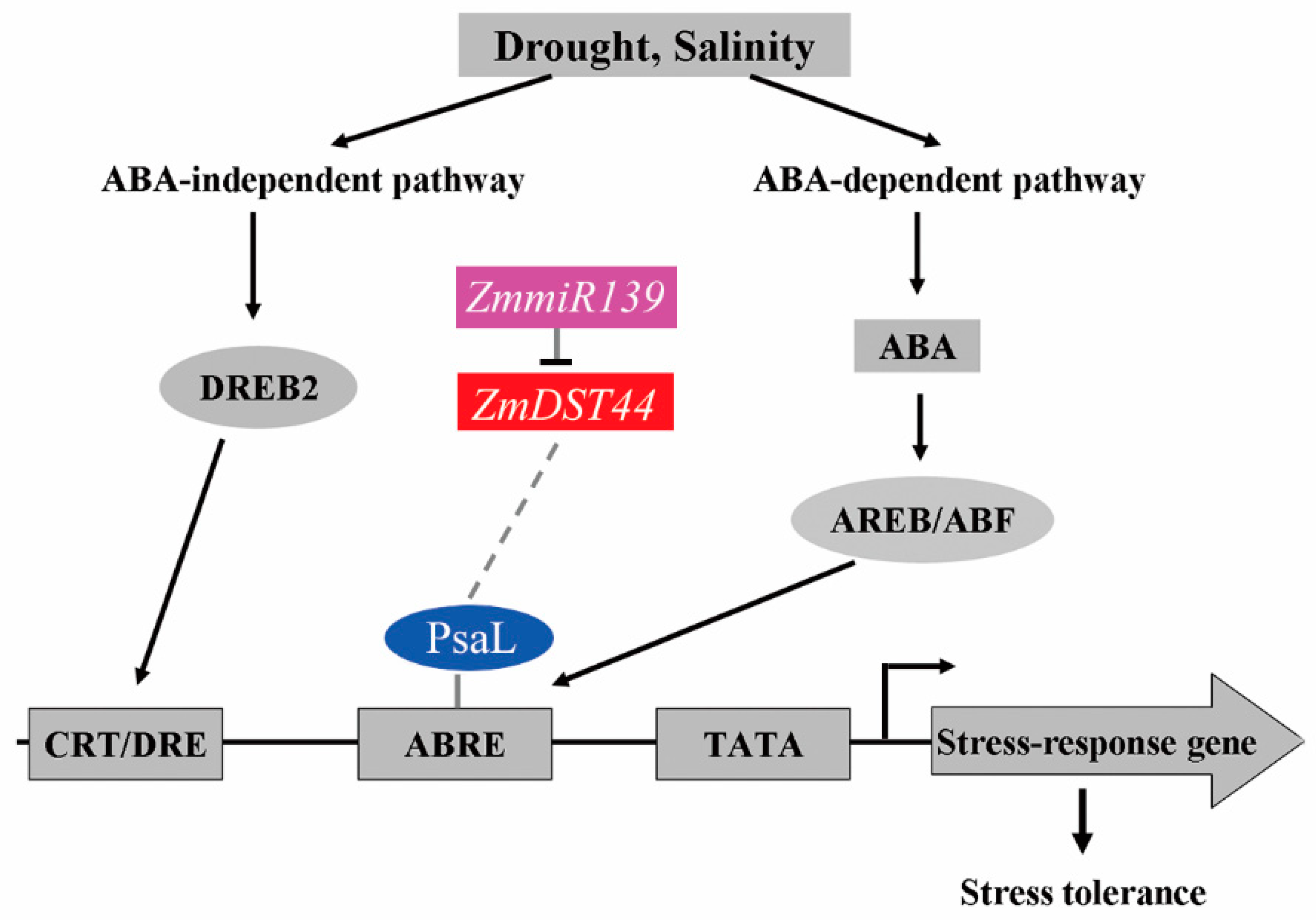
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. 5′ RACE Identification of the miRNA Target Cleavage Site
2.3. Construction of Overexpression Vectors
2.4. Transformation of Rice and Maize
2.5. Germination and Root Length Assays
2.6. Drought Stress Treatments on Transgenic Plants
2.7. RNA Extraction, Quantitative RT-PCR
2.8. Determination of Malondialdehyde (MDA) Content
2.9. Relative Electrolyte Leakage Estimation
2.10. Determination of Proline Content
2.11. DAB Staining
2.12. Statistical Analysis
3. Results
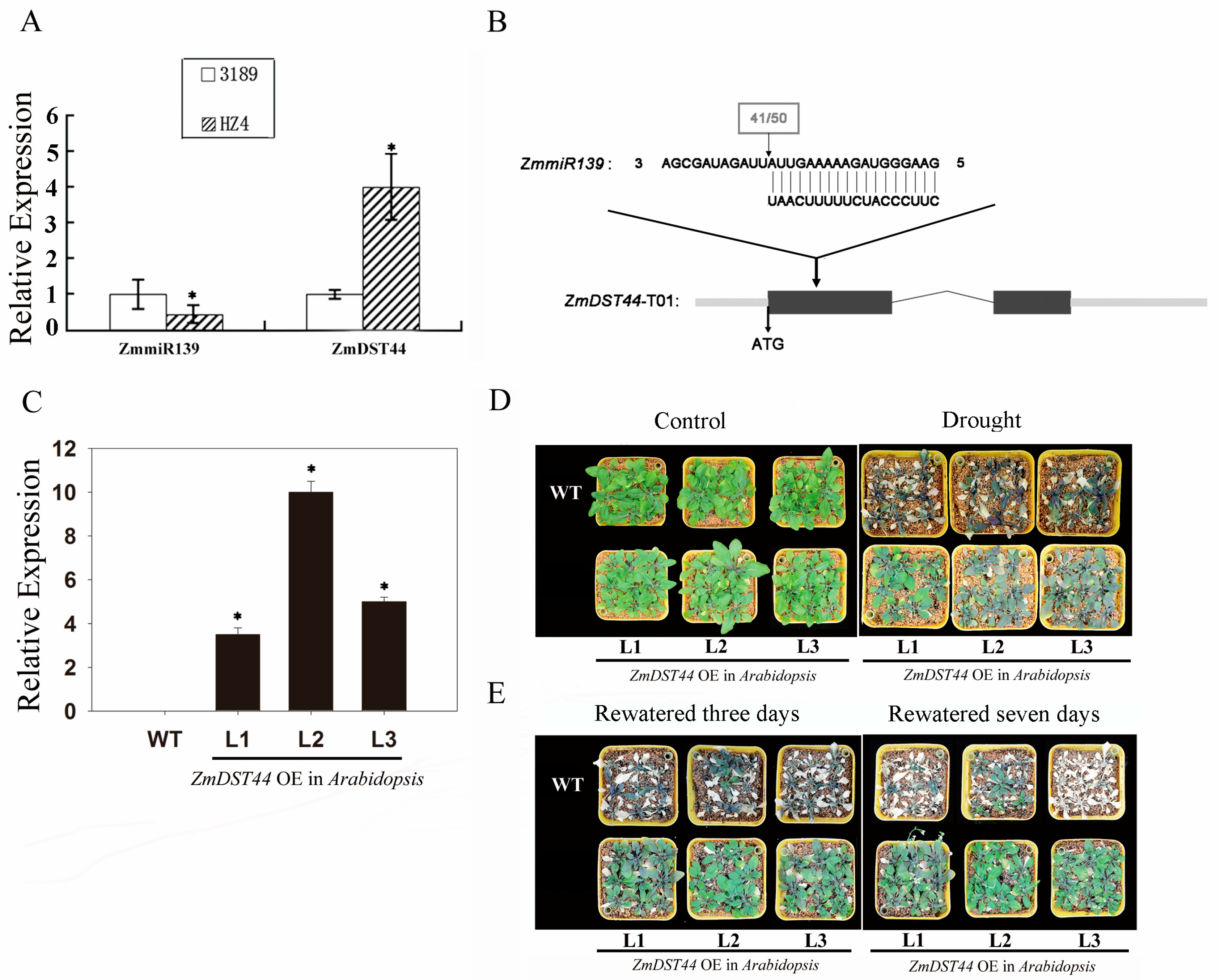
3.1. ZmDST44 mRNA Is Cleaved by ZmmiR139
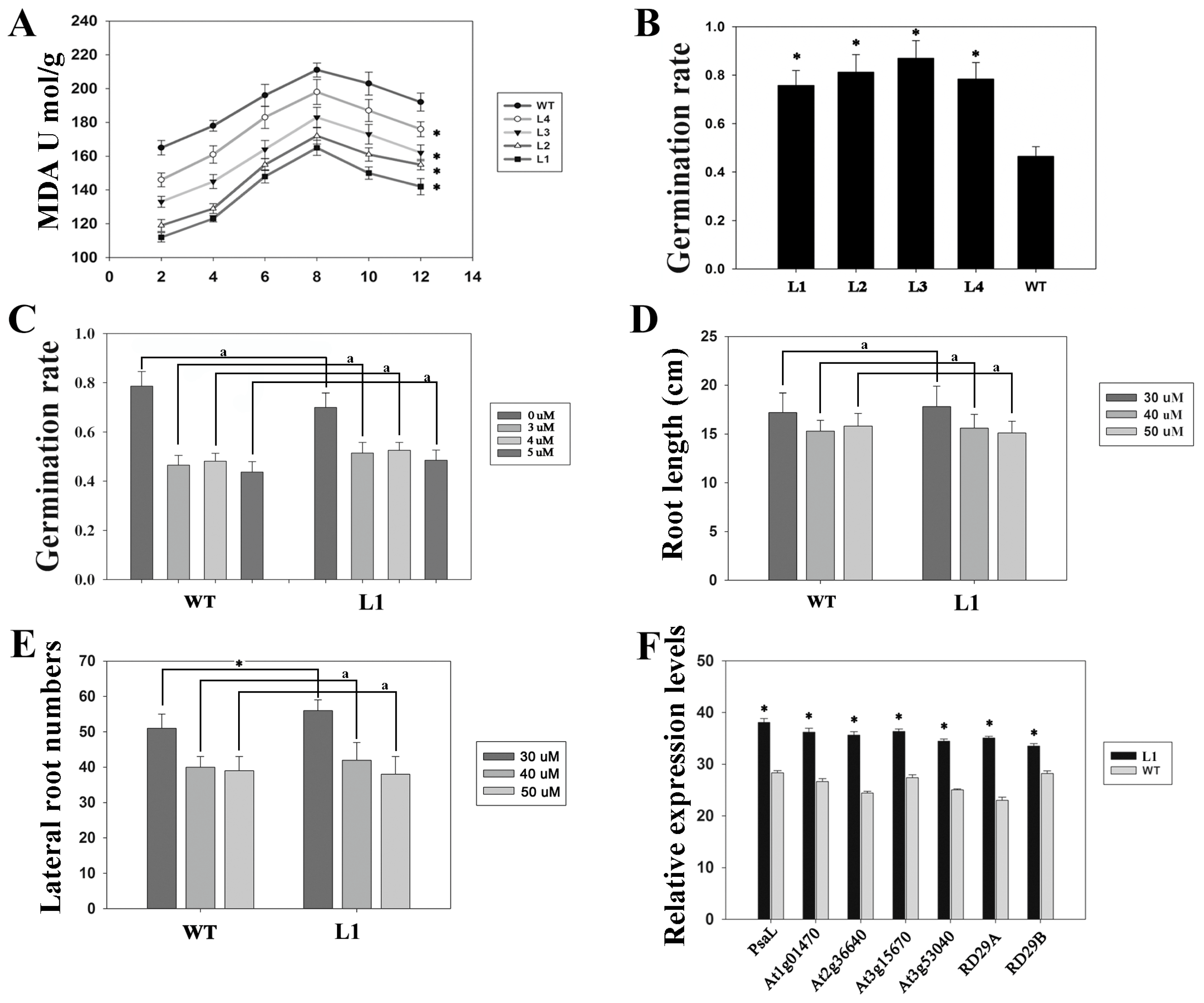
3.2. Overexpression of ZmDST44 Confers Drought Tolerance in Arabidopsis
3.3. Drought-Related Genes Are Highly Expressed in Plants Overexpressing of ZmDST44
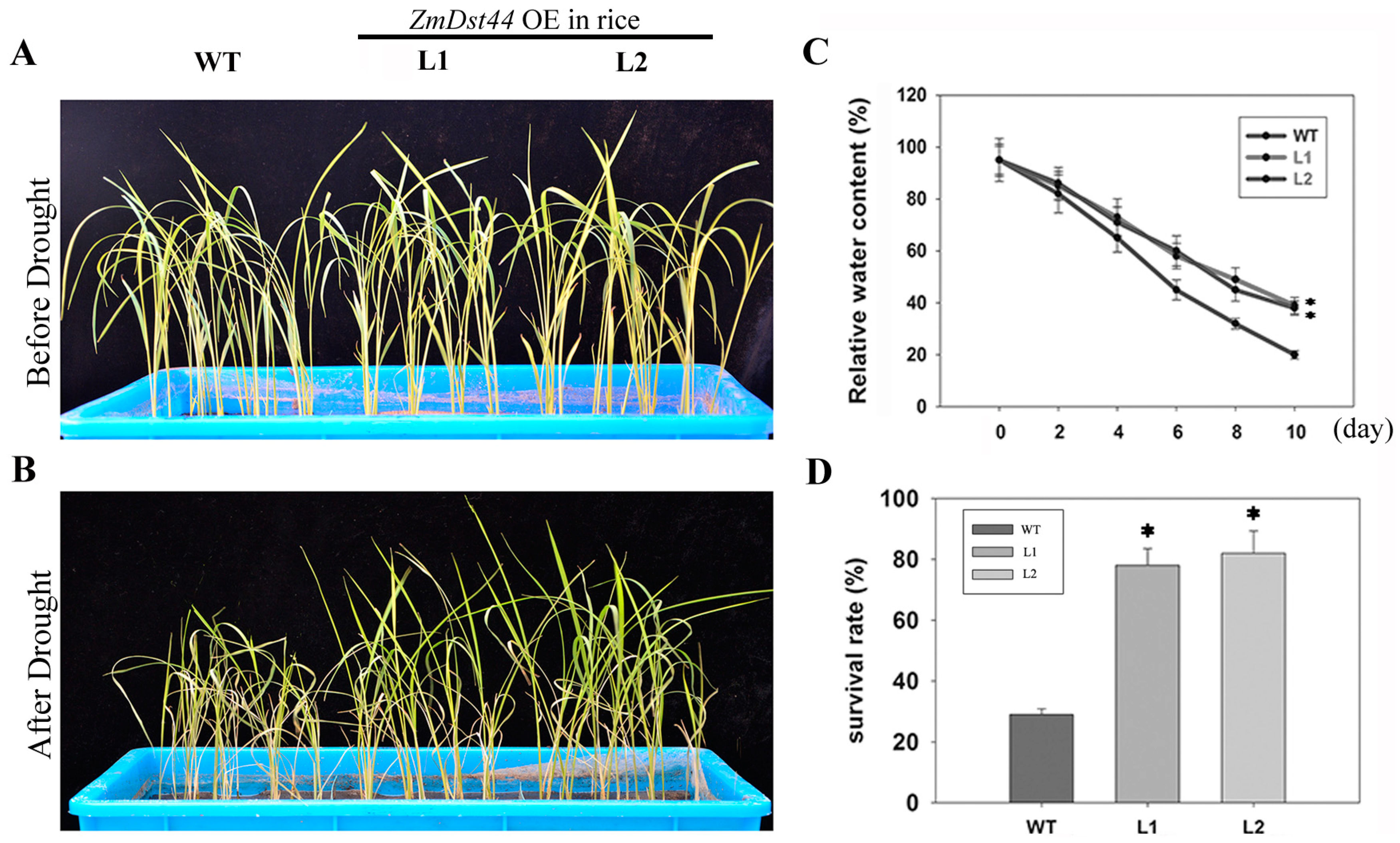
3.4. ZmDST44 Transgenic Rice Exhibits an Enhanced Tolerance to Drought
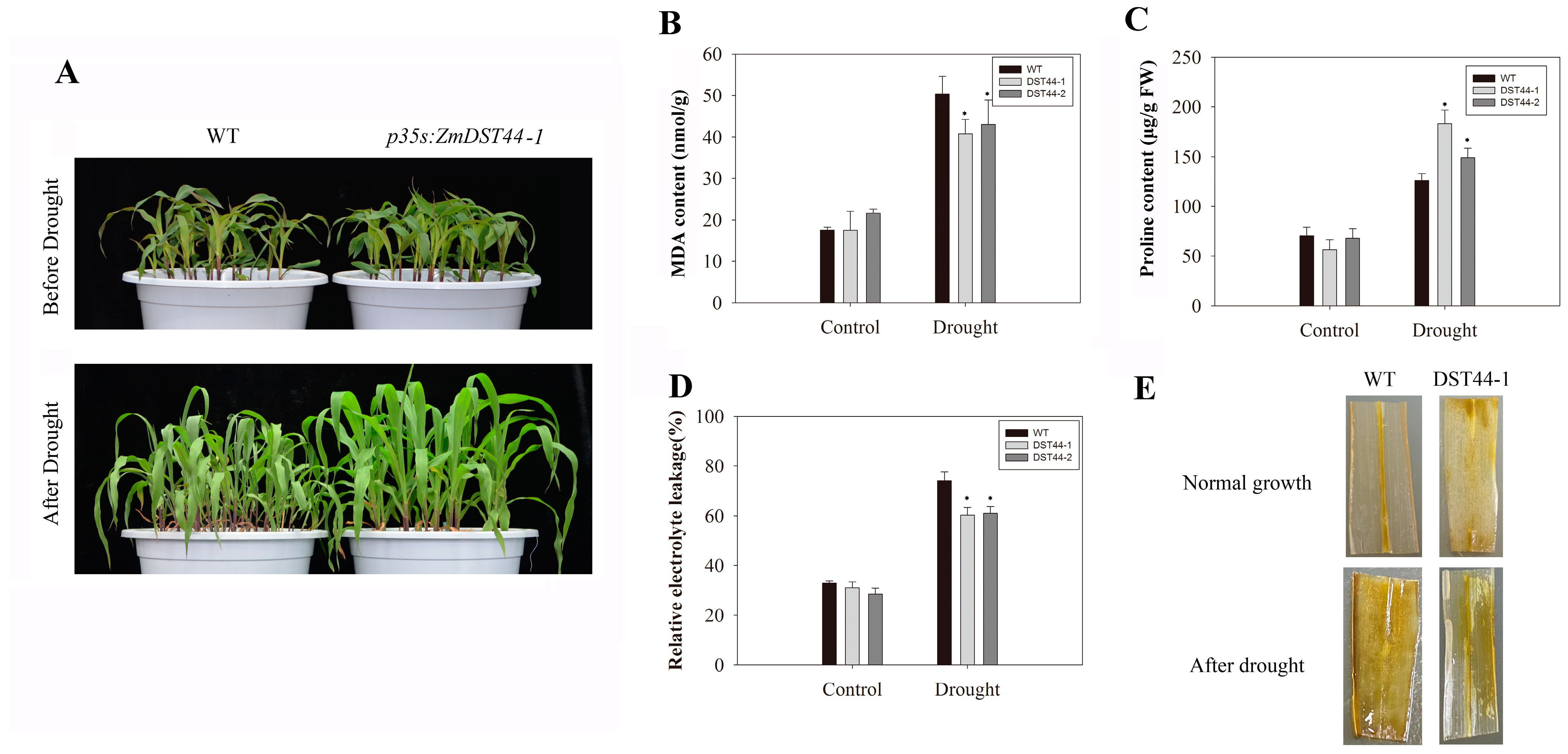
3.5. Overexpression of ZmDST44 Enhanced Maize Tolerance to Drought Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Edmeades, G.O.; Bolaños, J.; Hernàndez, M.; Bello, S. Causes for Silk Delay in a Lowland Tropical Maize Population. Crop Sci. 1993, 33, 1029–1035. [Google Scholar] [CrossRef]
- Farah, S.M. An examination of the effects of water stress on leaf growth of crops of field beans (Vicia faba L.). 2. Mineral content. J. Agric. Sci. 1981, 96, 337–346. [Google Scholar] [CrossRef]
- Zlatev, Z.; Lidon, F.C. An overview on drought induced changes in plant growth, water relations and photosynthesis. Emir. J. Food Agric. 2012, 24, 520–524. [Google Scholar] [CrossRef]
- Ellis, R.J. Most abundant protein in the world. Trends Biochem. Sci. 1979, 4, 241–244. [Google Scholar] [CrossRef]
- Prins, A.; Orr, D.J.; Andralojc, P.J.; Reynolds, M.P.; Carmo-Silva, E.; Parry, M.A.J. Rubisco catalytic properties of wild and domesticated relatives provide scope for improving wheat photosynthesis. J. Exp. Bot. 2016, 67, 1827–1838. [Google Scholar] [CrossRef] [PubMed]
- Madgwick, P.J.; Colliver, S.P.; Banks, F.M.; Habash, D.Z.; Dulieu, H.; Parry, M.A.J.; Paul, M.J. Genetic Manipulation of Rubisco: Chromatium vinosum rbcL is expressed in Nicotiana tabacum but does not form a functional protein. Ann. Appl. Biol. 2015, 140, 13–19. [Google Scholar] [CrossRef]
- Ford, D.M.; Shibles, R. Photosynthesis and Other Traits in Relation to Chloroplast Number during Soybean Leaf Senescence. Plant Physiol. 1988, 86, 108–111. [Google Scholar] [CrossRef]
- Crafts-Brandner, S.J. Phosphorus nutrition influence on leaf senescence in soybean. Plant Physiol. 1992, 98, 1128–1132. [Google Scholar] [CrossRef] [PubMed]
- Taylor, T.C.; Andersson, I. Structural transitions during activation and ligand binding in hexadecameric Rubisco inferred from the crystal structure of the activated unliganded spinach enzyme. Nat. Struct. Biol. 1996, 3, 95–101. [Google Scholar] [CrossRef]
- Maliga, K.P. Relocation of the plastid rbcL gene to the nucleus yields functional ribulose-1,5-bisphosphate carboxylase in tobacco chloroplasts. Proc. Natl. Acad. Sci. USA 1994, 91, 1969–1973. [Google Scholar] [CrossRef]
- Ambros, V.; Bartel, B.; Bartel, D.P.; Burge, C.B.; Carrington, J.C.; Chen, X.; Dreyfuss, G.; Eddy, S.R.; Griffiths-Jones, S.; Marshall, M.; et al. A uniform system for microRNA annotation. Rna-A Publ. Rna Soc. 2003, 9, 277. [Google Scholar] [CrossRef] [PubMed]
- Humphreys, D.T.; Preiss, T. MicroRNAs Control Translation Initiation by Inhibiting Eukaryotic Initiation Factor 4E/Cap and Poly(A) Tail Function. Proc. Natl. Acad. Sci. USA 2005, 102, 16961–16966. [Google Scholar] [CrossRef]
- Jing, Q.; Huang, S.; Guth, S.; Zarubin, T.; Motoyama, A.; Chen, J.; Di Padova, F.; Lin, S.-C.; Gram, H.; Han, J. Involvement of MicroRNA in AU-Rich Element-Mediated mRNA Instability. Cell 2005, 120, 623. [Google Scholar] [CrossRef] [PubMed]
- Vaucheret, H. Post-transcriptional small RNA pathways in plants: Mechanisms and regulations. Genes Dev. 2006, 20, 759. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Stellwag, E.J.; Pan, X. Large-scale genome analysis reveals unique features of microRNAs. Gene 2009, 443, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.M.; Bernstein, E.; Beach, D.; Hannon, G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 2000, 404, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Hutvágner, G.; Zamore, P.D. RNAi: Nature abhors a double-strand. Curr. Opin. Genet. Dev. 2002, 12, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Schott, G.; Mari-Ordonez, A.; Himber, C.; Alioua, A.; Voinnet, O.; Dunoyer, P. Differential effects of viral silencing suppressors on siRNA and miRNA loading support the existence of two distinct cellular pools of ARGONAUTE1. Embo J. 2012, 31, 2553–2565. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, R.; Vaucheret, H.; Trejo, J.; Bartel, D.P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006, 20, 3407–3425. [Google Scholar] [CrossRef]
- Sunkar, R.; Jagadeeswaran, G. In silico identification of conserved microRNAs in large number of diverse plant species. Bmc Plant Biol. 2008, 8, 37. [Google Scholar] [CrossRef]
- Licausi, F.; Weits, D.A.; Pant, B.D.; Scheible, W.; Geigenberger, P.; van Dongen, J.T. Hypoxia responsive gene expression is mediated by various subsets of transcription factors and miRNAs that are determined by the actual oxygen availability. New Phytol. 2011, 190, 442–456. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Ding, H.; Zhu, J.-K.; Zhang, F.; Li, W.-X. Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol. 2011, 190, 906. [Google Scholar] [CrossRef]
- Voinnet, O. Origin, Biogenesis, and Activity of Plant MicroRNAs. Cell 2009, 136, 669. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19. [Google Scholar] [CrossRef]
- Opitz, N.; Marcon, C.; Paschold, A.; Malik, W.A.; Lithio, A.; Brandt, R.; Piepho, H.-P.; Nettleton, D.; Hochholdinger, F. Extensive tissue-specific transcriptomic plasticity in maize primary roots upon water deficit. J. Exp. Bot. 2015, 67, 1095. [Google Scholar] [CrossRef]
- He, Z.; Zhong, J.; Sun, X.; Wang, B.; Terzaghi, W.; Dai, M. The Maize ABA Receptors ZmPYL8, 9, and 12 Facilitate Plant Drought Resistance. Front. Plant Sci. 2018, 9, 422. [Google Scholar] [CrossRef]
- Wu, X.; Feng, H.; Wu, D.; Yan, S.; Zhang, P.; Wang, W.; Zhang, J.; Ye, J.; Dai, G.; Fan, Y.; et al. Using high-throughput multiple optical phenotyping to decipher the genetic architecture of maize drought tolerance. Genome Biol. 2021, 22, 185. [Google Scholar] [CrossRef]
- Sheng, L.; Chai, W.; Gong, X.; Zhou, L.; Cai, R.; Li, X.; Zhao, Y.; Jiang, H.; Cheng, B. Identification and Characterization of Novel Maize Mirnas Involved in Different Genetic Background. Int. J. Biol. Sci. 2015, 11, 781. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, K.B.; Hulm, J.L.; Young, L.W.; Bonham-Smith, P.C. A rapid Agrobacterium-mediated Arabidopsis thaliana transient assay system. Plant Mol. Biol. Rep. 2004, 22, 53–61. [Google Scholar] [CrossRef]
- Hiei, Y.; Komari, T.; Kubo, T. Transformation of rice mediated by Agrobacterium tumefaciens. Plant Mol. Biol. 1997, 35, 205–218. [Google Scholar] [CrossRef]
- Patel, M.; Dewey, R.E.; Qu, R. Enhancing Agrobacterium tumefaciens -mediated transformation efficiency of perennial ryegrass and rice using heat and high maltose treatments during bacterial infection. Plant Cell Tissue Organ. Cult. 2013, 114, 19–29. [Google Scholar] [CrossRef]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil. 1973. [Google Scholar] [CrossRef]
- Seok, M.S.; You, Y.N.; Park, H.J.; Lee, S.S.; Aigen, F.; Luan, S.; Ahn, J.C.; Cho, H.S. AtFKBP16-1, a chloroplast lumenal immunophilin, mediates response to photosynthetic stress by regulating PsaL stability. Physiol. Plant. 2014, 150, 620–631. [Google Scholar] [CrossRef]
- Xiong, L.; Schumaker, K.S.; Zhu, J.-K. Cell Signaling during Cold, Drought, and Salt Stress. Plant Cell 2002, 14 (Suppl. S1), S165. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Sairam, R.K.; Saxena, D.C. Oxidative stress and antioxidants in wheat genotypes: Possible mechanism of water stress tolerance. J. Agron. Crop Sci. 2000, 184, 55–61. [Google Scholar] [CrossRef]
- Khanna, R.; Lee, E.J.; Papazian, D.M. Transient calnexin interaction confers long-term stability on folded K+ channel protein in the ER. J. Cell Sci. 2004, 117 Pt 14, 2897. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chai, W.; Li, H.; Xu, H.; Zhu, Q.; Li, S.; Yuan, C.; Ji, W.; Wang, J.; Sheng, L. ZmDST44 Gene Is a Positive Regulator in Plant Drought Stress Tolerance. Biology 2024, 13, 552. https://doi.org/10.3390/biology13080552
Chai W, Li H, Xu H, Zhu Q, Li S, Yuan C, Ji W, Wang J, Sheng L. ZmDST44 Gene Is a Positive Regulator in Plant Drought Stress Tolerance. Biology. 2024; 13(8):552. https://doi.org/10.3390/biology13080552
Chicago/Turabian StyleChai, Wenbo, Hongtao Li, Hanyuan Xu, Qing Zhu, Shufen Li, Chao Yuan, Wei Ji, Jun Wang, and Lei Sheng. 2024. "ZmDST44 Gene Is a Positive Regulator in Plant Drought Stress Tolerance" Biology 13, no. 8: 552. https://doi.org/10.3390/biology13080552
APA StyleChai, W., Li, H., Xu, H., Zhu, Q., Li, S., Yuan, C., Ji, W., Wang, J., & Sheng, L. (2024). ZmDST44 Gene Is a Positive Regulator in Plant Drought Stress Tolerance. Biology, 13(8), 552. https://doi.org/10.3390/biology13080552



